Abstract
Giant cell tumor of bone (GCTB), is a rare intermediate malignant bone tumor with high local infiltrative ability, and is genetically characterized by mutation in the H3-3A gene. Standard treatment is curative surgical tumor resection. GCTB demonstrates both local recurrence and pulmonary metastasis after surgical treatment, and effective systematic chemotherapy is yet to be established. Therefore, development of novel chemotherapies for GCTB is necessary. Although patient-derived tumor cell lines are potent tools for preclinical research, 15 GCTB cell lines have been reported to date, and only four are publicly available. Thus, this study aimed to establish and characterize a novel GCTB cell line for preclinical studies on GCTB. Herein, we described the establishment of a cell line, NCC-GCTB5-C1, from the primary tumor tissue of a patient with GCTB. NCC-GCTB5-C1 was shown to harbor a mutation in the H3-3A gene, which is typical of GCTB; thus, it has useful properties for in vitro studies. We conducted the largest integrated screening analysis of 214 antitumor agents using NCC-GCTB5-C1 along with four GCTB cell lines. Romidepsin (a histone deacetylase inhibitor), camptothecin, and actinomycin D (topoisomerase inhibitors) demonstrated remarkable antitumor effects, suggesting that these antitumor agents are potential therapeutic candidates for GCTB treatment. Therefore, the NCC-GCTB5-C1 cell line could potentially contribute to the elucidation of GCTB pathogenesis and the development of novel GCTB treatments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A giant cell tumor of bone (GCTB) is an intermediate malignant bone tumor with high local infiltrative ability beyond the cortical bone and surrounding soft tissue [1, 2]. GCTB is histologically defined by the presence of reactive multinuclear osteoclast-like giant cells and neoplastic mononuclear stromal cells [3,4,5]; it is genetically characterized by mutation in the H3-3A gene, which encodes replication-independent histone H3.3 [6,7,8,9]. GCTBs are rare; they account for 5% of all primary bone tumors. GCTBs typically affect adults between 20 and 50 years of age. Most lesions develop in long bones (75–90%), with the majority (50–65%) occurring in the knee [1, 10]. Surgical tumor resection with wide excision or intralesional curettage is the only curative treatment [2, 10, 11]. However, local recurrence and pulmonary metastases after surgical treatment were observed; the local recurrence rate was 15–50% [1, 11, 12], and pulmonary metastases occurred in 2.1–6.6% of cases [13,14,15]. Therefore, it is imperative to develop systematic chemotherapy for GCTB.
Patient-derived cell lines are fundamental tools for preclinical research as they offer the advantage of being easily grown, inexpensive, and amenable to high-throughput testing of therapeutic agents [16, 17]. However, very few sarcoma cell lines exist [18]. To the best of our knowledge, 15 GCTB cell lines have been reported to date, and only four are publicly available (Supplementary Table 1). Numerous cell lines are required for drug screening, and cell lines of rare cancers such as GCTB should be actively established.
Herein, we established a novel GCTB cell line from surgically resected tumor tissues and named it NCC-GCTB5-C1. We then determined its molecular and phenotypic characteristics. Furthermore, we conducted drug screening on NCC-GCTB5-C1 cells along with four previously established GCTB cell lines to evaluate the utility of a panel of cell lines.
Materials and methods
Patient data
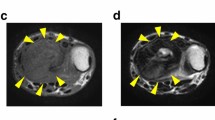
The donor patient was a 52-year-old man with GCTB, and had no relevant medical history. He had experienced pain in his right knee without any onset and visited a hospital previously. The patient was initially treated conservatively. However, the pain gradually worsened, and he underwent further inspection. X-ray inspection and computed tomography revealed an osteolytic lesion in the distal femur without destructive changes in the marginal bone cortex (Fig. 1a, b). Magnetic resonance imaging (MRI) revealed a neoplastic lesion within the femur (Fig. 1c, d). The patient was referred to the National Cancer Center Hospital, Tokyo, Japan for further treatment. The tumor was diagnosed as a GCTB on open biopsy. Metastases were undetected. The patient underwent aggressive curettage of the lesion with reinforcement using autologous bone graft implantation and plate fixation. Tumor tissues obtained at the time of surgery were used to establish cell lines. Histologically, the tumor partly consisted of a classic histology showing uniform mononuclear cells with scattered osteoclast-like giant cells (Fig. 1e). Furthermore, atypical epithelioid cells were occasionally identified, exhibiting enlarged lobulated nuclei and/or nuclear pseudo-inclusions (Fig. 1f). Neoplastic cells were diffusely positive for H3.3 G34W on immunohistochemistry (Fig. 1g). Neither local recurrence nor lung metastases were reported postoperatively. The use of clinical materials for this study was approved by the ethical committee of the National Cancer Center and written informed consent was obtained from the donor patient.
Clinical and pathological data of NCC-GCTB5-C1. a, b X-ray inspection and computed tomography depict osteolytic lesions within the femur delineated by yellow arrows. c, d Magnetic resonance imaging shows a mass in the distal femur that exhibits homogeneously high intensity with a short inversion time inversion recovery image and low intensity with T1-weighted image. e H&E staining depicts a classic histology of GCTB, showing uniform mononuclear cells with scattered osteoclast-like giant cells. f Atypical epithelioid cells are occasionally identified, exhibiting enlarged lobulated nuclei and/or nuclear pseudoinclusions. g Neoplastic cells are diffusely positive for H3.3 G34W immunohistochemistry
Histological analysis
Histological examination was performed on 4 μm-thick sections of a representative formalin-fixed paraffin-embedded tumor sample. The sections were deparaffinized and subjected to morphological observation using hematoxylin and eosin (H&E) staining. They were stained with an H3.3 G34W antibody (1:1000, RM263, RevMAb Biosciences USA, South San Francisco, CA, USA) to investigate H3.3 G35W expression.
Cell line establishment
Specimens derived from surgically resected tumors were used to establish cell lines. The primary tissue culture was done using Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) (Gibco, Grand Island, NY, USA) supplemented with 5% heat-inactivated fetal bovine serum (FBS) (Gibco), 100 μg/mL penicillin, 100 μg/mL streptomycin (Nacalai Tesque, Kyoto, Japan), 0.4 μg/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO, USA), 10 ng/mL bFGF (Sigma-Aldrich), 5 μg/mL insulin (Sigma-Aldrich), and 10 μM Y-27632 (Selleck Chemicals, Houston, TX, USA: Rock inhibitor) in 37 ℃ humidified environment with 5% CO2. The cells were cultured in the environment described above for more than three months and passaged at least 20 times.
Short tandem repeat analysis for authentication
We conducted short tandem repeat (STRs) analysis of the DNA extracted from the original tumor tissue and cell line for authentication. DNA extraction was performed using an AllPrep DNA/RNA Mini Kit (Qiagen, Venlo, Netherlands). STR analysis of DNA was performed using GenePrint 10 (Promega, Madison, WI, USA), and the amplified DNA fragments were detected using a 3500XL Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). The data were analyzed using GeneMapper 5 (Applied Biosystems); the STR profiles were compared with those recorded in the public cell bank for reference matching with a standard match threshold of 80%, according to the Tanabe formula [19, 20].
Mycoplasma contamination inspection
DNA derived from cells was evaluated for Mycoplasma contamination using the standard procedure of cell line establishment, according to the international cell line authentication committee (The International Cell Line Authentication Committee 2018) [21]. DNA extracted from cells was amplified using an e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Gyeonggi-do, Korea). The amplified DNA was separated by electrophoresis on a 1.5% agarose gel, stained with SYBR Safe DNA gel stain (Invitrogen, Waltham, MA, USA), and evaluated using Amersham Imager 600 (GE Healthcare Biosciences, Uppsala, Sweden).
Genetic analysis
To reveal the H3-3A gene mutation in the established cell line, total RNA was extracted from GCTB cells using QIAzol Lysis Reagent (Qiagen) and miRNeasy Mini Kit (Qiagen). Subsequently, the extracted RNA was reverse-transcribed to complementary DNA using Superscript III reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. The H3-3A gene was amplified by PCR using the H3-3A forward primer H3-3A_F (5’- TAAAGCACCCAGGAAGCAAC-3’), H3-3A reverse primer H3-3A_R (5’- CAAGAGAGACTTTGTCCCATTTTT-3’), and Platinum Taq DNA Polymerase High Fidelity (Life Technologies Co., Carlsbad, CA, USA). The PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega Co.), and direct sequencing was performed using the BigDye v3.1 Cycle Sequencing Kit (Applied Biosystems) and the Applied Biosystems 3130xL Sequencer (Thermo Fisher Scientific Inc., Waltham, MA, USA) by GENEWIZ. Sequence results were analyzed using ApE v2.0.61.
Cell proliferation assay
The cells were seeded at a density of 1.25 × 104 cells/well in 24-well culture plates at day 1. The number of cells was measured using the Cell Counting Kit 8 (DOJINDO LABORATORIES, Kumamoto, Japan) every 24-h for four days. Doubling time was calculated based on the growth curve.
Spheroid formation assay
The cells (1 × 104) were seeded in 96-well plates (96-well Clear Flat Bottom Ultra Low Attachment Microplate; Corning, Inc., Corning, NY, USA) in DMEM/F12 containing 10% FBS and maintained in a 37 ℃ humidified environment with 5% CO2. Spheroid formation was confirmed by microscopy (Keyence, Osaka, Japan).
Invasion assay using real-time cell analyzer
Invasiveness was evaluated using a real-time cell analyzer (xCELLigence, Agilent, Santa Clara, CA, USA), according to the manufacturer’s instructions. The cells (4 × 104) were plated in the upper chamber with serum-free medium and BD Biocoat Matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA). DMEM/F12 with 10% FBS was set in the lower chamber. Cell invasion into the lower chamber was evaluated using the impedance change detected by the electronic device, which was recorded every 15 min for 72 h. MG63 osteosarcoma cells were used as control [22].
Tumorigenesis assessment in nude mice
Tumorigenesis in mice was evaluated according to the guidelines of the Institute for Laboratory Animal Research (https://www.nationalacademies.org/ilar/institute-for-laboratory-animal-research). The mixture comprising 1 × 106 cells, 100 μL D-PBS, and 100 μL BD Matrigel matrix was injected subcutaneously into four different locations of female BALB/c nude mice (CLEA Japan, Inc., Tokyo, Japan). Tumor size was measured weekly with volume defined by the following formula: volume = length × width2/2 as previously described [23]. Two months after inoculation, the tumor was surgically excised and subjected to morphological inspection using H&E staining. All animal experiments were performed in accordance with the guidelines for animal experiments of the National Cancer Center and were approved by the Institutional Committee for Ethics of Animal Experimentation.
Drug screening test
Antitumor effects of the 214 antitumor agents were assessed (Supplementary Table 2). The cells were seeded at 5 × 104 cells/well in 384-well plates and incubated for one day. On the second day, each agent was disseminated into two wells of the plate and incubated for three days. Cell viability was calculated using the Cell Counting Kit-8 (DOUJINDO LABORATORIES). The acquired data were integrated with the results obtained for NCC-GCTB1-C1 [24], NCC-GCTB2-C1 [25], NCC-GCTB3-C1 [25], and NCC-GCTB4-C1 [26], which we previously reported. Quantile normalization was performed using R (version 4.0.3, limma package version 3.46.0, Bioconductor) and unsupervised hierarchical clustering was performed using the gplots package (version 3.1.0, CRAN, https://cran.r-project.org).
To estimate the half-maximal inhibitory concentration (IC50), we chose 23 agents that demonstrated superior antiproliferative effects based on results of the preceding drug screening test. Cells were exposed to each agent at ten different concentrations. The IC50 value was calculated using logistic regression analysis applied to the relationship between the common logarithm of concentration and cell viability for each agent.
Results
Authentication of the established cell line
Authentication of the NCC-GCTB5-C1 cell line was performed by analyzing the STR at 10 loci (Table 1, Supplementary Fig. 1). The match ratio of STR between the cells and the original tumor tissue was 97%. Mycoplasma contamination was investigated by detecting its DNA sequence in the cellular DNA specimen; we confirmed that the contamination was negative (data not shown).
Characteristic of NCC-GCTB5-C1 cells
NCC-GCTB5-C1 harbored a p.Gly35-Trp mutation in the H3-3A gene, resulting from a GGG > TGG nucleotide alteration (Fig. 2). NCC-GCTB5-C1 cells exhibited pleomorphic spindle-shaped morphology under 2D-culture condition (Fig. 3a, b). NCC-GCTB5-C1 cells formed spheroids under 3D-culture condition, and HE staining of the spheroids showed densely proliferating pleomorphic spindle cells (Fig. 3c). NCC-GCTB5-C1 cells showed constant but slower proliferation than NCC-GCTB1-C1, NCC-GCTB3-C1, and NCC-GCTB4-C1 cells, with a population doubling time of 32 h (Fig. 3d). The RTCA invasion assay showed that NCC-GCTB5-C1 cells were more invasive than the MG63 cells in vitro (Fig. 3e). Tumorigenesis of NCC-GCTB5-C1 cells was not observed in the nude mice (data not shown).
Characterization of NCC-GCTB5-C1. a, b NCC-GCTB5-C1 (passage 36) showing pleomorphic spindle cell morphology under 2D culture conditions. c The H&E-stained spheroids section of NCC-GCTB5-C1 (passage 26). d Growth curve of NCC-GCTB5-C1 (passage 34) along with previously established NCC-GCTB-C1 cell lines. e Invasive capability of NCC-GCTB5-C1 (passage 33) was observed using Real Time Cell Analyzer. MG63 osteosarcoma cell lines were shown as control cells
Sensitivity to 214 antitumor agents
We examined the antitumor effects of 214 antitumor agents on NCC-GCTB5-C1 cells. The viability of NCC-GCTB5-C1 cells after exposure to each antitumor agent is summarized in Supplementary Table 3, along with the cell viabilities of NCC-GCTB1-C1 [24], NCC-GCTB2-C1 [25], NCC-GCTB3-C1 [25], and NCC-GCTB4-C1 [26]. The antitumor agents were categorized into two groups according to their antitumor effect: cluster A, effective group; and cluster B, ineffective group (Fig. 4a, Supplementary Table 3). The proportion of cytotoxic agents, particularly topoisomerase inhibitors, was higher in cluster A than in cluster B (Fig. 4b, c, Supplementary Table 3, 4). As for tyrosine kinase, the agent subtype was equally distributed between clusters A and B (Fig. 4d, Supplementary Table 5), while all of the histone deacetylase (HDAC) inhibitors belonged to cluster A (Fig. 4e, Supplementary Table 6).
Drug screening tests on GCTB cell lines. a The antitumor agents were categorized into two groups according to their antitumor effect: Cluster A as the effective group and Cluster B as the ineffective group. b-e The proportion of each agent type belonging to each cluster. The graphs are depicted after normalization of the number of agents. Data regarding NCC-GCTB1-C1, NCC-GCTB2-C1, NCC-GCTB3-C1, and NCC-GCTB4-C1 were previously reported [24,25,26]
The IC50 values were calculated for antitumor agents that were identified in the screening study; they are summarized in Supplementary Table 7, along with those of NCC-GCTB1-C1 [24], NCC-GCTB2-C1 [25], NCC-GCTB3-C1 [25], and NCC-GCTB4-C1 [26]. The HDAC and topoisomerase inhibitors showed remarkable antitumor effects in GCTB cell lines, and their IC50 values and growth curves are shown in Table 2 and Fig. 5, respectively. Notably, romidepsin demonstrated the strongest antitumor effect toward the NCC-GCTB5-C1 cell line among the HDAC inhibitors, while actinomycin D and camptothecin showed the strongest antitumor effect among the topoisomerase inhibitors.
Cell viability of GCTB cell lines at different concentrations of each antitumor agent. a The anti-proliferative effects of the nine antitumor agents on NCC-GCTB1-C1, NCC-GCTB2-C1, NCC-GCTB3-C1, NCC-GCTB4-C1, and NCC-GCTB5-C1 cells are shown. The IC50 values are summarized in Table 2
Discussion
Patient-derived cancer cell lines facilitate fundamental research for elucidating pathogenesis and searching for novel antitumor agent candidates. However, only four GCTB cell lines have been reported in public cell banks to date, and the paucity of GCTB cell lines hinders the development of chemotherapy. In this study, we established a novel GCTB cell line, NCC-GCTB5-C1, from a surgical specimen of GCTB and assessed its characteristics. We performed a drug screening analysis of five GCTB cell lines: NCC-GCTB5-C1, NCC-GCTB1-C1[24], NCC-GCTB2-C1 [25], NCC-GCTB3-C1 [25], and NCC-GCTB4-C1 [26], which is the largest analysis that has been conducted. We reported antitumor agents that showed remarkable effects on these five GCTB cell lines.
The NCC-GCTB5-C1 cell line recapitulated the typical characteristics of GCTB in multiple aspects. First, the donor patient exhibited the typical clinical course of GCTB, considering its epidemiology. The original tumor occurred in the knee, which was the most common location of GCTB. Radiological and histological images of the original tumor showed typical GCTB features. Second, NCC-GCTB5-C1 cells harbored a typical gene mutation of GCTB. The mutation observed in NCC-GCTB5-C1 was the p.Gly35-Trp mutation of H3-3A gene, which occurs in more than 90% of GCTB cases. Third, the morphology of NCC-GCTB5-C1 cells were not inconsistent with the characteristics of the original tumor. NCC-GCTB5-C1 showed both pleomorphic spindle-shaped cells under tissue culture conditions, which reflected the morphology of the original tumor that comprised a heterogeneous population of cells. Thus, NCC-GCTB5-C1 cells retain the typical features of GCTB and serve as representative GCTB cell lines.
NCC-GCTB5-C1 cells exhibited multiple characteristics suitable for in vitro experiments, and showed constant proliferative ability, which was comparable to that of previously established GCTB cell lines. Furthermore, they demonstrated invasive abilities and formed spheroids under 3D culture conditions. They did not demonstrate tumorigenesis in the skin of nude mice and were unsuitable for xenograft experiments.
We performed an integrated drug-screening assay using NCC-GCTB5-C1 cells and four previously reported GCTB cell lines. Notably, romidepsin, an HDAC inhibitor, exerted remarkable antitumor effects on all GCTB cell lines except NCC-GCTB2-C1, suggesting its effectiveness in GCTB treatment. HDAC regulates gene expression by controlling histone acetylation, which is one of the most extensively studied post-translational covalent modifications of histones [27]. Actinomycin D and camptothecin, which are topoisomerase inhibitors, also demonstrated remarkable antitumor effects in GCTB cell lines, except NCC-GCTB2-C1. Topoisomerase is a ubiquitous enzyme that plays a vital role in replication, transcription, recombination, DNA repair, and chromatin remodeling by regulating the DNA helix construction [27, 28]. Neither HDAC inhibitors nor topoisomerase inhibitors have been clinically investigated for use in GCTB treatment according to the clinical trial database Clinical Trials.gov [29]; thus, they are worth investigating further. Although our results should be validated in other cancer models, such as patient-derived xenografts, the results of the present study indicate the utilization of our cell lines in the search for novel antitumor agent candidates.
Our study had limitations. First, although this is the largest drug screening analysis conducted for GCTB, the number of cell lines used is still insufficient compared to that used for common cancers. For example, more than 100 cancer cell lines have been used in drug screening studies for lung cancer [16]. Second, to achieve maximal clinical benefit, the discovery of a synergistic combination of antitumor agents is required [30]. For example, a combination of HDAC inhibitors and topoisomerase inhibitors has been shown effective in preclinical studies [27, 28, 31, 32]; thus, this should be further examined in GCTB cell lines.
In conclusion, we successfully established a novel GCTB cell line, NCC-GCTB5-C1, using a typical GCTB case. NCC-GCTB5-C1 cells exhibited continuous proliferation, spheroid formation, and aggressive invasiveness. We also revealed the remarkable antitumor effect of HDAC inhibitors and topoisomerase inhibitors, especially romidepsin, actinomycin D, and campothecin, on GCTB cell lines. These results indicate that NCC-GCTB5-C1 has the potential to facilitate numerous advances in preclinical and basic research on GCTB.
Change history
18 October 2022
A Correction to this paper has been published: https://doi.org/10.1007/s13577-022-00805-2
References
van der Heijden L, Dijkstra PD, van de Sande MA, et al. The clinical approach toward giant cell tumor of bone. Oncologist. 2014;19:550–61.
López-Pousa A, Martín Broto J, Garrido T, Vázquez J. Giant cell tumour of bone: new treatments in development. Clin Transl Oncol. 2015;17:419–30.
Tsukamoto S, Mavrogenis AF, Kido A, Errani C. Current concepts in the treatment of giant cell tumors of bone. Cancers (Basel). 2021;13:3647.
Basu Mallick A, Chawla SP. Giant cell tumor of bone: an update. Curr Oncol Rep. 2021;23:51.
Liede A, Bach BA, Stryker S, et al. Regional variation and challenges in estimating the incidence of giant cell tumor of bone. J Bone Jt Surg Am. 2014;96:1999–2007.
Cleven AH, Höcker S, Briaire-de Bruijn I, Szuhai K, Cleton-Jansen AM, Bovée JV. Mutation analysis of H3F3A and H3F3B as a diagnostic tool for giant cell tumor of bone and chondroblastoma. Am J Surg Pathol. 2015;39:1576–83.
Presneau N, Baumhoer D, Behjati S, et al. Diagnostic value of H3F3A mutations in giant cell tumour of bone compared to osteoclast-rich mimics. J Pathol Clin Res. 2015;1:113–23.
Amary F, Berisha F, Ye H, et al. H3F3A (Histone 3.3) G34W immunohistochemistry: a reliable marker defining benign and malignant giant cell tumor of bone. Am J Surg Pathol. 2017;41:1059–68.
Behjati S, Tarpey PS, Presneau N, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82.
Chakarun CJ, Forrester DM, Gottsegen CJ, Patel DB, White EA, Matcuk GR Jr. Giant cell tumor of bone: review, mimics, and new developments in treatment. Radiographics. 2013;33:197–211.
Errani C, Ruggieri P, Asenzio MA, et al. Giant cell tumor of the extremity: a review of 349 cases from a single institution. Cancer Treat Rev. 2010;36:1–7.
Jamshidi K, Gharehdaghi M, Hajialiloo SS, Mirkazemi M, Ghaffarzadehgan K, Izanloo A. Denosumab in patients with giant cell tumor and its recurrence: a systematic review. Arch Bone Jt Surg. 2018;6:260–8.
Chan CM, Adler Z, Reith JD, Gibbs CP Jr. Risk factors for pulmonary metastases from giant cell tumor of bone. J Bone Jt Surg Am. 2015;97:420–8.
Dominkus M, Ruggieri P, Bertoni F, et al. Histologically verified lung metastases in benign giant cell tumours–14 cases from a single institution. Int Orthop. 2006;30:499–504.
Wang J, Liu X, Yang Y, et al. Pulmonary metastasis of giant cell tumour: a retrospective study of three hundred and ten cases. Int Orthop. 2021;45:769–78.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Tseng YY, Boehm JS. From cell lines to living biosensors: new opportunities to prioritize cancer dependencies using ex vivo tumor cultures. Curr Opin Genet Dev. 2019;54:33–40.
Hattori E, Oyama R, Kondo T. Systematic review of the current status of human sarcoma cell lines. Cells. 2019;8:157.
Hideyuki T, Yoko T, Daisuke M, Miharu K, Tohru M, Hiroshi M. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tissue Cult Res Commun. 1999;18:329–38.
Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98:8012–7.
Drexler HG, Dirks WG, MacLeod RA, Uphoff CC. False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017;140:1209–14.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Cox C, Teknos TN, Barrios M, Brewer GJ, Dick RD, Merajver SD. The role of copper suppression as an antiangiogenic strategy in head and neck squamous cell carcinoma. Laryngoscope. 2001;111:696–701.
Noguchi R, Yoshimatsu Y, Ono T, et al. Establishment and characterization of NCC-GCTB1-C1: a novel patient-derived cancer cell line of giant cell tumor of bone. Hum Cell. 2020;33:1321–8.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of novel patient-derived cell lines from giant cell tumor of bone. Hum Cell. 2021;34:1899–910.
Ono T, Noguchi R, Yoshimatsu Y, et al. Establishment and characterization of the NCC-GCTB4-C1 cell line: a novel patient-derived cell line from giant cell tumor of bone. Hum Cell. 2022;35:392–9.
Suraweera A, O’Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front Oncol. 2018;8:92.
Skok Ž, Zidar N, Kikelj D, Ilaš J. Dual inhibitors of human DNA topoisomerase II and other cancer-related targets. J Med Chem. 2020;63:884–904.
Zarin DA, Fain KM, Dobbins HD, Tse T, Williams RJ. 10-year update on study results submitted to ClinicalTrials.gov. N Engl J Med. 2019;381:1966–74.
Shinn P, Chen L, Ferrer M, et al. High-throughput screening for drug combinations. Methods Mol Biol. 2019;1939:11–35.
Marchion DC, Bicaku E, Turner JG, Daud AI, Sullivan DM, Munster PN. Synergistic interaction between histone deacetylase and topoisomerase II inhibitors is mediated through topoisomerase IIbeta. Clin Cancer Res. 2005;11:8467–75.
Marchion DC, Bicaku E, Daud AI, Richon V, Sullivan DM, Munster PN. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–37.
Acknowledgements
We thank Drs. E. Kobayashi, S. Iwata, K.Ogura, S.Osaki, K.Sato, C.Sato, S.Ishihara (Department of Musculoskeletal Oncology), and the National Cancer Center Hospital for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Mrs. Y. Kuwata (Division of Rare Cancer Research) and technical support provided by Mrs. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for providing English language editing services and for their constructive comments on this manuscript. This study was technically assisted by the Fundamental Innovative Oncology Core of the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant number: 20ck0106537h0002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number: 2004-050). Animal experiments were conducted in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute.
Informed consent
Written informed consent was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2022_724_MOESM1_ESM.tiff
Supplementary file1 Short tandem repeat patterns of NCC-GCTB5-C1 and the original tumor tissue (a) Short tandem repeat patterns of the original tumor tissue of NCC-GCTB5-C1 and (b) short tandem repeat patterns of NCC-GCTB5-C1 cells (passage 25) (TIFF 3925 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akiyama, T., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-GCTB5-C1: a novel cell line of giant cell tumor of bone. Human Cell 35, 1621–1629 (2022). https://doi.org/10.1007/s13577-022-00724-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00724-2