Abstract
Purpose
Although it has been reported that up-regulation of phosphofructokinase (PFK) expression is a major feature of malignant tumors, the role of platelet type PFK (PFKP) in tumor initiation and progression is as yet poorly understood. The objective of this study was to evaluate PFKP expression in lung cancer and its effect on glycolysis, and to explore correlations between PFKP expression levels and clinical lung cancer patient features.
Methods
PFKP mRNA expression levels in cancer tissues and adjacent normal tissues were compared using The Cancer Genome Atlas (TCGA) database. PFKP mRNA and protein expression levels in fresh lung cancer tissues and cell lines were assessed using quantitative real-time PCR and Western blotting. Immunohistochemistry (IHC) was used to assess PFKP expression in 150 archival lung adenocarcinoma samples, after which follow-up data and their correlations with clinical features and patient prognosis were evaluated. A retroviral shRNA-mediated method was used to construct stable cell lines expressing low levels of PFKP. Glucose, lactate and adenosine triphosphate concentrations in the cell culture supernatants were determined using enzymatic, spectrophotometric and enzyme-linked immunosorbent (ELISA) assays, respectively. The effect of PFKP expression on the proliferation of lung cancer cells was assessed using colony forming, MTT and flow cytometry assays, respectively. Finally, data from tissue samples of 533 patients with lung adenocarcinoma and 502 patients with lung squamous cell carcinoma were downloaded from the TCGA database, after which pathway and gene correlation information was retrieved using gene set enrichment analyses.
Results
We found that PFKP was highly expressed in lung cancer tissues and cell lines. Using IHC we found that PFKP was highly expressed in primary lung adenocarcinoma tissues and that a high expression was associated with a poor prognosis. Clinical data analysis revealed that the PFKP expression levels correlated with tumor size and patient survival. Lung cancer cell lines with decreased PFKP expression levels showed significant decreases in glucose uptake rates, lactate levels and adenosine triphosphate concentrations. They also exhibited significantly decreased proliferation rates, colony forming abilities and increased G2/M cell cycle phase percentages. Gene set enrichment analysis revealed that multiple pathways, including glycolytic pathways, may be involved in the regulation PFKP.
Conclusions
Our data indicate that PFKP is highly expressed in lung cancer tissues and cell lines and is associated with tumor size and patient prognosis. As such, PFKP may serve as a prognostic biomarker. We also found that PFKP regulates the level of glycolysis in lung cancer cells and is associated with lung cancer cell proliferation. These data may be instrumental for the design of new lung cancer treatment options.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with an average 5-year survival rate of 16% [1]. Non–small cell lung cancer (NSCLC) accounts for 80% of all lung cancer cases and has a high incidence in underdeveloped countries [2]. NSCLC local invasion (T), lymph node metastasis (N) and distant metastasis (M) significantly affect the prognosis of patients, and tumor progression is inseparable from energy supply. Unlike normal cells, in which oxidative phosphorylation occurs in mitochondria, tumor cells primarily acquire energy through glycolysis, which results in the production of large amounts of lactic acid and small amounts of adenosine triphosphate (ATP). This phenomenon is known as the Warburg effect [3,4,5]. The Warburg effect occurs in various tumor tissues and promotes the proliferation, invasion and metastasis of tumor cells and their resistance to radiotherapy and chemotherapy. Although this hampers the efficacy of current treatment regimens, it may also indicate directions for the development of new treatment options. Several studies have shown that the Warburg effect and the degree of dependence of tumor cells on it are determined by both intracellular and extracellular environments in which lactic acid, glycolytic enzyme and hypoxia-inducible factor-1 levels and tumor proliferation and metastasis are closely related [6,7,8]. Therefore, it is of major clinical significance to thoroughly study the involvement of glycolysis in lung cancer development and to identify new clues to inhibit its proliferation and growth.
The conversion of fructose 6-phosphate to fructose 1,6-diphosphate, which is catalyzed by phosphofructokinase 1 (PFK-1), is the first key step in glycolysis. PFK-1 is a complex tetrameric enzyme that contains three subtypes: platelet type (PFKP), muscle type (PFKM) and liver type (PFKL) [9, 10]. Although up-regulation of PFK expression is a major feature of malignant tumors, the role of PFKP in tumor initiation and progression, as well as cancer-related phenotypic changes, are poorly understood. Previous studies have shown that PFKP expression may be associated with tumor type, i.e., its expression has been found to be low in seminomas and embryonal carcinomas [11] and to be high in human breast tumor cells [12]. It has also been found that PFKP regulates the glucose metabolism and phenotypic reorganization of glioma stem cells [13]. As yet, however, it is not clear whether alterations in PFKP expression and glucose metabolism are associated with lung cancer. Here, we measured the PFKP mRNA and protein expression levels in primary lung cancer tissues and cell lines and evaluated correlations between PFKP expression and clinicopathologic features including the prognosis of lung cancer patients. We also downregulated the expression of PFKP in lung cancer cells and assessed its effect on glycolysis in these cells. Additionally, we investigated the effect of PFKP expression on lung cancer cell colony formation, proliferation and cell cycle progression.
2 Materials and methods
2.1 Cell lines and culture
The normal human bronchial epithelial cell line HBE and the lung adenocarcinoma cell lines NCI-1975, A549, NCI-H1650, H358, H827 and H2228 were obtained from the Guangzhou Institute of Respiratory Diseases, State Key Laboratory of Respiratory Diseases. The cells were cultured in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics.
2.2 Clinical specimens and data collection
Thirty fresh lung adenocarcinoma specimens excised by thoracic surgery from July 2014 to June 2015 were randomly collected and divided into cancerous tissues and adjacent non-cancerous tissues (more than 5 cm from the tumors) for protein extraction. Paraffin-embedded lung cancer specimens (150 cases from June 2010 to June 2012) that were confirmed pathologically were collected from the tissue bank of Taizhou Hospital of Zhejiang Province. For all cases complete follow-up data were available. The patient data are listed in Table 1. Normal lung tissues were derived from benign lung lesion specimens. Written informed consent was obtained from all patients or their family members.
2.3 TCGA data retrieval and analysis
PFKP mRNA expression data from cancer tissues and para-cancerous tissues were downloaded from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), after which data analyses were performed using the R language.
2.4 Quantitative real time PCR (qRT-PCR)
Total RNA of cells and tissues was extracted using the TRIzol method and reverse-transcribed into cDNA using a cDNA synthesis kit (TaKaRa, Japan). Primers were designed using Primer 5.0 software and synthesized by Invitrogen. PCR was performed with SYBR GREEN on an ABI 7300 plus real-time system. The primer sequences used were as follows: GAPDH-forward AATCCCATCACCATCTTCCA, GAPDH-reverse CCTGCTTCACCACCTTCTTG, PFKP-forward TCGGTCTCTCCAAGTTTGTGC, PFKP- reverse TTCCGTTGTCCACCAATCTGA. The data were normalized using the 2-ΔΔCt method. The PFKP mRNA expression levels in HBE cells and control samples were set at 1, after which relative PFKP abundances were calculated.
2.5 Western blot analysis
Tumor tissue samples were rapidly grinded using a grinding rod until powdered with continuous liquid nitrogen addition. Protein concentrations were determined using a BCA protein kit (Biyuntian Bio-technology Co., Shanghai, China). Next, the proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. After blocking with 5% bovine serum albumin, the membranes were incubated with a specific primary antibody overnight at 4 °C, washed with tris-buffered saline and incubated with a secondary antibody for 1 h. Finally, proteins were visualized by LAS 500 using an enhanced chemiluminescent chromogenic reagent. The Western blot results were processed using Image J software. The relative PFKP protein expression level was determined by the gray value of PFKP versus the gray value of the internal reference α-tubulin. The antibodies used were anti-PFKP (ab204131, Abcam) and anti-α-tubulin (ab210797, Abcam).
2.6 Immunohistochemistry (IHC)
Paraffin-embedded specimens were dewaxed with xylene, hydrated using an ethanol gradient and exposed to microwave antigen retrieval. Next, a hydrogen peroxide methanol solution was added (one drop at a time), after which the specimens were incubated with a primary antibody at 4 °C overnight. After rewarming, a horseradish peroxidase conjugated secondary antibody was added and DAB was developed. The intensity and extent of IHC staining were independently assessed by two pathologists, and their mean values were recorded. The target protein expression results were evaluated by combining the staining intensity with the staining range. The staining intensity was scored from 1 to 4: negative, 1 point; weakly positive, 2 points; positive, 3 points; strongly positive, 4 points. The staining range was scored as follows: Four different fields of view were randomly selected under a high-power microscope (200×), after which the total number of cells and the number of cytoplasmic positive cells were counted and the percentages of positive cells were scored from 1 to 4: 0% to 25%, 1 point; 26% to 50%, 2 points; 51% to 75%, 3 points; 76% to 100%, 4 points. The final staining score (expressed in p) was calculated by multiplying the staining intensity of the section by the staining range, defined as four levels: –, p ≤ 4; +, 4 > p ≤ 8; ++, 8 > p ≤ 12; +++, 12 > p ≤ 16. The symbols – and + indicate negative or weakly positive expression, whereas ++ and +++ indicate positive expression.
2.7 Construction of stable cell lines expressing low levels of PFKP
The NSCLC cell lines NCI-H1650 and A549 were infected with the retroviral vector pSUPER that harbored a PFKP-shRNA. After infection, cell lines stably expressing low levels of PFKP were selected in culture medium containing puromycin (0.5 μg/ml).
For glucose oxidase-peroxidase (GOD-POD), spectrophotometric, enzyme-linked immunosorbent assay (ELISA), colony formation and MTT procedures: see Supplementary Material.
2.8 Cell cycle analysis by flow cytometry
A single-cell suspension was added to a 2 ml round-bottom centrifuge tube and washed by centrifugation (1500 rpm, 5 min), after which the supernatant was discarded. Next, 2 ml pre-cooled 70% alcohol was added and the cells were fixed at 4 °C for 30 min. Subsequently, RNase A (working concentration 20 μg/ml) was added to degrade the RNA and propidium iodide (PI) solution was added (working concentration 50 μg/ml), followed by incubation at room temperature for 30 min in the dark. Finally, the cells were mixed, passed through a 300-mesh screen and added to a flow tube for cytometric detection.
2.9 TCGA data analysis
The HTSeq-FPKM datasets of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) were downloaded from the cancer genome atlas (TCGA) database. Information on RNA expression levels in 533 LUAD and 502 LUSC specimens was retrieved. We used gene set enrichment analysis (GSEA) 4.0.3 software for our analyses. To this end, HTSeq-FPFM mRNA expression data of the 533 LUAD cases were imported into the GSEA software. According to the expression profile data, we set the first 50% of the samples of PFKP expression as high-expression group and the last 50% of the samples as low-expression group. The GSEA enrichment analysis-specific parameter settings were: c5.bp.v7.0.symbols.gmt; number of permutations: 1000; collapse dataset to gene symbols: false; enhancement statistic: weighted; and metric for ranking genes: signal-to-noise metric. The same method was used to analyze the data of the 502 LUSC cases.
2.10 Statistical analyses
For all statistical analyses the SPSS 16.0 software package was used. Measured data were expressed as χ2 ± SD. A paired sample t-test was used to compare groups. A χ2 test was used to count data. Survival curves were drawn using the Kaplan-Meier method. A log-rank test was used to compare survival rates of two groups. Statistical significance was indicated by p < 0.05.
3 Results
3.1 PFKP mRNA is highly expressed in lung cancer tissues and cell lines
Data from 31 patients with lung adenocarcinoma (LUAD) and 17 patients with lung squamous cell carcinoma (LUSC) were retrieved from the TCGA database and PFKP mRNA expression levels were compared in lung cancer tissues and normal adjacent tissues. Paired t-test analysis revealed that the PFKP mRNA levels in LUAD (Fig. 1a) and LUSC (Fig. 1b) tissues were higher than those in the adjacent normal tissues. This difference was statistically significant (p < 0.05).
To verify whether the expression of PFKP was also up-regulated in lung cancer cell lines, the PFKP mRNA levels in a series of lung cancer cell lines and a normal human bronchial epithelial (HBE) cell line were measured by qRT-PCR. We found that compared to the normal HBE cell line, the PFKP mRNA levels were significantly up-regulated in the following lung cancer cell lines: PC-9, NCI-H1650, NCI-H520, NCI-H460, H1975, HCC827 and A549 (Fig. 2a). This result was statistically significant (p < 0.05). In addition, we found that the PFKP mRNA expression levels were higher in eight fresh LUAD tissue samples compared to their adjacent normal tissue samples (Fig. 2b). These results are consistent with those of the TCGA, i.e., the expression of PFKP in lung cancer tissues is up-regulated.
3.2 PFKP protein is highly expressed in lung cancer tissues and cell lines
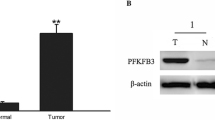
Western blot analyses were used to detect PFKP protein expression in lung cancer tissues and cell lines; α-tubulin was used as an internal reference. We found that the PFKP expression levels were significantly higher in the lung cancer cell lines PC-9, NCI-H1650, NCI-H520, NCI-H460, H1975, HCC827 and A549, compared to that in normal HBE cells (Fig. 3a). In order to confirm the high PFKP expression level in lung cancer tissues, we examined PFKP protein expression levels in 30 fresh lung adenocarcinoma tissues and adjacent normal tissues. By doing so, we found that the PFKP expression levels were significantly increased in the lung adenocarcinoma tissues compared to those in the adjacent normal tissues (Fig. 3b).
3.3 PFKP protein expression levels correlate with clinicopathological features and prognosis
In total 150 archival lung adenocarcinoma specimens (cancerous and para-cancerous tissues) with clinical follow-up data were assessed by IHC. We found that 89 cases were positive for PFKP expression, with a positivity rate of 66%. Of these, 45 were strongly positive, 34 were positive and 10 were weakly positive. In contrast, we found that PFKP expression in the adjacent tissues was weakly positive or negative, indicating that the expression of PFKP was significantly higher in the lung adenocarcinomas than in the adjacent normal tissues (Fig. 4a), which was statistically significant (p < 0.05).
High expression of PFKP in lung adenocarcinomas as measured by IHC (a). The expression of PFKP in lung adenocarcinomas was associated with prognosis: the PFKP high-expression group had a poor prognosis (overall survival) compared to the low-expression group (b), p = 0.001. A positive correlation was found between PFKP and Ki67 expression in lung adenocarcinomas (c and d), p = 0.001
A χ2 test was used to assess whether the PFKP expression levels (strongly positive and medium positive results were placed in a high-expression group; weakly positive and negative results were placed in a low-expression group) were associated with clinical and pathologic features of the lung cancer patients. We found that the PFKP expression level was correlated with tumor size and survival, which was statistically significant (p < 0.05) (Table 1). Conversely, we found that the expression level was not related to age, gender or regional lymph node metastasis status (p > 0.05).
In order to assess correlations between PFKP protein expression levels and patient prognosis, complete follow-up data of 109 patients with lung adenocarcinomas were analyzed. We found that 72 patients (66.06%) survived and 37 patients (32.94%) died during the follow-up period. The χ2 test revealed that the PFKP protein expression level was correlated with the survival status of the patients, which was statistically significant (p = 0.033). The 5-year survival rate was 78.4% (40/51) for the low-expression group and 48.3% (28/58) for the high-expression group. Survival curves were plotted using the Kaplan-Meier method, and the log-rank test was used to compare differences in survival rates between the two groups (Fig. 4b). The results indicated that the survival time of patients with a low PFKP protein expression was significantly longer than that of patients with a high PFKP protein expression (5-year survival rates were 67% and 36%, respectively), suggesting that patients with a low expression have a good prognosis, whereas those with a high expression have a poor prognosis. This difference was statistically significant (p = 0.001).
To further study the relationship between PFKP protein expression and the development of lung cancer, IHC was used to analyze the expression of PFKP in conjunction with the cell proliferation marker Ki67 in patient-derived lung cancer tissues (Fig. 4c and d). We found that there was a positive correlation between PFKP and Ki67 expression, suggesting that a high PFKP protein expression may be associated with a high proliferation rate in lung cancer cells.
3.4 Establishment of stable cell lines exhibiting a low PFKP expression
Stable NCI-H1650 and A549 cell lines with a low PFKP expression were generated using viral shRNA transfections. Next, RNA and protein were extracted from stably transfected and control cells after which the PFKP levels were detected by qRT-PCR and Western blotting, respectively. We found that the PFKP expression in the stably transfected lung cancer cell lines (PFKP-RNAi#1 and PFKP-RNAi#2) was significantly lower at both the mRNA (Fig. 5a) and protein (Fig. 5b) levels compared to those in the negative control cells. The difference was statistically significant (p < 0.05).
The expression of PFKP at the mRNA (a) and protein (b) levels was significantly lower in the PFKP-RNAi#1 and PFKP-RNAi#2 cell lines than the wild-type NCI-H1650 and A549 cell lines (negative controls). In the PFKP-RNAi#1 and PFKP-RNAi#2 cell lines the glucose uptake rates (c), lactic acid levels (d) and ATP concentrations (e) were decreased compared to the negative controls, p < .05
3.5 Effect of PFKP expression on glucose metabolism in lung cancer cells
In the stably transfected lung cancer cell lines the glucose uptake rate was found to be significantly decreased in the PFKP low-expression group compared to the negative control group (Fig. 5c). The glucose uptake rates of the NCI-H1650-derived cell lines PFKP-RNAi#1 and PFKP-RNAi#2 were 0.52 and 0.45 times those of the negative control group, respectively. A paired t-test revealed that there was a statistically significant difference in glucose uptake rates between the low PFKP expression group and the negative control group (p = 0.01 and 0.008, respectively). Similar results were obtained with the A549-derived cell lines with a low PFKP expression, i.e., the glucose uptake rates of the PFKP-RNAi#1 and PFKP-RNAi#2 cells were 0.61 and 0.5 times that of the negative control group, respectively. A paired t-test revealed that there was a statistically significant difference in glucose uptake rates between the low PFKP expression group and the negative control group (p = 0.04 and 0.02, respectively).
We also found that the lactic acid levels in the culture media of the low PFKP expression groups were significantly reduced compared to those in the negative control groups (Fig. 5d) (p < 0.05). The lactic acid levels of the NCI-H1650-derived cell lines PFKP-RNAi#1 and PFKP-RNAi#2 were 0.62 and 0.4 times those of the negative control group, respectively. A paired t-test revealed that these differences were statistically significant (p = 0.01 and 0.003, respectively). Similar results were obtained with the low PFKP expression A549-derived cell lines, i.e., the lactic acid levels of PFKP-RNAi#1 and PFKP-RNAi#2 cells were 0.63 and 0.5 times those of the negative control group, respectively. A paired t-test revealed that these differences were statistically significant (p = 0.01 and 0.005, respectively).
Additionally, we found that the ATP concentrations in the PFKP low-expression groups were significantly reduced compared to those in the negative control groups (Fig. 5e). The ATP concentrations of the NCI-H1650-derived cell lines PFKP-RNAi#1 and PFKP-RNAi#2 were 0.43 and 0.42 times those of the negative control group, respectively. A paired t-test revealed that these differences were statistically significant (p = 0.004 for both groups). Similar results were obtained with the PFKP low-expression A549-derived cell lines. The ATP concentrations of PFKP-RNAi#1 and PFKP-RNAi#2 were 0.71 and 0.52 times those of the negative control group, respectively. A paired t-test revealed that these differences were a statistically significant (p = 0.042 and 0.005, respectively).
3.6 PFKP expression affects colony formation, proliferation and cell cycle progression of lung cancer cells
We found that the colony forming abilities of the stably transfected lung cancer cell lines PFKP-RNAi#1 and PFKP-RNAi#2 exhibiting PFKP silencing were reduced compared to those of the negative control groups (Fig. 6). A paired t-test revealed that the differences were statistically significant (p < 0.05). Subsequent plate colony formation experiments revealed that PFKP expression silencing inhibited the colony forming ability of both lung cancer cell lines (Fig. 7), i.e., compared to the negative control group fewer PFKP-RNAi#1 and PFKP-RNAi#2 colonies in the PFKP low-expression groups of both the NCI-H1650 and A549 cell lines were noted. A paired t-test revealed that these differences were statistically significant (p < 0.05).
Using a MTT assay, we found that PFKP expression silencing also inhibited the proliferation of lung cancer cells. The stably transfected NCI-H1650 (Fig. 8a) and A549 (Fig. 8b) cell lines exhibited reduced abilities to proliferate compared to the respective negative control groups. A paired t-test revealed that these differences were statistically significant for both the NCI-H1650- and A549-derived cell lines (p < 0.05).
To further investigate correlations between low PFKP expression and lung cancer cell cycle parameters, flow cytometry was used. We found that the proportion of G2/M cells in the lung cancer cell lines NCI-H1650 (Fig. 9a) and A549 (Fig. 9b) exhibiting a low PFKP expression was significantly reduced compared to those in the negative control groups. This result indicates inhibition of tumor cell proliferation. The G2/M cell cycle phase percentages were 32.05% and 29.97% in the NCI-H1650-derived PFKP-RNAi#1 and PFKP-RNAi#2 cell lines, respectively, whereas the G2/M cell cycle phase percentage in the negative control cells was 44.26%.
3.7 Identification of pathways involved in the regulation of PFKP expression
GSEA pathway enrichment was used to analyze mRNA expression data of the top 50% and bottom 50% of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) cells exhibiting PFKP expression. We found that multiple pathways may be involved in the regulation of PFKP expression (Table 2). In LUAD cells, we found that the GLYCOLYTIC PROCESS THROUGH FRUCTOSE 6 PHOSPHATE and other pathways were significantly enriched in the PFKP high-expression group (Table 2). Genes related to the GLYCOLYTIC PROCESS THROUGH FRUCTOSE 6 PHOSPHATE pathway, i.e., PFKP, GAPDH, TPI1, ALDOA, GPI, PKM, PGAM1, PGK1, HK2, ENO2, ENO1, PFKM and GALK1, were more abundantly expressed in the PFKP high-expression group than in the low-expression group (Table 3). In LUSC cells, the GLYCOLYTIC PROCESS and other signaling pathways were significantly enriched in the PFKP high-expression group (Table 2). Genes related to the GLYCOLYTIC PROCESS pathway, i.e., PFKP, PFKFB3, ALDOA, ENO1, PGAM1, ENO2, PFKFB4, HK2, GPI, PKM, PGAM4, PGK1, DHTKD1, ALDOC, LDHA, HK1, HIF1A, TPI1, PFKL, ENTPD5, GAPDH, NUP188, HKDC1, PFKM, NUP205, PGM2L1, NUP210, PRKAA2, PGM1, ZBTB7A, NUP93, ENO3, POM121C, POM121, PRKAG3, EIF6, OGDH, GCK and MYOG were more abundantly expressed in the PFKP high-expression group than in the low-expression group (Table 3).
4 Discussion
PFKP is known to be an important mediator of cell metabolism and to play an important role in the processes of tumorigenesis and metastasis [14]. Wang et al. [15] conducted a study evaluating the role of PFKP mRNA expression and the occurrence of single-nucleotide polymorphisms and found that PFKP may function as an oncogene in lung cancer. Here, we found that PFKP mRNA and protein were highly expressed in lung cancer tissues and cell lines, suggesting that PFKP may promote their growth. We also found that in lung adenocarcinoma cells that stably expressed reduced levels of PFKP, the ability of the cells to metabolize glucose was decreased. Additionally, we used a large number of clinical lung cancer tissue samples to assess correlations between PFKP expression and clinicopathologic features, including patient prognosis and survival rates. Our results indicate that PFKP may serve as a new prognostic marker for lung cancer. We also performed pathway enrichment and gene correlation analyses based on TCGA data and found that both lung adenocarcinomas and lung squamous cell carcinomas may be enriched in PFKP through glycolysis-related pathways.
PFKP has been reported to promote aerobic glycolysis, tumor cell proliferation and tumor growth. In human glioblastoma, for example, phosphorylation of PFKP S386 has been found to increase the expression level of PFKP, thereby promoting aerobic glycolysis, cell proliferation and growth of brain tumors [16]. Also, phosphorylation of PFKP Y64 has been found to increase GLUT1 expression, thereby promoting the Warburg effect as well as brain tumor formation and cell proliferation [17]. Another study revealed that the PFKP mRNA expression level was significantly up-regulated in patients with renal clear cell carcinoma [18]. Our current study shows that the PFKP mRNA and protein levels are significantly increased in lung cancer tissues and cell lines. Furthermore, we observed a positive correlation between PFKP protein expression and expression of the cell proliferation marker protein Ki67 in lung adenocarcinomas.
Changes in energy metabolism are known to play important roles in the development of cancer [4, 19, 20]. The ability to accelerate the uptake and oxidation of glucose is a characteristic of most malignancies. Glycolysis is the main energy supply route for rapidly proliferating tumor cells, it allows them to adapt to hypoxia and to increase their malignant potential [21]. Silencing of PFKP has been found to inhibit hunger-induced autophagy, glycolysis and epithelial mesenchymal transition (EMT) in oral squamous cell carcinoma [22]. In fact, a 18F-deoxyglucose-PET/CT method is widely used clinically to measure glucose uptake rates in order to discriminate between benign and malignant lesions [23, 24]. We found that a reduced PFKP expression resulted in decreased glucose concentrations, lactic acid levels and ATP concentrations in the cell culture supernatants. This finding underscores the notion that PFKP can inhibit glycolysis in lung cancer cells, thus inhibiting tumor growth. Presently, increasing attention is focusing on the role of glycolysis in the ability of tumors to escape from the immune system. Using glycolysis as the main mode of energy supply, as in lung cancer, provides favorable conditions for cancer growth by selective regulation of the tumor microenvironment (TME) [25]. Lactic acid produced by glycolysis, for example, accumulates outside the cell thereby altering the TME by generating an acidic pH that is detrimental to normal cells. Note that this acidic TME confers resistance to cancer treatment and is a major obstacle to its success [26]. The above studies indicate that the expression of PFKP is related to glycolysis in lung cancer cells, suggesting that PFKP may affect the occurrence and development of lung cancer by affecting the glucose metabolic level of lung cancer cells. In addition, through GSEA enrichment analysis, we identified PFKP-mediated pathways and genes involved in glycolytic pathways.
We also found that the proliferation rate of lung cancer cells expressing low levels of PFKP was significantly decreased and that the colony forming ability of the cells was significantly reduced. We also found that a low PFKP expression significantly reduced the percentage of G2/M cells, indicative of proliferation inhibition of lung cancer cells. A previous study of renal clear cell carcinoma showed that reducing the expression of PFKP not only inhibited the proliferation of cancer cells by inducing cell cycle arrest and apoptosis, but also led to obstruction of glycolysis, both in vitro and in vivo [18]. Recent studies also showed that PFKP-mediated metabolic reprogramming enhanced the invasiveness and metastasis of hypoxic breast cancer cells [27]. Our results suggest that PFKP expression inhibition can reduce the proliferative ability of lung cancer cells. Therefore, PFKP may be a potential therapeutic target for lung cancer.
PFKP is not only related to glycolysis and tumor proliferation, but it also affects tumor development and prognosis. A recent study suggested that abnormal PKFP hypermethylation may be a prognostic biomarker for prostate cancer [28] Phosphorylation of PFKP S386 has also been associated with a poor prognosis in human glioblastoma [16], and breast cancer patients with high PFKP expression have been reported to have a poor prognosis [29]. Through the analysis of polymorphisms in non-small cell lung cancer (NSCLC) glycolytic-related genes, Lee et al. [30] found that PFKM rs11168417C > T and PFKP rs1132173C > T variants were significantly associated with the prognosis of NSCLC patients. We found that a high PFKP expression was correlated with tumor size and patient survival, i.e., patients with a high PFKP expression had larger tumor lesions and exhibited higher mortality rates by the end of the follow-up time. These results indicate that PFKP protein levels may serve as prognostic markers in patients with lung adenocarcinoma.
Abbreviations
- IHC:
-
Immunohistochemistry
- DMEM:
-
Dulbecco’s Modified Eagle’s Medium
- FBS:
-
Fetal Bovine Serum;
- qRT-PCR:
-
Quantitative Reverse Transcriptase PCR
- NSCLC:
-
Non-Small Cell Lung Carcinoma
- PFK-1:
-
Phosphofructokinase-1
- ELISA:
-
Enzyme-Linked Immunosorbent Assay
- TCGA:
-
The Cancer Genome Atlas
- GSEA:
-
Gene Set Enrichment Analysis
References
R. Siegel, D. Naishadham, A. Jemal, Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013)
C.M. North, D.C. Christiani, Women and lung cancer: What is new? Semin Thorac Cardiovasc Surg 25, 87–94 (2013)
P.P. Hsu, D.M. Sabatini, Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008)
R.A. Cairns, I.S. Harris, T.W. Mak, Regulation of cancer cell metabolism. Nat Rev Cancer 11, 85–95 (2011)
G. van Niekerk, A.M. Engelbrecht, Role of PKM2 in directing the metabolic fate of glucose in cancer: A potential therapeutic target. Cell Oncol 41, 343–351 (2018)
I. Mor, E.C. Cheung, K.H. Vousden, Control of glycolysis through regulation of PFK1: Old friends and recent additions. Cold Spring Harb Symp Quant Biol 76, 211–216 (2011)
R. Moreno-Sanchez, E. Saavedra, Energy metabolism in tumor cells. FEBS J 274, 1393–1418 (2007)
P. Nilendu, S.C. Sarode, D. Jahagirdar, I. Tandon, S. Patil, G.S. Sarode, J.K. Pal, N.K. Sharma, Mutual concessions and compromises between stromal cells and cancer cells: Driving tumor development and drug resistance. Cell Oncol 41, 353–367 (2018)
C. Sanchez-Martnez, J.J. Aragon, Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine mammary gland. FEBS Lett 409, 86–90 (1997)
G. Wang, Z. Xu, C. Wang, F. Yao, J. Li, C. Chen, S. Sun, Differential phosphofructokinase-1 isoenzyme patterns associated with glycolytic efficiency in human breast cancer and paracancer tissues. Oncol Lett 6, 1701–1706 (2013)
R.J. Shaw, Glucose metabolism and cancer. Curr Opin Cell Biol 18, 598–608 (2006)
R.A. Gatenby, R.J. Gillies, Why do cancers have high aerobic glycolysis? Nat Rev Cancer 4, 891–899 (2004)
K. Zhou, Y.L. Yao, Z.C. He, C. Chen, X.N. Zhang, K.D. Yang, Y.Q. Liu, Q. Liu, W.J. Fu, Y.P. Chen, Q. Niu, Q.H. Ma, R. Zhou, X.H. Yao, X. Zhang, Y.H. Cui, X.W. Bian, Y. Shi, Y.F. Ping, VDAC2 interacts with PFKP to regulate glucose metabolism and phenotypic reprogramming of glioma stem cells. Cell Death Dis 9, 988 (2018)
L. Lang, R. Chemmalakuzhy, C. Shay, Y. Teng, PFKP signaling at a glance: An emerging mediator of Cancer cell metabolism. Adv Exp Med Biol 1134, 243–258 (2019)
Y. Wang, Q. Mei, Y.Q. Ai, R.Q. Li, L. Chang, Y.F. Li, Y.X. Xia, W.H. Li, Y. Chen, Identification of lung cancer oncogenes based on the mRNA expression and single nucleotide polymorphism profile data. Neoplasma 62, 966–973 (2015)
J.H. Lee, R. Liu, J. Li, C. Zhang, Y. Wang, Q. Cai, X. Qian, Y. Xia, Y. Zheng, Y. Piao, Q. Chen, J.F. de Groot, T. Jiang, Z. Lu, Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun 8, 949 (2017)
J.H. Lee, R. Liu, J. Li, Y. Wang, L. Tan, X.J. Li, X. Qian, C. Zhang, Y. Xia, D. Xu, W. Guo, Z. Ding, L. Du, Y. Zheng, Q. Chen, P.L. Lorenzi, G.B. Mills, T. Jiang, Z. Lu, EGFR-phosphorylated platelet isoform of phosphofructokinase 1 promotes PI3K activation. Mol Cell 70, 197–210 (2018)
J. Wang, P. Zhang, J. Zhong, M. Tan, J. Ge, L. Tao, Y. Li, Y. Zhu, L. Wu, J. Qiu, X. Tong, The platelet isoform of phosphofructokinase contributes to metabolic reprogramming and maintains cell proliferation in clear cell renal cell carcinoma. Oncotarget 7, 27142–27157 (2016)
C.V. Dang, Links between metabolism and cancer. Genes Dev 26, 877–890 (2012)
G. Kroemer, J. Pouyssegur, Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13, 472–482 (2008)
C.V. Dang, M. Hamaker, P. Sun, A. Le, P. Gao, Therapeutic targeting of cancer cell metabolism. J Mol Med 89, 205–212 (2011)
G. Chen, H. Liu, Y. Zhang, J. Liang, Y. Zhu, M. Zhang, D. Yu, C. Wang, J. Hou, Silencing PFKP inhibits starvation-induced autophagy, glycolysis, and epithelial mesenchymal transition in oral squamous cell carcinoma. Exp Cell Res 370, 46–57 (2018)
J.D. Gordan, C.B. Thompson, M.C. Simon, HIF and c-Myc: Sibling rivals for control of Cancer cell metabolism and proliferation. Cancer Cell 12, 108–113 (2007)
A. Ramanathan, C. Wang, S.L. Schreiber, Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A 102, 5992–5997 (2005)
S. Ganapathy-Kanniappan, Linking tumor glycolysis and immune evasion in cancer: Emerging concepts and therapeutic opportunities. Biochim Biophys Acta Rev Cancer 1868, 212–220 (2017)
L. Baitsch, P. Baumgaertner, E. Devêvre, S.K. Raghav, A. Legat, L. Barba, S. Wieckowski, H. Bouzourene, B. Deplancke, P. Romero, N. Rufer, D.E. Speiser, Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest 121, 2350–2360 (2011)
M. Peng, D. Yang, Y. Hou, S. Liu, M. Zhao, Y. Qin, R. Chen, Y. Teng, M. Liu, Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis 10, 228 (2019)
M.T. Bjerre, S.H. Strand, M. Nørgaard, H. Kristensen, A.K. Rasmussen, M.M. Mortensen, J. Fredsøe, P. Mouritzen, B. Ulhøi, T. Ørntoft, M. Borre, K.D. Sørensen, Aberrant DOCK2, GRASP, HIF3A and PKFP hypermethylation has potential as a prognostic biomarker for prostate cancer. Int J Mol Sci 20, pii: E1173 (2019)
Y. He, F. Deng, S. Zhao, S. Zhong, J. Zhao, D. Wang, X. Chen, J. Zhang, J. Hou, W. Zhang, L. Ding, J. Tang, Z. Zhou, Analysis of miRNA-mRNA network reveals miR-140-5p as a suppressor of breast cancer glycolysis via targeting GLUT1. Epigenomics 11, 1021–1036 (2019)
S.Y. Lee, C.C. Jin, J.E. Choi, M.J. Hong, D.K. Jung, S.K. Do, S.A. Baek, H.J. Kang, H.G. Kang, S.H. Choi, W.K. Lee, Y. Seok, E.B. Lee, J.Y. Jeong, K.M. Shin, S. Cho, S.S. Yoo, J. Lee, S.I. Cha, C.H. Kim, Y.M. Lee, I.K. Lee, S. Jheon, J.Y. Park, Genetic polymorphisms in glycolytic pathway are associated with the prognosis of patients with early stage non-small cell lung cancer. Sci Rep 6, 35603 (2016)
Acknowledgements
The authors thank Taizhou Hospital of Zhejiang Province for providing the research environment.
Availability of data and materials
All data generated or analyzed in this study are included in this published article.
Funding
This study was funded by Zhejiang Provincial Natural Fund (grant number LQ18H160029 and Y19H160116).
Author information
Authors and Affiliations
Contributions
JF, ZJ, & HL searched the literature and drafted and revised the manuscript. KJ & KBJ processed the data. CZ and BC revised and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The experimental protocol was established according to the ethical guidelines of the Helsinki Declaration, which was approved by the Medical Ethics Committee of Taizhou Hospital, Zhejiang Province, China. Written informed consent to participate in the study was obtained from all individual participants or their guardians.
Consent for publication
Written informed consent for publication was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOC 26.0 kb)
Rights and permissions
About this article
Cite this article
Shen, J., Jin, Z., Lv, H. et al. PFKP is highly expressed in lung cancer and regulates glucose metabolism. Cell Oncol. 43, 617–629 (2020). https://doi.org/10.1007/s13402-020-00508-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-020-00508-6