Abstract
Giant cell tumor of bone (GCTB) is a rare osteolytic intermediate bone tumor that harbors a pathogenic H3F3A gene mutation and exhibits characteristic histology. The standard curative treatment for GCTB is complete surgical resection, but it frequently results in local recurrence and, more rarely, metastasis. Therefore, effective multidisciplinary treatment is needed. Although patient-derived tumor cell lines are promising tools for preclinical and basic research, there are only four available cell lines for GCTB in public cell banks. Thus, the aim of this study was to establish a novel GCTB cell line. Using surgically resected tumor tissues from a patient with GCTB, we established a cell line named NCC-GCTB4-C1. The cells harbored the typical H3F3A gene mutation and exhibited constant proliferation and invasive capabilities. After characterizing NCC-GCTB4-C1 cell behaviors, we conducted high-throughput screening of 214 anti-tumor drugs and identified seven effective drugs. Comparing the results of high-throughput screening using NCC-GCTB4-C1 cell line with the results using NCC-GCTB1-C1, NCC-GCTB2-C1, and NCC-GCTB3-C1 cell lines that we previously established, four drugs were in common effective. This study showed potential drugs for the treatment of GCTB. These data indicate that NCC-GCTB4-C1 has the potential to be a powerful tool in preclinical and basic research on GCTB.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Giant cell tumor of bone (GCTB) is a locally aggressive intermediate bone tumor comprising multinucleated giant cells, similar to osteoclast stromal tumors of the bone [1, 2]. GCTB is genetically characterized by a mutation in the histone tail of histone variant H3.3, which is encoded by H3F3A on chromosome 1 [3,4,5,6]. GCTB cases account for 5% of all primary bone tumor cases [7, 8]. The incidence of GCTB is slightly higher in women than in men [2, 8, 9]. Most patients show GCTB occurrence between 20 and 50 years of age with less than 3% of patients showing occurrence before the age of 14 years, and only 13% of patients showing occurrence after the age of 50 years [10]. GCTB is typically observed in the ends of long bones such as the distal femur, proximal tibia, distal radius, and proximal humerus [1]. GCTB has a high risk of destroying the bone and extending into the surrounding soft tissue, causing pain [11].
For patients with GCTB, surgical removal, such as curettage or resection, is a standard curative treatment [12,13,14]. However, it is not amenable to resect some sites (e.g., the skull and spine), and 15%–50% of patients exhibit local recurrence at any site after surgery usually within 2 years, with a higher prevalence after curettages [15]. When GCTB is not cured by surgical removal, treatment options become scarce and may not be curative or could be associated with substantial morbidity. Thus, there is a need to develop novel therapeutic strategies for GCTB.
Currently, hundreds of targeted treatments are under clinical evaluation and outweigh the number of patients with tumors who can be enrolled in such studies [16,17,18]. Preclinical models have historically been a useful tool in research to develop therapies for various diseases, including tumor [19,20,21]. Mouse models, such as cell line-derived models, patient-derived xenograft models, environmentally induced models, and genetically engineered models are time consuming, cost intensive, and exhibit low throughput for drug screening [20]. Since the establishment of the first patient-derived tumor cell line, HeLa, in 1951 [22], cell lines have played a critical role in tumor research and facilitated the development of new anti-tumor therapeutics through high-throughput screening of anti-tumor drugs [23,24,25,26]. Using patient-derived cell lines, the Bodmer laboratory constructed a panel of more than 120 colorectal tumor cell lines. This colorectal tumor cell line panel revealed that 5-fluorouracil showed sensitivity in a subset of 77 cell lines that had mismatch repair status with a high-throughput drug screening [27]. Similarly, in the study of GCTB, the use of cell lines could provide valuable information.
It is not easy for researchers to utilize GCTB cell lines. According to the cell line database Cellosaurus [28], 12 cell lines of GCTB have been reported (Supplementary Table 1). However, six cell lines were not registered for the cell bank and two cell lines had discontinued distribution. Therefore, only four GCTB cell lines registered for cell banks are currently available for research. This may be due to the lack of clinical materials for both basic and translational research and the intermediate characteristics of GCTB tumors. Considering these observations, it is imperative to establish more cell lines from patients with GCTB who exhibit different clinical features.
Here, we report a new GCTB cell line, NCC-GCTB4-C1, established from the surgically resected tumor tissue of a patient with GCTB. To demonstrate the usefulness of this cell line, we studied its characteristics such as proliferation, spheroid formation, and invasion. Furthermore, we revealed the effectivity of this cell line in drug screening.
Materials and methods
Patient history
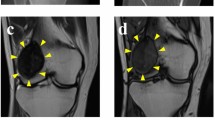
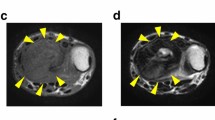
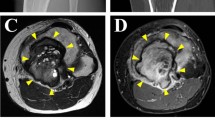
The patient was a 52-year-old male with a GCTB. The patient had no particular symptoms, and an X-ray taken by chance at a previous hospital indicated a bone tumor on the left proximal tibia. For further examination and treatment, the patient was referred to the Tochigi Cancer Center (Utsunomiya, Tochigi, Japan). X-ray showed a soap bubble appearance, which is typical of GCTB (Fig. 1A). Computed tomography revealed a well-demarcated and translucent lesion (Fig. 1B). Magnetic resonance imaging revealed a 52 mm × 43 mm × 42 mm tumor (Fig. 1C, D). An open biopsy was performed, and the tumor contained osteoclast-like giant cells scattered between the mononuclear neoplastic cells (Fig. 1E). The mononuclear neoplastic cells showed diffuse H3.3G34W immunoreactivity (Fig. 1F). Based on these pathological findings, the tumor was diagnosed as GCTB. A part of the curetted tumor at the open biopsy was used to establish the cell line described in this study. The patient then underwent surgery, and no local recurrence or metastasis was observed. The use of clinical materials for this study was approved by the ethical committee of the National Cancer Center (2004-050) and Tochigi Cancer Center (A-493). Written informed consent was obtained from the patient.
Clinical and pathological data. A X-ray showing soap bubble appearance in left proximal tibia. B Computed tomography showing well-demarcated and translucent lesion. Magnetic resonance imaging showing tumor with C low intensity in T1-weighted image and D low to iso intensity in T2-weighted image. Yellow arrows indicate the tumor. E Hematoxylin and eosin staining showing osteoclast-like giant cells scattered between mononuclear neoplastic cells. F Immunohistochemistry showing mononuclear neoplastic cells diffusely positive for H3.3G34W
Histological analysis of tumor tissue
Histological examination was performed on 4-μm-thick sections from a representative paraffin-embedded tumor sample. After deparaffinization of the tissue sections with xylene and ethanol, the sections were stained with hematoxylin and eosin (HE). Endogenous peroxidase activity was inhibited by adding 3% hydrogen peroxide to the sections. The epitope retrieval procedure was performed to reverse the loss of antigenicity that occurs with some epitopes in formalin-fixed, paraffin-embedded tissues. Primary antibody against histone H3.3G34W mutant (RM263, 1:400, RevMAb Biosciences USA Inc., South San Francisco, CA, USA) was used for immunohistochemical analysis of the tissues. The I-VIEW DAB universal kit (F. Hoffmann-La Roche Ltd, Basal, Switzerland) and the Histofine simple stain-MAX-PO (multi) kit (Nichirei Biosciences Inc., Chuo-ku, Tokyo, Japan) were used to detect immunoreactivity. We used hematoxylin, a nuclear stain, as a counterstain. BZ-X710 (KEYENCE Corp., Osaka, Osaka, Japan) was applied for phase-contrast inverted microscopic observation to visualize the stained cell morphology.
Primary cell isolation and culture
Primary GCTB tumor cells were obtained as previously described [29]. In brief, surgically resected tumor tissue was mechanically dissected into small pieces and digested with 1 mg/mL collagenase type II (Worthington Biochemical Corp., Lakewood, NJ, USA) for 30 min at 37 °C. The cells obtained from the digested tissue were seeded on a collagen type I-coated culture plate (Sumitomo Bakelite Co. Ltd., Shinagawa-ku, Tokyo, Japan) and maintained in DMEM/F12 supplemented with GlutaMAX (Thermo Fisher Scientific Inc., Waltham, MA, USA), 5% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific Inc), 10 µM Y-27632 (ROCK inhibitor; Selleck Chemicals, Houston, TX, USA), 10 ng/mL bFGF (Sigma-Aldrich Co. LLC, St Louis, MO, USA), 5 ng/mL EGF (Sigma-Aldrich Co. LLC), 5 µg/mL insulin (Sigma-Aldrich Co. LLC), 0.4 µg/mL hydrocortisone (Sigma-Aldrich Co. LLC), 100 μg/mL penicillin, and 100 µg/mL streptomycin (Nacalai Tesque Inc., Nakagyo-ku, Kyoto, Japan). The culture medium was changed every 2–3 days. The status of the cells was confirmed through microscopic observations (Carl Zeiss AG, Oberkochen, Germany), and when the cultured cells reached sub-confluency, they were washed with PBS (−) (Nacalai Tesque Inc), which was followed by dissociation with Accutase (Nacalai Tesque Inc); they were then transferred to another tissue culture plate. The cells were continuously incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Cell line authentication and quality control
Authentication and quality control of the established cell line were conducted following a previously reported protocol [29]. In brief, we extracted DNA from the obtained tumor tissues and established a cell line using the Qiagen DNeasy Blood and Tissue Kit (QIAGEN N.V., Hilden, Germany). To confirm the authentication, we conducted short tandem repeat (STR) analysis for 10 loci using the GenePrint 10 system (Promega Co., Madison, WI, USA) and a 3500xL Genetic Analyzer (Thermo Fisher Scientific Inc.). STR profiles were analyzed using GeneMapper software (Thermo Fisher Scientific Inc.) and matched to the data in the public cell banks with the Cellosaurus 38.0 STR similarity search tool, CLASTR 1.4.4 [28] with a standard match threshold of 80% [30]. To confirm the quality of the established cell line, we performed a mycoplasma contamination test with the DNA fragmentation of mycoplasma using the e-Myco Mycoplasma PCR Detection Kit (iNtRON Biotechnology Inc., Seongnam-si, Gyeonggi-do, Korea). Using agarose gel electrophoresis, DNA fragments amplified using PCR were separated and stained with SYBR Safe DNA gel stain (Invitrogen Corp., Waltham, MA, USA).
Genetic analysis
To reveal the H3F3A gene mutation in the established cell line, total RNA was extracted using QIAzol Lysis Reagent (QIAGEN N.V.) and miRNeasy Mini Kit (QIAGEN N.V.) from the GCTB cells. Subsequently, the extracted RNA was reverse-transcribed to complementary DNA using Superscript III reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. The H3F3A gene was amplified using PCR with the H3F3A forward primer H3F3A_F (5′- TAAAGCACCCAGGAAGCAAC-3′), H3F3A reverse primer H3F3A_R (5′- CAAGAGAGACTTTGTCCCATTTTT-3′), and Platinum Taq DNA Polymerase High Fidelity (Life Technologies Co., Carlsbad, CA, USA). The PCR products were purified using Wizard SV Gel and PCR Clean-Up System (Promega Co.), and direct sequencing was performed using the BigDye v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA) and the Applied Biosystems 3130xL Sequencer (Thermo Fisher Scientific Inc.) by GENEWIZ. The sequence result was analyzed using ApE v2.0.61.
Spheroid formation assay
Spheroid formation capability was assessed as previously described [29]. In brief, the established cells were seeded at a density of 1 × 105 cells/well in a 96-well Clear Round Bottom Ultra Low Attachment Microplate (Corning Inc., NY, USA), and spheroid formation was confirmed through microscopic observation (KEYENCE Co.). After 3 days of culture, the spheroids picked up from the plate were covered in gel using iPGell (GenoStaff Co. Ltd., Bunkyo-ku, Tokyo, Japan) and fixed with 10% formalin neutral buffer solution. To prepare a paraffin section of the fabricated spheroids, the gel-covered spheroids were embedded in paraffin and sliced into 4-micrometer-thick paraffin sections. The sectioned spheroids were subjected to hematoxylin and eosin staining followed by microscopic observations.
Tumor cell proliferation assay
To assess the potential for proliferation, established cells were seeded at a density of 2.5 × 104 cells/well in a 24-well culture plate (Corning Inc.). The number of cells was counted at multiple time points for 96 h, and the doubling time was calculated based on the growth curve. All experiments were performed in triplicates.
Tumor cell invasion assay
To examine the invasive potential, we utilized the Real Time Cell Analyzer, xCELLigence (Agilent Technologies Inc., Santa Clara, CA, USA). The MG63 osteosarcoma cell line (Japanese Collection of Research Bioresources Cell Bank, Ibaraki, Osaka, Japan), which has a constant invasiveness, was used as a control [31]. Subsequently, Matrigel Basement Membrane Matrix (Corning Inc.), which has been generally used for in vitro tumor invasion assay, at a protein concentration of 9.3 mg/mL was layered on the membrane in the upper chamber, and 1 × 104 cells were seeded on it. DMEM/F12 supplemented with GlutaMAX, 5% FBS, 10 µM Y-27632, 10 ng/mL bFGF, 5 ng/mL EGF, 5 µg/mL insulin, 0.4 µg/mL hydrocortisone, 100 μg/mL penicillin, and 100 µg/mL streptomycin were added to the lower chamber. The upper chamber was filled with DMEM/F12 without FBS. The cells cultured on the upper chamber with a Matrigel-coated membrane migrated to the bottom chamber and adhered to the electronic sensors on the underside of the membrane. The attached cells influenced the electrical impedance of the electronic sensors. The invasion capability of the cells was estimated based on the positive correlation between the impedance and the number of cells. The impedance was monitored every 15 min for 120 h and plotted as a function of time after seeding.
Tumorigenicity assay in nude mice
The animal experiment was conducted in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. Briefly, 50 μL of cells (1 × 106 cells) mixed with an equal volume of Matrigel (21.2 mg/ml) was injected subcutaneously into BALB/c nude mice (CLEA Japan Inc., Meguro-ku, Tokyo, Japan) using a 5 ml syringe (Terumo Corp., Shibuya-ku, Tokyo, Japan) and 26G needle (Terumo Corp.). The tumor size was then measured weekly.
Screening for anti-tumor drugs
Drug screening was conducted using the established cell line with 214 anti-tumor drugs, as previously described [29]. Using a Bravo automated liquid handling platform (Agilent Technologies Inc.), the cells were seeded in a 384-well plate (Thermo Fisher Scientific Inc.) at a concentration of 5 × 103 cells/well in DMEM/F12 supplemented with GlutaMAX, 5% heat-inactivated FBS, 10 µM Y-27632, 10 ng/mL bFGF, 5 ng/mL EGF, 5 µg/mL insulin, and 0.4 µg/mL hydrocortisone and incubated at 37 °C in a humidified atmosphere containing 5% CO2. On the day after cell seeding, using the Bravo automated liquid handling platform, a drug library that included 214 anti-tumor drugs (Selleck Chemicals) (Supplementary Table 2), was applied at a concentration of 10 µM, to the cells and incubated for 72 h. After incubation, cell viability was determined using the CCK-8 reagent (Dojindo Molecular Technologies Inc., Kamimashiki-gun, Kumamoto, Japan) according to the manufacturer’s protocol. The response readout was calculated relative to the DMSO-treated control in terms of % relative growth inhibition.
IC50 values, which are the concentrations required to inhibit cell growth by 50% compared to the growth of control cells, were calculated from curves fabricated by plotting cell survival (%) versus drug concentration (μM). Using the Bravo automated liquid handling platform, cell suspensions (5 × 103 cells) were dispensed into 384-well plates. Subsequently, 21 drugs were selected according to the 214 anti-tumor drug screening. Additionally, we selected pazopanib HCl, eribulin and doxorubicin which were often use for the treatment of soft tissue sarcomas. These 24 drugs were added to the 384-well plates at a serial dilution of 0.1–100,000 nM. The cells were maintained for 72 h, and then cell viability was assessed using the CCK-8 reagent. The readout was plotted against the concentrations of drugs and examined with GraphPad Prism 9.1.1 software (GraphPad Software Inc., San Diego, CA, USA). This anti-tumor drug screening test was conducted in duplicate.
Results
Establishment and authentication of NCC-GCTB4-C1 cell line
Using surgically resected tumor tissues from a patient with GCTB, we established a cell line named NCC-GCTB4-C1. The cell line was maintained for over 30 passages for approximately six months. To authenticate the established cell line, we examined 10 STRs and found that the STR patterns of the cell line were identical to those of the original tumor tissues (Table 1, Supplementary Fig. 1). A database search using the Cellosaurus 38.0 STR similarity search tool, CLASTR 1.4.4, revealed that there are no existing cell lines in public cell banks in which the STR patterns were identical to the NCC-GCTB4-C1 cell line. These data show that NCC-GCTB4-C1 is a novel GCTB cell line. Mycoplasma contamination test showed that NCC-GCTB4-C1 cells were negative for mycoplasma, as mycoplasma-specific DNA was not detected in the NCC-GCTB4-C1 cell line (data not shown).
Characterization of NCC-GCTB4-C1 cells
The G34W (p.Gly34Trp) mutation, which is the most typical mutation in GCTB, was detected in NCC-GCTB4-C1 cells. The G34W mutation in the cell line was confirmed by Sanger sequencing (Fig. 2). NCC-GCTB4-C1 cells exhibited spindle morphology under 2D monolayer cultures (Fig. 3A, B). We found that NCC-GCTB4-C1 cells possessed the ability to form spheroids when they were cultured in a low-attachment round plate. The HE staining of the spheroids showed that NCC-GCTB4-C1 cell line comprised spindle and polygonal cells under 3D cultures (Fig. 3C). In the spheroids, a small number of multinucleated cells were observed (Fig. 3C). Constant proliferation of NCC-GCTB4-C1 cell line was observed and population doubling time of 65.78 h was calculated according to the growth curves of the cells (Fig. 3D). Furthermore, the RTCA invasion assay showed that NCC-GCTB4-C1 cells were more invasive than MG63 cells in vitro (Fig. 3E). Tumorigenesis in nude mice injected with NCC-GCTB4-C1 cells was not observed in this study (data not shown).
Characterization of NCC-GCTB4-C1 cells. A, B NCC-GCTB4-C1 cells (passage 19) showing spindle cell morphology under two-dimensional culture conditions. C Hematoxylin and eosin-stain showing spindle and polygonal cells in the spheroid of NCC-GCTB4-C1 cells (passage 19). D Growth curve of NCC-GCTB4-C1 cells (passage 25). Y-axis indicates the relative cell proliferation of NCC-GCTB4-C1 cells, and X-axis represents the day after seeding. E Real-time cell analyzer invasion assay showing the invasive ability of NCC-GCTB4-C1 cells (passage 23) compared with that of MG63 osteosarcoma cells
Sensitivity to anti-tumor drugs
High-throughput screening with 214 anti-tumor drugs (Supplementary Table 2) evaluated by the anti-proliferative effects on NCC-GCTB4-C1 cells showed the effects of anti-tumor drugs at a concentration of 10 μM (Supplementary Table 3) and ranked the 214 drugs according to cell viability after treatment. The IC50 values were calculated using GraphPad Prism 9.1.1 software (Supplementary Table 4), and the growth curves of the seven drugs with the lowest IC50 values (< 100 nM) (Table 2) were shown in Fig. 4.
Discussion
We established a novel GCTB cell line, NCC-GCTB4-C1. While complete resection is a standard treatment for GCTB, it is often challenging due to postoperative recurrence [12,13,14,15]. The effectiveness of conventional chemotherapy has not been proven in GCTB because of the difficulty of large-scale randomized clinical trials. Therefore, novel treatment methods are required. In recent years, multiple large-scale drug screening analyses have been conducted for major tumors using a number of cell lines [26, 32, 33]. However, only four GCTB cell lines in public cell banks have been reported to date, indicating that GCTB cannot benefit from these analyses.
According to the patient background of the NCC-GCTB4-C1 cell line, the original tumor was found in the left proximal tibia, which is a typical site of GCTB. With respect to patient age, the NCC-GCTB4-C1 cell line was established from a 52-year-old male patient with GCTB. Only 13% of cases occur in patients over the age of 50 years, and females show a higher incidence of GCTB than men [2, 8,9,10]. Therefore, only one GCTB cell line from a male patient aged > 50 years has been reported to date (Supplementary Table 1). Thus, the successfully established NCC-GCTB4-C1 cell line was derived from patients with different clinical features from those of previously established GCTB cell lines.
Genetically, at least 95% of giant cell tumors of bone harbor pathogenic H3F3A gene mutations, approximately 90% of which are H3.3 p.Gly34Trp resulting from a GGG > TGG nucleotide alteration [6, 34]. Rare variants (for example, p.Gly34Leu, p.Gly34Met, p.Gly34Arg, and p.Gly34Val) have also been reported [35]. The most common p.Gly34Trp mutation in the H3F3A gene was also detected in our previously established GCTB cell lines, NCC-GCTB1-C1 [36], NCC-GCTB2-C1 [29] and NCC-GCTB3-C1 [29]. Considering the genetic diversity of GCTB, the number of GCTB cell lines remains inadequate. Therefore, continuous efforts to establish GCTB cell lines are required.
We found that the morphology of NCC-GCTB4-C1 cells was mainly spindle-shaped under culture conditions. HE staining showed that osteoclast-like multinucleated giant cells were not included in the spheroid of NCC-GCTB4-C1, and the main components of the spheroid were spindle and polygonal cells. Considering that HE-stained tumor tissues of GCTB contain multinucleated cells, the NCC-GCTB4-C1 cell line is a clonal cell line of the spindle and polygonal population. NCC-GCTB4-C1 cells also demonstrated constant growth and were capable of invasion and spheroid formation. While these characteristics are suitable for in vitro studies, such as drug screening, tumorigenesis in nude mice injected with NCC-GCTB4-C1 cells was not observed. These results indicate that NCC-GCTB4-C1 cells may not be suitable for xenograft experiments. Additional GCTB cell lines are needed for in vivo research using GCTB cell lines.
In drug screening using 214 anti-tumor drugs, seven drugs with the lowest IC50 values (< 100 nM) were doxorubicin, bortezomib, camptothecin, homoharringtonine, mitoxantrone, romidepsin and vinblastine sulfate. Notably, NCC-GCTB1-C1, NCC-GCTB2-C1, and NCC-GCTB3-C1 cell lines also showed high sensitivity to doxorubicin, homoharringtonine, mitoxantrone, and romidepsin [29, 36]. Among these drugs, romidepsin, a histone deacetylase (HDAC) inhibitor, which was approved by the U.S. Food and Drug Administration for the treatment of cutaneous T-cell lymphoma [37] is the most effective drug for NCC-GCTB1-C1, NCC-GCTB2-C1, NCC-GCTB3-C1, and NCC-GCTB4-C1. However, according to the clinical trial database ClinicalTrials.gov [38], there are no studies that use HDAC inhibitor for the treatment of GCTB. Recently, Yafei et al. reported the anti-tumor effects of the HDAC inhibitor by the drug screening in the patient-derived xenograft model of GCTB [39], and our findings were concordant with their results. While frequent point mutation have been reported in H3F3A histone drivers of GCTB [3,4,5], the mutation is not targeted therapeutically for GCTB [40], and the drug screening using the models may be the effective approach. With GCTB cell lines, this study showed potential drugs for the treatment of GCTB.
In conclusion, we established a novel GCTB cell line, NCC-GCTB4-C1, which exhibited continuous proliferation, spheroid formation ability, and aggressive invasiveness. From the genomic analysis, we identified point mutation in NCC-GCTB4-C1. We also revealed the anti-tumor effects of doxorubicin, homoharringtonine, mitoxantrone, and romidepsin on GCTB cell lines. These results indicate that NCC-GCTB4-C1 has the potential to facilitate a numerous advances in preclinical and basic research on GCTB.
References
Antonescu CR, Who, Classification of Tumours Editorial B. Soft tissue and bone tumours. Lyon: International Agency for Research on Cancer; 2020.
Beebe-Dimmer JL, Cetin K, Fryzek JP, Schuetze SM, Schwartz K. The epidemiology of malignant giant cell tumors of bone: an analysis of data from the Surveillance, Epidemiology and End Results Program (1975–2004). Rare Tumors. 2009;1:e52.
Behjati S, Tarpey PS, Presneau N, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82.
Presneau N, Baumhoer D, Behjati S, et al. Diagnostic value of H3F3A mutations in giant cell tumour of bone compared to osteoclast-rich mimics. J Pathol Clin Res. 2015;1:113–23.
Cleven AH, Höcker S, Briaire-de Bruijn I, Szuhai K, Cleton-Jansen AM, Bovée JV. Mutation analysis of H3F3A and H3F3B as a diagnostic tool for giant cell tumor of bone and chondroblastoma. Am J Surg Pathol. 2015;39:1576–83.
Amary F, Berisha F, Ye H, et al. H3F3A (Histone 3.3) G34W immunohistochemistry: a reliable marker defining benign and malignant giant cell tumor of bone. Am J Surg Pathol. 2017;41:1059.
Xu SF, Adams B, Yu XC, Xu M. Denosumab and giant cell tumour of bone-a review and future management considerations. Curr Oncol. 2013;20:e442–7.
Palmerini E, Picci P, Reichardt P, Downey G. Malignancy in giant cell tumor of bone: a review of the literature. Technol Cancer Res Treat. 2019;18:1533033819840000–00.
Muheremu A, Niu X. Pulmonary metastasis of giant cell tumor of bones. World J Surg Oncol. 2014;12:261.
Chakarun CJ, Forrester DM, Gottsegen CJ, Patel DB, White EA, George R, Matcuk J. Giant cell tumor of bone: review, mimics, and new developments in treatment. Radiographics. 2013;33:197–211.
López-Pousa A, Broto JM, Garrido T, Vázquez J. Giant cell tumour of bone: new treatments in development. Clin Transl Oncol. 2015;17:419–30.
Balke M, Schremper L, Gebert C, et al. Giant cell tumor of bone: treatment and outcome of 214 cases. J Cancer Res Clin Oncol. 2008;134:969–78.
Zhang S, Zhang J, Wang X. Comparison of tumor curettage and resection for treatment of giant cell tumor of the bone around the knee joint. Pak J Med Sci. 2016;32:662–6.
Rutkowski P, Ferrari S, Grimer RJ, et al. Surgical downstaging in an open-label phase II trial of denosumab in patients with giant cell tumor of bone. Ann Surg Oncol. 2015;22:2860–8.
Chawla S, Blay J-Y, Rutkowski P, et al. Denosumab in patients with giant-cell tumour of bone: a multicentre, open-label, phase 2 study. Lancet Oncol. 2019;20:1719–29.
Zhong L, Li Y, Xiong L, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther. 2021;6:201.
Lopez JS, Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol. 2017;14:57–66.
Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–92.
Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480.
Gengenbacher N, Singhal M, Augustin HG. Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat Rev Cancer. 2017;17:751–65.
Su D, Zhang D, Jin J, et al. Identification of predictors of drug sensitivity using patient-derived models of esophageal squamous cell carcinoma. Nat Commun. 2019;10:5076.
Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710.
Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–53.
Kodack DP, Farago AF, Dastur A, et al. Primary patient-derived cancer cells and their potential for personalized cancer patient care. Cell Rep. 2017;21:3298–309.
Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Can Res. 2014;74:2377.
Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Bracht K, Nicholls AM, Liu Y, Bodmer WF. 5-Fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency. Br J Cancer. 2010;103:340–6.
Bairoch A. The Cellosaurus, a cell-Line knowledge resource. J Biomol Tech. 2018;29:25–38.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of novel patient-derived cell lines from giant cell tumor of bone. Hum Cell. 2021;34:1899–910.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
He M, Jiang L, Ren Z, Wang G, Wang J. Noscapine targets EGFRp-Tyr1068 to suppress the proliferation and invasion of MG63 cells. Sci Rep. 2016;6:37062.
Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Lee J-C, Liang C-W, Fletcher CDM. Giant cell tumor of soft tissue is genetically distinct from its bone counterpart. Mod Pathol. 2017;30:728–33.
Divisato G, di Carlo FS, Pazzaglia L, et al. The distinct clinical features of giant cell tumor of bone in pagetic and non-pagetic patients are associated with genetic, biochemical and histological differences. Oncotarget. 2017;8(38):63121.
Noguchi R, Yoshimatsu Y, Ono T, et al. Establishment and characterization of NCC-GCTB1-C1: a novel patient-derived cancer cell line of giant cell tumor of bone. Hum Cell. 2020;33:1321–8.
Grant C, Rahman F, Piekarz R, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10:997–1008.
Zarin DA, Fain KM, Dobbins HD, Tse T, Williams RJ. 10-Year update on study results submitted ClinicalTrials.gov. N Engl J. 2019;381:1966–74.
Yafei J, Haoran M, Wenyan J, et al. Personalized medicine modality based on patient-derived xenografts from a malignant transformed GCTB harboring H3F3A G34W mutation. J Orthop Transl. 2021;29:106–12.
Wagner MJ, Livingston JA, Patel SR, Benjamin RS. Chemotherapy for bone sarcoma in adults. J Oncol Pract. 2016;12:208–16.
Acknowledgements
We would like to thank the Tochigi Cancer Center Operating Room Nurse Team and the Secretary of the Medical Office for their assistance in processing and transporting the samples. We also appreciate the technical assistance provided by Mrs. Y. Kuwata (Division of Rare Cancer Research), and the technical support provided by Mrs. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for providing English language editing services and for their constructive comments on the manuscript. This research was technically assisted by the Fundamental Innovative Oncology Core of the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The ethical committee of the Tochigi Cancer Center and the National Cancer Center approved the use of clinical materials for this study. The animal experiment was conducted in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute.
Informed consent
Written informed consent for publication was provided by the patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2021_639_MOESM1_ESM.tif
Supplementary Fig. 1 Short tandem repeat patterns of NCC-GCTB4-C1 and original tumor tissue. (A) Short tandem repeat patterns of NCC-GCTB4-C1 (B) Short tandem repeat patterns of original tumor tissue
Rights and permissions
About this article
Cite this article
Ono, T., Noguchi, R., Yoshimatsu, Y. et al. Establishment and characterization of the NCC-GCTB4-C1 cell line: a novel patient-derived cell line from giant cell tumor of bone. Human Cell 35, 392–399 (2022). https://doi.org/10.1007/s13577-021-00639-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00639-4