Abstract
Chronic kidney disease (CKD) disrupts mineral homeostasis and its representative pathosis is defined as secondary hyperparathyroidism (SHPT). SHPT occurs during the early course of progressive renal insufficiency, and is associated with mortality and cardiovascular events. SHPT results in reduction of calcium-sensing receptor (CaSR) and vitamin D receptor (VDR) in the parathyroid glands during CKD. However, the precise mechanism of CaSR and VDR reduction is largely unknown. CKD was induced through two-step 5/6 nephrectomy, and then CKD rats and sham-operated rats were maintained for 8 weeks on diets containing 0.7 % phosphorus (normal phosphate) or 1.2 % phosphorus (high phosphate). In gene expression analysis, TaqMan probes were used for quantitative real-time polymerase chain reaction. Finally, CaSR and VDR protein expressions were analyzed using immunohistochemistry. DNA methylation analysis was performed using a restriction digestion and quantitative PCR. CaSR and VDR mRNA were reduced only in CKD rats fed the high-phosphorus diets (CKD HP), then CaSR and VDR immunohistochemical expressions were compatible with gene expression assay. SHPT was then confirmed only in CKD HP rats. Furthermore, sole CKD HP rats showed the hypermethylation in CaSR and VDR genes; however, the percentage methylation of both genes was low. Although CaSR and VDR hypermethylation was demonstrated in PTGs of CKD HP rats, the extent of hypermethylation was insufficient to support the relevance between hypermethylation and down-regulation of gene expression because of the low percentage of methylation. Consequently, our data suggest that mechanisms, other than DNA hypermethylation, were responsible for the reduction in mRNA and protein levels of CaSR and VDR in PTGs of CKD HP rats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parathyroid hormone (PTH) secreted by the parathyroid glands (PTGs) is a crucial regulator of mineral homeostasis. Elevated serum PTH levels are known to be caused by vitamin D deficiency, rickets, and chronic kidney disease (CKD). CKD is a global public health problem that shortens lifespan, primarily by increasing the risk of cardiovascular disease [1]. With CKD, secondary hyperparathyroidism (SHPT) is a common disorder that occurs early in progressive renal insufficiency as an adaptive mechanism to help maintain mineral homeostasis [2]. SHPT induces phosphate retention, hyperphosphatemia, elevated PTH levels, elevated fibroblast growth factor 23 (FGF23) and subsequent 1,25-dihydroxyvitamin (1,25(OH)2D) deficiency, hypocalcemia, intestinal calcium malabsorption, and reduced calcium-sensing receptor (CaSR) and vitamin D receptor (VDR) expression in the PTGs [3, 4]. Then SHPT largely contributes to death and cardiovascular events in CKD patients. Although many regulatory factors are involved in mineral homeostasis, CaSR and VDR are regarded as major determinants of parathyroid function in CKD. CaSR directly modulates PTH secretion, whereas both CaSR and VDR signaling affect pre-pro-PTH gene transcription, PTH mRNA expression, and parathyroid hyperplasia.
During long-term and end-stage renal failure, PTG growth accelerates along with a shift in the hyperplasia pattern from a diffuse to a nodular type. Reduction in CaSR and VDR occurs throughout this process, although the underlying mechanism remains largely unknown. Our previous study focused on the epigenetics, particularly DNA hypermethylation, because gene hypermethylation is a commonly reported mechanism for reducing gene expression [5]. Reports of epigenetic studies for various diseases, such as metabolic syndrome, heart failure, cancer, allergy, and chronic inflammatory disease, have increased in recent years. Some studies have also assessed the epigenetics of CKD, such as the relevance of DNA hypermethylation and cardiovascular mortality in patients with CKD [6]. Epigenetic modifications in patients with CKD are under active investigation. However, there are few reports relating these changes to mineral homeostasis, particularly in parathyroid diseases.
Here, we investigated the methylation status of CaSR and VDR in the PTGs of CKD rats fed normal-phosphate diets, high-phosphate diets, and sham-operated rats fed normal-phosphate diets, high-phosphate diets.
Materials and methods
Animals
All experiments were performed in accordance with the Animal Care and Use Committee of The Jikei University School of Medicine. Seven-week-old adult male Wistar rats (Japan-CREA, Japan) were fed a standard diet (CE-II) with free access to food and water until the completion of the second surgery.
Nephrectomy and dietary intake of phosphorus
Chronic kidney disease was induced by a two-step 5/6 nephrectomy. In the first surgery, 8-week-old rats were anesthetized by peritoneal injection of pentobarbital (30–50 mg/kg). The right kidney (approximately 600 mg) was exposed and excised. Following a 1-week recovery period, the second surgery was performed, during which the left kidney was removed. Control animals (shams) were similarly anesthetized and subjected to surgical procedures without kidney excision and removal. On the day after the second surgery, the sham surgery with normal-phosphorus diet group (sham NP) was fed a standard diet that contained 0.9 % calcium, 0.7 % phosphorus, and 1000 IU/kg vitamin D, the sham surgery with high-phosphorus diet group (sham HP) was fed a high-phosphorus diet that contained 0.9 % calcium, 1.2 % phosphorus, and 1000 IU/kg vitamin D, and the CKD with normal-phosphorus diet group (CKD NP) was fed a standard diet, and CKD with high-phosphorus diet group (CKD HP) was fed a high-phosphorus diet, with free access to food and water. The rats were maintained under these conditions for 8 weeks [7].
Parathyroidectomy
The PTGs of 17-week-old control and CKD rats were removed by microsurgery and immediately frozen in liquid nitrogen [7].
Serum determinations
Blood urea nitrogen(BUN), creatinine (Cr), calcium (Ca), and phosphate (Pi) levels were analyzed by SPOTCHEM D-00 and D-02 (ARKRAY, Japan) and PTH levels were evaluated using a rat intact PTH (iPTH) enzyme-linked immunosorbent assay (ELISA) kit (Immutopics, San Clemente, CA, USA).
Immunohistochemistry
All tissue specimens were fixed in 4 % paraformaldehyde (PFA), processed, and embedded in paraffin using routine methods. PFA-fixed, paraffin-embedded, 3-μm-thick tissue sections were serially cut and mounted on MAS-coated glass slides. Slides were deparaffinized in xylene and dehydrated through an ethanol series. Before incubation with primary antibodies, tissue sections were treated with HistoVT one (Nacalai Tesque, Kyoto, Japan) (Ki67) and 10 mM citrate, pH 6.0 (CaSR, VDR), first at 105 °C for 15 min (Ki67, VDR) and then at 121 °C for 20 min (CaSR) for antigen retrieval in a high-pressure steam sterilizer (TOMY, Tokyo, Japan). Non-specific binding was blocked using goat serum in 1 % Tween 20/phosphate-buffered saline (PBST) for 30 min at room temperature. The tissue sections were then incubated with anti-VDR (SC-13133; Santa Cruz Biotechnology Inc., CA, USA), anti-CaSR (ab79038; Abcam, UK), or anti-Ki67 (RM-9106; LabVision, CA, USA) primary antibodies overnight at 4 °C. Bound primary antibodies were detected with biotinylated goat anti-rabbit IgG (Vector Laboratories, CA, USA) for Ki67, CaSR and biotinylated goat anti-mouse IgG (Vector Laboratories, CA, USA) for VDR, followed by the Vectastain Elite ABC standard kit (Vector Laboratories, CA, USA) according to the manufacturer’s instructions. After the final slide wash in PBST, tissue sections were stained using DAB-H2O2 as a substrate followed by counterstaining with hematoxylin. The distribution of immunoreactive proteins was analyzed by quantifying nuclear staining in randomly selected areas on each slide. The number of positive immunoreactive nuclei per 1000 parenchymal cells (Ki67) was expressed as the labeling index (LI).
RNA isolation and quantitative real-time PCR
Total RNA from each of the four unilateral PTGs from the four groups were pooled. Total RNA was extracted using the RiboPure Kit (Ambion) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 300 ng of RNA using the High-Capacity RNA-to-cDNA kit (ABI). Rotor-Gene 6000 (QIAGEN) and TaqMan Gene Expression Master Mix (ABI) were used for quantitative real-time PCR. The TaqMan probe identifiers are listed in Table 1. All samples were run in triplicate. Gene activity was normalized to the mean with the housekeeping gene β-actin.
Quantitative analysis of DNA methylation using real-time PCR (qAMP)
Genomic DNA from each of the six unilateral PTGs from the four groups was pooled. Genomic DNA was extracted using DNeasy blood and tissue kit (QIAGEN) according to the manufacturer’s protocol. Next, the extracted genomic DNA was digested with EcoRI (TaKaRa), followed by inactivation of the restriction enzyme using ethanol precipitation. Then 1 μg of each DNA sample was digested with HapII (TaKaRa) and HhaI (TaKaRa). Restriction enzymes were heat inactivated by incubating the reactions at 70 °C for 15 min. qPCR was performed to amplify and quantify the methylated genome using the Rotor-Gene 6000 (QIAGEN) and Rotor-Gene SYBR green PCR kit (QIAGEN). The following formula was used to describe the relationship of ΔC t to percent methylation: %methylation = 100 \((2^{{ - \Delta C_{\text{t}} }} )\). A previous study reported variable methylation patterns at the restriction sites [8]. Therefore, we defined the hypermethylation status was the only situation in which significant increases were confirmed in both the HapII and HhaI assays. All primer sequences used in this study are listed in Table 2.
Statistical analysis
The Kruskal–Wallis test followed by the Mann–Whitney test with Bonferroni correction were used for post hoc analysis in relation to biochemical examination, with p < 0.05 in Kruskal–Wallis test and 0.0083 in post hoc analysis considered statistically significant. The Mann–Whitney test was also performed in conjunction with other analyses, with p < 0.05 considered statistically significant.
Results
CKD model with SHPT induced by 5/6 nephrectomy and high phosphorus intake
The biochemical parameters of male sham-operated and nephrectomized rats with two types of phosphorus diets are shown in Table 3. A significant decrease in Ca levels and the significant increases in serum Pi, iPTH, BUN, and Cr levels were confirmed in the CKD HP rats, as compared with the other three groups. In CKD NP rats, BUN and Cr levels significantly increased compared to sham groups and there is no significant difference in Ca, Pi levels compared to sham NP rats. The severity of CKD and chronic kidney disease-mineral and bone disorder (CKD-MBD) was greater in the CKD HP rats compared with the CKD NP rats.
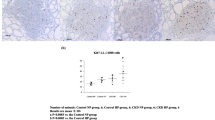
Next, we analyzed the SHPT status and CKD-MBD status. Elevations in both serum iPTH and PTG were confirmed using a stereoscopic microscope (data not shown) in CKD HP rats. SHPT, the hyperplastic PTGs were in proliferative state [9]; therefore, we analyzed Ki67 expression in four groups. The increase in Ki67 immunohistochemical expression was observed in sham HP and CKD HP rats (Fig. 1a), and a significant increase in the Ki67 expression cells in the CKD HP rats relative to that in sham NP rats was confirmed (Fig. 1b). Ki67 expression levels were significantly increased in the sham HP rats as compared with the sham NP rats (Fig. 1b). From the above, SHPT and severe CKD-MBD status correspond to only CKD HP rats.
Representative immunohistochemical findings of Ki67 and hematoxylin counterstain (×100) (a), showing differences in the mean labeling index (LI) of Ki67 protein expression among the four groups. a Nuclear Ki67 expression. Many nuclei in CKD HP were positively stained for the Ki67 protein. b In CKD HP rats, Ki67 protein expression was significantly increased compared with that of the other groups. Number of animals: sham NP, 3; sham HP, 3; CKD NP, 3; and CKD HP, 3. Results are presented as the mean ± SD. *P < 0.05 vs. the CKD HP group, **P < 0.05 vs. the sham NP group
CaSR and VDR reduction was observed in CKD HP rats
SHPT reduced CaSR and VDR levels in both humans and rats [3, 4], then the changes in PTGs CaSR and VDR mRNA expressions, and protein expressions were quantified using real-time RT-PCR and were analyzed by immunohistochemistry. These results are shown in Fig. 2. CaSR expression was significantly decreased in the CKD HP rats compared with the sham NP rats as previously reported [9], then sham HP and CKD NP expressions were not significantly different compared to sham NP rats (Fig. 2a). Furthermore, CaSR protein expressions in PTGs were compatible with gene expression assay (Fig. 2c–f). In the CKD HP rats, VDR expression was also significantly decreased compared to sham NP rats (Fig. 2b), then the expressions in sham HP rats was significantly increased compared with sham NP rats and the expressions in CKD NP rats was significantly increased compared to sham NP rats. Moreover, VDR immunohistochemical expressions in these groups’ PTGs were same as gene expressions (Fig. 2g–j). We verified the consistency of antibodies using Western blot analysis and immunohistochemical analyses (Supplemental figure S1, S2).
Gene expression of CaSR (a) and VDR (b) in PTGs and typical immunohistochemical findings of CaSR (c–f) and VDR (g–j) protein expression in PTGs resected from each of four groups. a, b Effects of 5/6 nephrectomy and high phosphorus intake on CaSR and VDR expression, showing that relative mRNA levels were significantly decreased. All gene expression data were measured using real-time RT-PCR and normalized to β-actin. c–j Representative immunohistochemical findings of CaSR and VDR and hematoxylin counterstain (×100). c–f CaSR and g–j VDR protein expressions are shown. These results were in agreement with those of gene expression analysis. Number of animals: in gene expression assay sham NP, 4; sham HP, 4; CKD NP, 4; CKD HP, 4; and in protein expression assay sham NP, 3; sham HP, 3; CKD NP, 3; CKD HP, 3. Results are presented as the mean ± SD. *P < 0.05 vs. the CKD HP group, **P < 0.05 vs. the sham NP group
Hypermethylation of CaSR and VDR genes in PTGs were observed in CKD rats with high phosphorus intake
To elucidate the reasons for the reduced expression of CaSR and VDR, we focused on the epigenetic mechanism. Gene hypermethylation is a commonly reported mechanism for reducing gene expression [5]. qAMP was used to evaluate changes in CaSR and VDR DNA methylation in four groups. CaSR CpG islands are located in the vicinity of exons 1, 4, 6, and 9. Our assessment targeted the CpG island approximately 460-bp upstream of CaSR exon 1. Analysis results showed CaSR hypermethylation in CKD HP rats (Fig. 3a, b).
Analysis of the DNA methylation of CaSR (a, b) and VDR (c, d) using qAMP. Methylation status in PTGs of the four groups was analyzed using the restriction enzymes HapII (a, c) and HhaI (b, d). Hypermethylation was observed in CKD HP rats. Number of animals: sham NP, 6; sham HP, 6; CKD NP, 6; and CKD HP, 6. Results are mean ± SD. *P < 0.05 vs. the CKD HP group, **P < 0.05 vs. the CKD NP group
Next, VDR CpG islands are located in the vicinity of exon 1 and introns 2 and 4. We targeted the VDR CpG island approximately 560-bp upstream of the first exon. We found hypermethylation in CKD NP rats’ and CKD HP rats’ DNA samples digested with HapII (Fig. 3c); however, the hypermethylation was observed in only CKD HP rats’ DNA samples digested with HhaI (Fig. 3d). Thus, it is only in CKD HP rats that the hypermethylation was confirmed in both HapII and HhaI assays. Accordingly, this VDR CpG island was hypermethylated specifically in CKD HP rats (Fig. 3c, d).
However, the extent of methylation of both genes was low.
Discussion
SHPT and decreased CaSR and VDR expression were only detected in CKD rats consuming a high-phosphorus diet
Previous reports have shown that CaSR mRNA and protein expression is significantly decreased in uremic rats consuming a high-phosphorus diet [9, 10], and that VDR mRNA is significantly decreased in uremic rats consuming either a normal or a high-phosphorus diet [10]. In the present study, CaSR mRNA was significantly decreased only in CKD HP rats; however, the reduction in VDR expression was also only significant in CKD HP rats (Fig. 2a, b). SHPT and decreased CaSR and VDR expression were only found in CKD HP rats. Meanwhile, VDR expression was significantly increased in the sham HP rats and CKD NP rats relative to that in the sham NP rats. Calvo et al. reported that a high-phosphorus diet in a rodent model elevated serum FGF23 levels and a subsequent decrease in serum 1,25(OH)2D levels [11]. Hence, it was conceivable that the VDR expression in PTGs of sham HP rats was increased to compensate for the reduction in serum 1,25(OH)2D levels. Regarding the increase in VDR expression in PTGs of CKD NP rats, Nitta et al. reported that a decrease in 1,25(OH)2D levels is an early event in CKD and levels are progressively reduced along with the decline in renal function [12]. Serum 1,25(OH)2D was expected to be decreased in CKD NP rats; hence, it was conceivable that the VDR expression in PTGs of CKD NP rats was increased to compensate for the reduction in serum 1,25(OH)2D levels. Immunohistochemical evaluation of CaSR and VDR protein expression was in line with the results of CaSR and VDR expression analyses (Fig. 2c–j). However, we did not determine 1,25(OH)2D levels, which was a limitation to this study.
Epigenetic changes of CaSR and VDR genes in PTGs of CKD rats with high phosphorus intake
Many recent studies have reported a relationship between chronic diseases and altered epigenetics [5, 6]. Klotho is an important gene in CKD-MBD, and transcriptional suppression of Klotho by indoxyl sulfate and p-cresyl sulfate is correlated with Klotho CpG hypermethylation [13]. Moreover, multiple reports have correlated hypermethylation with reduced gene expression [14]. Accordingly, we suggest that during CKD, CaSR and VDR expression is suppressed by direct hypermethylation of CaSR and VDR, as we confirmed. A previous study reported no change in CaSR and VDR methylation in SHPT rats [15]. This discrepancy from our results may be explained by the fact that our experimental SHPT rats had more severe disease and by differences in study analytical methods. The mean ± SD intact PTH levels in our SHPT rats were 2645.0 ± 249.8 pg/ml, whereas the other study reported levels of 1851 ± 343 pg/ml. Melting curve analysis after bisulfite conversion was used to detect methylation in the previous study; however, we used qAMP methods.
We showed the hypermethylation in CaSR and VDR, however, DNA methylation percentages were low. Although additional studies are required to assess DNA methylation of other regions of the CaSR and VDR genes and histone modifications, our data suggest that other mechanisms are responsible for the decreased expression of CaSR and VDR in PTGs of CKD HP rats.
In conclusion, although CaSR and VDR hypermethylation was demonstrated in PTGs of CKD HP rats, the extent of hypermethylation was insufficient to support the relevance between hypermethylation and down-regulation of gene expression because of the low percentage of methylation. Consequently, our data suggest that mechanisms, other than DNA hypermethylation, were responsible for the reduction in mRNA and protein levels of CaSR and VDR in PTGs of CKD HP rats.
References
Eckardt KU, Coresh J, Levin A. Evolving important of kidney disease: from subspecialty to global health burden. Lancet. 2013;382:15–169.
Parfitt AM. The hyperparathyroidism of chronic renal failure: a disorder of growth. Kidney Int. 1997;52:3–9.
Korkor AB. Reduced binding of [3H]1,25-dihydroxyvitamin D3 in the parathyroid glands of patients with renal failure. N Engl J Med. 1987;316:1573–7.
Ritter CS, Finch JL, Brown AJ. Parathyroid hyperplasia in uremic rats precedes down-regulation of the calcium receptor. Kidney Int. 2001;60:1737–44.
Wajapeyee N, Serra RW, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–74.
Dwivedi RS, Herman JG, Raj DS. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011;79:23–32.
Hofman-Bang J, Olgaard K, Lewin E. Increased parathyroid expression of klotho in uremic rats. Kidney Int. 2010;78:1119–27.
Jang SJ, Li M. Methylation patterns in human androgen receptor gene and clonality analysis. Cancer Res. 2000;60:864–6.
Lewin E, Garfia B, Olgaard K. Persistent downregulation of calcium-sensing receptor mRNA in rat parathyroids when severe secondary hyperparathyroidism is reversed by an isogenic kidney transplantation. J Am Soc Nephrol. 2002;13:2110–6.
Brown AJ, Ritter CS, Slatopolsky EA. Decreased calcium-sensing receptor expression in hyperplastic parathyroid glands of uremic rats: role of dietary phosphate. Kidney Int. 1999;55:1284–92.
Calvo MS, Uribarri J. Public health impact of dietary phosphorus excess on bone and cardiovascular health in the general population. Am J Clin Nutr. 2013;98:6–15.
Nitta K, Nagano N, Tsuchiya K. Fibroblast growth factor 23/klotho axis in chronic kidney disease. Nephron Clin Pract. 2014;128:1–10.
Sun CY, Chang SC, Wu MS. Suppression of Klotho expression by protein-bound uremic toxins is associated with increased DNA methyltransferase expression and DNA hypermethylation. Kidney Int. 2012;81:640–50.
Song JZ, Stirzaker C, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–61.
Hofman-Bang J, Gravesen E, Lewin E. Epigenetic methylation of parathyroid CaR and VDR promoters in experimental secondary hyperparathyroidism. Int J Nephrol. 2012;2012:123576.
Acknowledgments
This work was supported by a grant for pathophysiological research conference in chronic kidney disease (Grant No. JKFB11-14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors have declared no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure S1. Typical immunohistochemical findings of CaSR, VDR protein expression in wild rat kidney and hematoxylin counterstain. Left panel is low magnification (× 25), right panel is high magnification (× 100). In the wild rat kidney, (a) the CaSR was primarily present in the cell membrane and cytoplasm of distal tubules and (b) the VDR was in the nucleus of renal tubular cells.
Supplemental Figure S2. The protein expressions of CaSR (a) and VDR (b) were found using western blot analysis on cell lysates from rat PTG. (a, b) the CaSR-specific and VDR-specific bands (arrows) were not present in HEK 293 cells. Therefore, the observed bands are specific for the CaSR and VDR. (c) Beta-actin (ActB) was used as a loading control
Rights and permissions
About this article
Cite this article
Uchiyama, T., Tatsumi, N., Kamejima, S. et al. Hypermethylation of the CaSR and VDR genes in the parathyroid glands in chronic kidney disease rats with high-phosphate diet. Human Cell 29, 155–161 (2016). https://doi.org/10.1007/s13577-016-0143-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-016-0143-9