Abstract
Purpose
Exosomes play important roles in intercellular communication through signaling pathways affecting tumor microenvironment modulation and tumor proliferation, including those in glioblastoma (GBM). As yet, however, limited studies have been conducted on the inhibitory effect of human bone marrow-derived mesenchymal stem cell (hBMSC)-derived exosomes on GBM development. Therefore, we set out to assess the role of hBMSC secreted exosomes, in particular those carrying microRNA-34a (miR-34a), in the development of GBM.
Methods
Microarray-based expression analysis was employed to identify differentially expressed genes and to predict miRNAs regulating MYCN expression. Next, hBMSCs were transfected with a miR-34a mimic or inhibitor after which exosomes were isolated. Proliferation, apoptosis, migration, invasion and temozolomide (TMZ) chemosensitivity of exosome-exposed GBM cells (T-98G, LN229 and A-172) were measured in vitro. The mechanism underlying MYCN regulation was investigated using lentiviral transfections. The in vivo inhibitory effect of exosomal miR-34a was measured in nude mice xenografted with GBM cells through subcutaneous injection of hBMSCs with an upregulated miR34a content.
Results
We found that poorly-expressed miR-34a specifically targeted and negatively regulated the expression of MYCN in GBM cells. In addition we found that miR-34a was delivered to T-98G, LN229 and A-172 GBM cells via hBMSC-derived exosomes. Exogenous overexpression of miR-34a in hBMSC-derived exosomes resulted in inhibition of GBM cell proliferation, invasion, migration and tumorigenesis in vitro and in vivo, while promoting the chemosensitivity of GBM cells to TMZ by silencing MYCN.
Conclusions
From our data we conclude that hBMSC-derived exosomes overexpressing miR-34a may be instrumental for the therapeutic targeting and clinical management of GBM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Glioblastoma (GBM) has been identified as the most common primary malignant brain tumor in adults. It cannot be completely eliminated due to its tendency to invade [1]. Although the cause underlying GBM progression remains unclear, the surrounding tissues may play a role, i.e., interactions between GBM cells and micro- and macro-environmental components may create conditions that inhibit or enhance their growth [2, 3]. The poor prognosis usually results from, next to tumor invasiveness, tumor heterogeneity and drug resistance [4]. The treatment options for GBM are limited, and include radiotherapy, surgery and chemotherapy along with an alkylating agent, temozolomide (TMZ) [5, 6]. Previous attempts to improve the efficacy of existing treatment strategies for GBM have had little success due to combined actions of intricate phenotypes and organ-specific clinical manifestations [7]. Human bone marrow-derived mesenchymal stem cells (hBMSCs) have been found to be capable of homing to gliomas after systemic delivery and they can, therefore, potentially be applied to glioma treatment [8]. Additional emerging evidence indicates that hBMSCs may be crucial for GBM-induced neovascularization [9].
Exosomes secreted by the BMSCs have been reported to have therapeutic potential [10]. Exosomes are small intraluminal vesicles that are secreted by various cells and can deliver intracellular contents, such as microRNAs (miRNAs), messenger RNAs (mRNAs) and proteins [11,12,13]. Dysregulated miRNA activities are known to be closely related to the initiation and progression of several tumors, including GBM [14, 15]. Previously, it has been found that miR-34a is poorly expressed in GBMs, and that upregulation of miR-34a may inhibit GBM cell proliferation, migration and invasion, as well as in vivo GBM xenograft growth [16]. In addition, it has been found that miR-34a may facilitate the suppression of GBM growth by binding to Notch1 and that the expression of the latter is decreased in GBM cells [17]. As a potential oncogene, it has been found that MYCN can induce GBM development [18], and that it may serve as an independent biomarker for rapid tumor progression and a poor prognosis, regardless of age and/or clinical stage of the disease [19]. Moreover, MYCN has been reported to mediate the transcription of genes associated with proliferation and apoptosis in neuroblastoma [20]. Based on this information, we set out to explore the therapeutic effect of miR-34a delivered by hBMSC-derived exosomes on GBM cells, speculating that it might inhibit the proliferation, migration and invasion of GBM cells, and/or enhance their sensitivity to TMZ.
2 Materials and methods
2.1 Ethics statement
This study was approved by the Institutional Review Board and the Institutional Animal Care and Use Committee of Henan Province People’s Hospital, Zhengzhou University. All patients signed informed consent prior to the study.
2.2 Microarray-based expression analysis
The GSE42656 chip used was retrieved from the National Center for Biotechnology Information (NCBI) and included 4 normal samples and 5 GBM samples. Differentially expressed genes were identified using the empirical Bayesian method in conjunction with the Limma package of Bioconductor in R, and annotated using the annotation package. A p value < 0.05 was considered to be a statistically significant.
2.3 MYCN regulatory miRNA identification
Candidate MYCN expression regulating miRNAs were predicted using the miRNA target prediction websites microRNA (http://34.236.212.39/microrna/getGeneForm.do), miRsearch (https://www.exiqon.com/miRSearch) and miRDB (http://www.mirdb.org/), after which overlapping prediction results were retrieved.
2.4 Study subjects
Primary GBM tissues (n = 35) and normal brain tissues (n = 25) were obtained from the Department of Neurosurgery of Henan Province People’s Hospital, Zhengzhou University. After the tissue samples were obtained through surgery, they were immediately washed with phosphate buffered saline (PBS; 3 times), cut into small blocks, placed in a cryopreservation tube and preserved in liquid nitrogen for further use. All GBM cases were confirmed through histopathological examination. The World Health Organization (WHO) classification criteria for nervous system tumors were used as the basis for pathological diagnosis and grading [21]. All normal brain tissues were excised by intracranial decompression.
2.5 Cell cultures
The normal human glial cell line HEB and the GBM cell lines T-98G, BT325, LN229 and A172 were obtained from the Shanghai Institutes for Biological Sciences (Chinese Academy of Sciences, Shanghai, China) and passaged during less than 6 months. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technology, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Life Technology, Grand Island, NY, USA) in an incubator containing 5% CO2 at 37 °C.
2.6 hBMSC isolation and culture
The hBMSCs were isolated using the following procedure: a 20 ml syringe (containing 2000 IU heparin) was used to extract 10 ml bone marrow from the femoral shaft after which the bone marrow cells were mixed quickly with heparin. Next, the cells were spun down at 257×g for 10 min, after which the upper adipose tissues were removed, and the remaining cells were washed 3 times with DMEM and resuspended in 15 ml complete culture medium. The same volume of Ficoll-Paque™ Plus lymphocyte isolate (density 1.077 g/ml) was added to the tube after which the suspension was centrifuged at 715×g for 20 min. By doing so, the nucleated cells were located in the interface and upper liquid phase while most of the red blood cells were located at the bottom. The nucleated cells were taken out from the interface with a pipette and centrifuged at 178×g for 8 min. Next, the cells were resuspended in 5 ml culture medium and 10 μl of the suspension was mixed with 490 μl PBS. From this, 10 μl was taken for cell counting. Subsequently, the cells were seeded in culture flasks at a density of 1 × 105 cells/flask, after which 5 ml low-sugar medium was added for cell culturing in a humidified incubator with 5% CO2 at 37 °C. After fluorescence-activated cell sorting (FACS) analysis, the cells were found to be positive for CD71, CD44 and CD29 (Abcam Inc., Cambridge, CA, USA), but negative for HLA-DR (Abcam Inc., Cambridge, MA, USA) and the hematopoietic markers CD45 and CD34 (PE, eBiosciences).
2.7 Transfection and lentiviral transduction
GBM cells were seeded at a density of 2 × 105 cells/well (6-well plate) or 2 × 106 cells/dish (100-mm dish) or 5 × 105 cells/dish (60-mm dish) 1 day before transfection. When the cells reached a confluence of 60% - 80%, they were transfected with miR-34a/NC inhibitor, miR-34a/NC mimic, shMYCN/NC or MYCN/NC (GenePharma, Shanghai, China), respectively, in accordance with the Lipofectamine 2000 instructions of the supplier (Invitrogen, Carlsbad, CA, USA).
The pMD2G (1 μg), psPAX2 (3 μg) and pLenti6.3-Luciferase/miR-34a (miR-NC/miR-34a) (4 μg) plasmids were co-transfected into HEK-293 T cells in 60-mm dishes to allow lentivirus packaging. After 24 h of transfection, the supernatant was collected and fresh medium was added to the cells for another 24 h culture period. The supernatant was harvested again, mixed with the previous supernatant and used to infect target cells. For the lentiviral transduction of hBMSCs, these cells were seeded in a 24-well plate at a density of 5 × 104 cells/well and cultured overnight before transduction.
2.8 In vitro hBMSC differentiation assay
The 3rd generation hBMSCs with a good growth rate were resuspended at a concentration of 5 × 104 cells/ml and seeded into a 6-well plate, which was preset with a cover glass. After 24 h the cells were completely adhered to the bottom. The hBMSCs were subsequently cultured using an OriCell™ Balb/c Mouse Bone Marrow Mesenchymal Stem Cell Osteogenic Differentiation Medium Kit (Cyagen, Silicon Valley, CA, USA) and an Adipogenic Differentiation Medium Kit (Cyagen, Silicon Valley, CA, USA) for 4 weeks, respectively, followed by cell staining to determine the abilities of adipogenic and osteogenic differentiation in strict accordance with the instructions of the kits. Images were captured and photographed using an optical microscope (CK40, Olympus, Tokyo, Japan).
2.9 Dual-luciferase reporter assay
The targeting of MYCN by miR-34a was verified using a dual-luciferase reporter assay. Twenty four hours before transfection, GBM cells were seeded into a 24-well plate and allocated either to the wild-type (WT) plasmid group (GBM cells transfected with miR-34a mimic and the WT luciferase reporter plasmid MYCN 3′-untranslated region, 3’UTR), the mutant (MUT) plasmid group (GBM cells transfected with miR-34a mimic and the MUT luciferase reporter plasmid MYCN 3’UTR), and the negative control (NC) group (GBM cells transfected with miR-34a and the NC luciferase plasmid). After 48 h of transfection, the cells were collected and lysed after which the luciferase activity was detected using a dual-luciferase reporter system (Promega, Madison, WI, USA). Renilla luciferase activity was used as internal reference. The degree of activation of the reporter gene was estimated using the ratio of the firefly luciferase activity and the Renilla luciferase activity. The experiments were repeated 3 times independently, and 6 duplicated wells for each treatment were used.
2.10 hBMSC and GBM cell transfections
GBM cells were transfected with a green fluorescent protein expression vector pCDNA3.1-GFP, and hBMSCs were transfected with Cy3-labeled miR-34a (miR-34a-Cy3) (GenePharma, Shanghai, China). After 12 h of transfection, the cells were seeded in a 96-well plate at a density of 100 cells per well and co-cultured at a ratio of 1: 1 for 2 days. Next, flow cytometry was conducted in order to separate the cells. The cells were simultaneously analyzed under a fluorescence microscope to assess the effect of miR-34a on GBM cell proliferation.
2.11 Exosome isolation and inhibition of exosome secretion
Culture medium of the hBMSCs was collected and subsequently centrifuged at 300×g for 5 min, at 1500×g for 10 min and at 12000×g for 35 min at 4 °C, followed by filtration using a 0.22 m filter (Merck Millipore, Tullagreen, Ireland) and ultracentrifugation at 120000×g for 2 h to spin down the exosomes. The resulting exosome pellet was washed with 20 ml cold PBS and further purified through centrifugation at 120000×g for 2 h at 4 °C. The final pellet containing the exosomes was resuspended in 50–100 μl PBS and stored at −80 °C. Exosome release was blocked using the specific inhibitors GW4869 (Sigma, St Louis, MO, USA) and DMA (Santa Cruz, Paso Robles, CA, USA). To this end, the hBMSCs were treated with GW4869, DMA or dimethylsulphoxide (DMSO) in order to determine that the miRNAs were delivered via the exosomes. DMSO was used as the negative control. The hBMSCs were transfected with miRNA mimic in a 6-well plate and co-cultured with 10 M GW4869, 15 nM DMA and DMSO, respectively, for 48 h for the isolation of the exosomes.
2.12 Transmission electron microscopy
Exosomes were prepared in PBS and identified using transmission electron microscopy. To this end, the samples were adsorbed by carbon-coated nickel grids and negatively stained with 2% methylamine tungstate for 5 min. Subsequently, the dye was blotted from the grids with filter paper and the samples were washed twice with drops of distilled water. Next, the water was blotted and the samples were dried and examined under a JEM-1230 electron microscope (Nihon Denshi, Tokyo, Japan) at an accelerating voltage of 80 kV.
2.13 RNA isolation and quantification
Total RNA was extracted using a miRNeasy Mini Kit (217,004, QIAGEN, Duesseldorf, Germany). A260/A230 and A260/A280 values were determined using a Nanodrop 2000 ultraviolet spectrophotometer (1011 U, Nanodrop Technologies Inc., Wilmington, USA) to verify the concentration and purity, respectively. Next, reverse transcription was carried out to obtain complementary DNA (cDNA) in accordance with the instructions of the TaqMan MicroRNA Assays Reverse Transcription Kit (4,427,975, Applied Biosystems, Inc., Carlsbad, CA, USA). The cDNA was then diluted to 50 ng/μl and added to the reaction amplification system (25 μl) with 2 μl added each time. The miR-34a and MYCN primers were designed and synthesized by TaKaRa (Tokyo, Japan) (listed in Table 1). Real-time quantitative PCR (RT-qPCR) was conducted using an ABI7500 Quantitative PCR instrument (7500, ABI, USA). U6 served as internal reference for miR-34a and β-actin as internal reference for MYCN. The 2-ΔΔCt method was employed to determine the expression ratio of a gene in the experimental group relative to that in the control group, using the following formula: ΔΔCT = ΔCt (experimental group) - ΔCt (control group), in which ΔCt = Ct miRNA – Ct U6/β-actin. Ct refers to the number of amplification cycles when the real-time fluorescence intensity of the reaction reached the set threshold. Each experiment was conducted 3 times.
2.14 Western blotting
Total protein was extracted and quantified using a 2D Quant kit. 20 μg protein samples were used for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Next, the proteins were transferred to membranes that were subsequently blocked and incubated with the primary antibody, rabbit anti-MYCN (ab24193, 1: 1000, Abcam Inc., Cambridge, MA, USA) at 4 °C overnight, followed by oscillation and incubation with the secondary antibody, goat anti-rabbit immunoglobulin G (IgG) (A21020, 1: 1000, Abbkine, California, USA) at 37 °C for 1 h. After that, the samples were developed using electrochemiluminescence (ECL). The relative expression of the target protein was calculated using the formula: relative expression = the gray value of the target protein band/the gray value of the internal reference band of the same sample, with glyceraldehyde phosphate dehydrogenase (GAPDH) as internal reference. Each experiment was repeated 3 times.
2.15 Transwell invasion assay
Cell invasion was evaluated using a Transwell assay. Matrigel (YB356234, Shanghai YuBo Biological Technology Co., Ltd., Shanghai, China) preserved at −80 °C was melted at 4 °C overnight after which 200 μl was mixed with 200 μl serum-free medium at 4 °C. The apical chamber of the Transwell plate was coated with 50 μl Matrigel and incubated for 2–3 h until the gel was solidified. Next, the cells were re-suspended in medium containing 20% FBS and counted, after which 200 μl cell suspension was added to the apical chamber of each well and the basolateral chamber was supplemented with 800 μl medium containing 20% FBS, followed by incubation at 37 °C for 20–24 h. Next, the Transwell plate was taken out and soaked in formaldehyde for 10 min, after which the cells were stained with 0.1% crystal violet at room temperature for 30 min. The cells on the upper surface were wiped out using cotton balls. Finally, at least 4 visual fields were randomly selected and the cells were observed, photographed and counted under an inverted microscope. Evaluation of the relative cell migration capacity was carried out without Matrigel with an incubation period set to 16 h.
2.16 Colony formation assay
Cells in a logarithmic growth phase were detached by trypsin, resuspended and counted. The cell density was adjusted to 1 × 103 cells/ml in fresh complete medium. Next, 1 ml cell suspension was added to each well of a 6-well plate, followed by incubation in 5% CO2 and saturated humidity at 37 °C for 12 h. Next, 2 ml fresh complete medium was added to each well. The culture medium was changed every 3 days, and the culture was terminated after 14 days. After evident formation of clones in the culture dish, the culture was terminated and the culture medium was discarded. Next, the clones were fixed with formaldehyde for 15 min and stained with 0.1% gentian violet for 30 min. The number of clones was determined microscopically. Each experiment was repeated 3 times.
2.17 Temozolomide (TMZ) treatment, cell viability and apoptosis assay
The sensitivity of the T-98G, LN229 and A-172 cells to TMZ was evaluated. To this end, cells treated with miR-34a were cultured in a 96-well plate at a concentration of 5 × 103 overnight. Next, the cells were treated with 100 μM TMZ after which apoptosis was determined using a Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) kit, and viability was measured using a cell counting kit-8 (CCK-8) assay at 0 h, 12 h, 24 h, 36 h and 48 h, respectively.
2.18 Xenograft tumor model
Male athymic BALB/c nude mice (4–6 weeks old, weighing 18–22 g) were randomly selected. The left flank of each mouse was injected subcutaneously with 1 × 107 T-98G cells in 200 μl PBS. Tumor volumes were measured every 3 or 4 days (2 times a week) according to the formula: (π × length × width2)/6. When the tumors reached a size of 100 mm3, the mice were randomly divided into 3 groups (MSCs-miR-34a, MSCs-miR-NC and PBS) with 6 mice in each group. BMSC stable cell lines transfected with miR-34a or miR-NC were injected into the BALB/c nude mice via the tail vein once every 3 days (5 × 105 cells/mouse) after which the tumor volumes were measured. Following 7 rounds of treatment, the mice were sacrificed by spinal dislocation after which the tumors were recovered and weighed. Finally, the organs and tumors were snap-frozen in liquid nitrogen for further analysis or fixed in formalin for apoptosis and immunohistochemical (IHC) analyses.
2.19 Statistical analysis
Statistical analyses were performed using SPSS 21.0 (IBM Corp. Armonk, NY, USA). Normal distribution and variance homogeneity were determined for all data. According to normal distribution the data were expressed as mean ± standard deviation. Interquartile data were used to express a skewed distribution with or without variance homogeneity. Data between two groups were compared using an independent-sample t test. Data that did not conform to heterogeneity of variance were corrected using Welch’s. Multiple data sets were compared using one-way analysis of variance, and Turkey was used for back testing. Data at different time points were analyzed using repeated measurement analysis of variance. A p value < 0.05 indicated statistical significance.
3 Results
3.1 Selection of miR-34a as MYCN regulator
GBM-related gene expression data (GSE42656) were downloaded from the GEO database, after which 1638 differentially expressed genes were identified, 608 of which were up-regulated and 1030 of which were down-regulated. Since hBMSCs may play a therapeutic role through exosomes carrying miR-34a [10, 22], target genes regulated by miR-34a may play a pivotal role in cellular proliferation and/or apoptosis. miR-34a has been found to inhibit the proliferation and metastasis of GBM cells and to be significantly down-regulated in these cells [21, 23, 24]. The oncogene MYCN is highly expressed in GBM [18]. In neuroblastoma, miR-34a has been found to directly target and down-regulate the expression of MYCN, resulting in apoptosis [25]. MYCN is also known to play a role in human nerve cells and brain tumors. Since MYCN was among the differentially expressed genes identified (Fig. 1a), we set out to assess the relationship between miR-34a and MYCN in GBM cells. We found that MYCN was up-regulated in GBM cells (Fig. 1b) (p < 0.05). Upstream MYCN gene regulatory miRNAs were predicted through bioinformatic analysis and displayed using a Venn diagram. Four miRNAs were shared by the different prediction methods, including miR-34a (Fig. 1c). Since this miRNA has been found to have an inhibitory effect on among others GBM cells [24, 26, 27], it was selected for further analysis.
Selection of miR-34a-5p as a regulator of MYCN. a, heat map of differential gene expression using the GSE42656 chip, in which the horizontal coordinate represents the sample number, the longitudinal coordinate indicates the name of the gene, the color gradation histogram on the right indicates the level of gene expression where each box in the diagram indicates the expression level of a gene in a sample, and the left dendrogram shows the gene expression cluster. b, MYCN gene expression profile analysis, in which red represents gene expression in the normal group, and green represents gene expression in the GBM group. c, upstream regulatory miRNA regulating MYCN predicted by the Venn diagram, in which blue refers to the prediction result of the microRNA database, red refers to the prediction result of the miRsearch database, and green denotes the prediction result of the miRDB database. miRNA, microRNA; GBM, glioblastoma
3.2 The expression of miR-34a is low in GBM tissues and cells
The expression of miR-34a in primary GBM tissues and GMB-derived cell lines was quantified using RT-qPCR. In addition, the miR-34a expression levels in normal brain tissues (n = 25) and in GBM tissues (n = 35) were compared (Fig. 2a). By doing so, we observed lower miR-34a expression levels in GBM tissues compared to those in normal brain tissues (p < 0.0001). We also compared the miR-34a expression levels in the normal glial cell line HEB and the GBM-derived cell lines T-98G, BT325, LN229 and A172 and found that, compared to the HEB cell line, the expression was lower in all 4 GBM-derived cell lines. Among the GBM-derived cell lines tested, the lowest expression level was observed in T-98G (Fig. 2b). These results indicate that the expression level of miR-34a is relatively low in GBM tissues and cells.
Decreased expression of miR-34a in GBM. a, expression of miR-34a in primary GBM (n = 35) and normal human brain (n = 25) tissues determined by RT-qPCR. b, expression of miR-34a in normal glial cell line HEB and GBM cell lines BT-325, LN229, A172 and T98G measured by RT-qPCR. The data between two groups were compared using independent-sample t test, and one-way analysis of variance was used for multigroup comparison. The experiment was repeated 3 times. *, p < 0.05 vs. normal brain tissues/HEB cell line; RT-qPCR, reverse transcription quantitative polymerase chain reaction; GBM, glioblastoma; miR-34a, microRNA-34a
3.3 The combination of miR-34a and TMZ plays a therapeutic role in GBM
In order to investigate the effect of miR-34a on GBM cells, the invasion and migration abilities of T-98G, LN229 and A-172 cells were assessed using a Transwell assay. We found that compared to the NC group, overexpression of miR-34a significantly decreased the invasion and migration capacities of these cells (Fig. 3a). The growth capacity of T-98G, LN229 and A-172 cells was evaluated using a colony formation assay. We found that the number of clones generated by the T-98G, LN229 and A-172 cells transfected with miR-34a mimic was significantly suppressed compared to the corresponding NC group. Conversely, we found that inhibition of miR-34a resulted in a significant increase in GBM cell proliferation, invasion and migration (Fig. 3b). These results indicate that miR-34a may act as a tumor suppressor, since it has the potential to inhibit the proliferation, invasion and migration of T-98G, LN229 and A-172 cells. We also treated these GBM cells with TMZ to assess whether miR-34a may play a role in chemotherapy. A CCK-8 assay was used to detect cell viability. We found that the T-98G, LN229 and A-172 cells treated with TMZ alone exhibited a significantly decreased viability, while those treated with miR-34a mimic and TMZ exhibited an even more evident inhibition in viability (Fig. 3c). Next, a TUNEL assay was performed to detect a combined effect of miR-34a and TMZ on apoptosis. The results revealed that in T-98G, LN229 and A-172 cells, the combined treatment markedly enhanced apoptosis compared to TMZ treatment alone, which could subsequently be reversed by miR-34a inhibition (Fig. 3d). These results indicate that the combined action of miR-34a and TMZ enhances the therapeutic effect of TMZ on GBM cells.
MiR-34a inhibits GBM cell proliferation, invasion and migration, while enhancing chemosensitivity. a, invasion and migration capacities of T-98G, LN229 and A-172 cells determined by Transwell assay. b, clonality of T-98G, LN229 and A-172 cells detected by colony formation assay. c, viability of T-98G, LN229 and A-172 cells after treatment with 100 μM TMZ determined by CCK-8 assay. d, apoptosis of T-98G, LN229 and A-172 cells after treatment of miR-34a and TMZ detected by TUNEL assay (scale bar = 50 μM). The data among multiple groups were compared by one-way analysis of variance, and cell viability at different time points was analyzed by repeated measurement analysis of variance. *, p < 0.05 vs. the DMSO group; #, p < 0.05 vs. the corresponding NC group; the experiment was repeated 3 times. GBM, glioblastoma; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling; NC, negative control; DMSO, dimethylsulfoxide; CCK-8, cell counting kit-8; miR-34a, microRNA-34a; TMZ, temozolomide
3.4 MYCN is a direct target of miR-34a
Binding between MYCN and miR-34a was predicted using bioinformatic analysis (microRNA.Org; http://www.microrna.org/) (Fig. 4a). A dual-luciferase reporter assay was performed in order to confirm that MYCN acts as a direct target of miR-34a. We found that compared to the NC group, the luciferase activity of WT-MYCN 3’-UTR was significantly inhibited by miR-34a (p < 0.05), while that of MUT-MYCN 3’-UTR was not (Fig. 4b, c). In addition, we found that miR-34a expression restoration decreased the mRNA and protein expression levels of MYCN in GBM cells, whereas this trend was reversed by miR-34a inhibition (Fig. 4d, e). These findings indicate that MYCN serves a direct target of miR-34a, and that miR-34a can down-regulate the expression of MYCN.
miR-34a specifically binds to MYCN. a, binding sites of miR-34a and MYCN predicted by bioinformatics. b, predicted binding of miR-34a to the MYCN 3’UTR. c, relative luciferase activity measured by dual-luciferase reporter assay. d, mRNA expression of MYCN after miR-34a overexpression or inhibition determined by RT-qPCR. e, protein expression of MYCN after miR-34a overexpression or inhibition determined by Western blotting. *, p < 0.05 vs. the corresponding NC group; data among multiple groups were compared by one-way analysis of variance and the measurement data were expressed as mean ± standard deviation; n = 3; miR-34a, microRNA-34a; RT-qPCR, reverse transcription quantitative polymerase chain reaction; 3’UTR, 3′-untranslated regions
3.5 miR-34a inhibits the proliferation, invasion and migration of the GBM cells, and enhances their chemosensitivity through MYCN down-regulation
The effect of MYCN silencing on the proliferation, invasion and migration of T-98G, LN229 and A-172 cells was determined using a Transwell assay, a CCK-8 assay and a colony formation assay (Fig. 5). We found that compared to the NC group, the invasion, migration and colony forming capacities were markedly inhibited trough MYCN silencing (Fig. 5a, b, c). In addition, we found that when miR-34a mimic and MYCN were delivered together in T-98G, LN229 and A-172 cells, compared to the NC group, miR-34a mimic alone could inhibit the expression of MYCN (Fig. 5d) and significantly suppress the proliferation, invasion and migration of these cells (Fig. 5e–f). We also found that miR-34a restoration in combination with exogenous MYCN expression reversed the inhibitory effect of miR-34a on T-98G, LN229 and A-172 cells (Fig. 5d–f). We also treated T-98G, LN229 and A-172 cells with TMZ and found (Fig. 5g, h) that MYCN silencing in combination with TMZ treatment increased chemosensitivity, decreased cell viability and enhanced apoptosis. In addition, we found that treatment of miR-34a mimic and MYCN overexpression inhibited the enhanced chemosensitivity caused by miR-34a mimic in T-98G, LN229 and A-172 cells. These results indicate that the inhibitory effect of miR-34a on GBM cells may be achieved through MYCN down-regulation.
MiR-34a inhibits GBM cell proliferation, invasion and migration while enhancing their chemosensitivity by down-regulating MYCN. a, protein level of MYCN after MYCN silencing determined by Western blotting. b, invasion and migration abilities of T-98G, LN229 and A-172 cells evaluated by Transwell assay (scale bar = 100 μM). c, clonality of T-98G, LN229 and A-172 cells detected by colony formation essay. d, protein level of MYCN after delivery of miR-34a mimic or MYCN or NC determined by Western blotting. e, invasion and migration abilities of T-98G, LN229 and A-172 cells evaluated by Transwell assay (scale bar = 100 μM). f, clonality of T-98G, LN229 and A-172 cells detected by colony formation assay. g, viability of T-98G, LN229 and A-172 cells after treatment with 100 μM TMZ tested by CCK-8 assay. h, apoptosis of T-98G, LN229 and A-172 cells treated with 100 μM TMZ detected by TUNEL assay. *, p < 0.05, vs. the sh-NC group in Fig. a - c where data analysis was performed using t test; *, p < 0.05, vs. the miR-34a NC + NC group in Fig. d - f where data analysis was performed using one-way analysis of variance; *, p < 0.05, vs. the DMSO group in panel G where data at different time points were compared by repeated measurement analysis of variance; all experiments were repeated 3 times. miR-34a, microRNA-34a; GBM, glioblastoma; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling; NC, negative control; DMSO, dimethylsulfoxide; CCK-8, cell counting kit-8; TMZ, temozolomide
3.6 hBMSCs exhibit adipogenic and osteogenic differentiation capacities
hBMSCs were isolated and the expression of cell surface markers CD29, CD34, CD44, CD45, CD71 and HLA-DR was assessed by flow cytometry using fluorescein isothiocyanate (FITC)-labeled mouse anti-human antibodies. The percentages of cells expressing specific CD markers was as follows: CD29, 98.00%; CD44, 83.00%; CD71, 99.03%; CD34, 1.00%; CD45, 2.04%; HLA-DR, 1.03% (Fig. 6a). CD29, CD44 and CD71 are makers of BMSCs, CD34 and CD45 are makers of hemopoietic stem cells, and HLA-DR is mainly expressed in antigen presenting cells, including B lymphocytes, macrophages and T lymphocytes. Our findings indicate that the cultured cells are hBMSCs. Next, the in vitro induction of hBMSC differentiation was assessed. We found that after 2 weeks of adipose inducing culture using a specific medium (see Materials and methods), the hBMSCs exhibited a large number of lipid droplets. These droplets were verified to be lipid depositions using Oil Red O staining, thereby confirming that the hBMSCs exhibited adipogenic differentiation. After 4 weeks of osteogenic differentiation culture, plenty of red calcium deposits were detected using Alizarin red staining (Fig. 6b). From these combined results we conclude that the hBMSCs have adipogenic and osteogenic differentiation capacities.
BMSCs have the potential of adipogenic and osteogenic differentiation. a, analysis of BMSCs for CD29, CD44 and CD71 (positive markers), CD34, HLA-DR and CD45 (negative markers) expression by flow cytometry. b, adipogenesis (left) and osteogenesis (right) of BMSCs detected by Oil Red O staining and Alizarin Red staining, respectively (scale bar = 50 μM, 200×); BMSCs, bone marrow mesenchymal stem cells
3.7 miR-34a is delivered to GBM cells via hBMSC-derived exosomes
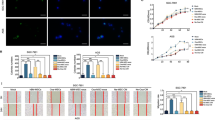
Next, T-98G, LN229 and A-172 cells transfected with pCDNA3.1-GFP and BMSCs transfected with miR-34a-Cy3 were co-cultured (Fig. 7a). Based on the assumption that miR-34a may be transferred to cancer cells via exosomes, the expression levels of miR-34a in BMSCs and BMSC-derived exosomes was measured by RT-qPCR. We found that compared to the miR-NC group, the miR-34a level in the miR-34a-transfected BMSCs and the BMSC-derived exosomes was significantly up-regulated (Fig. 7b). These results indicate that BMSCs can effectively release exosomes enriched in miR-34a. To verify whether miR-34a may be delivered to cancer cells via the BMSC-derived exosomes, the BMSCs were transfected with Cy3-labeled miR-34a mimic, after which the exosomes were isolated (Fig. 7c). The merged images subsequently showed that miR-34a could indeed be transferred from BMSCs (donor cells in red fluorescence) transfected with miR-34a-Cy3 into T-98G, LN229 and A-172 cells (receptor cells in green fluorescence) (Fig. 7d). In order to determine the role of the exosomes in this process, GW4869 (an inhibitor of neutral sphingomyelinase-2) and DMA (an inhibitor of H+/Na + and exchange of Na+/Ca2+) were used to reduce exosomal secretion [28, 29]. We found that co-culture with GW4869 and DMA resulted in a significant decrease in the content of miR-34a in the T-98G, LN229 and A-172 cells (Fig. 7e). RT-qPCR was subsequently performed to determine the mRNA level of MYCN in the GBM cells co-cultured with miR-34a-transfected BMSCs in order to test whether the transferred miR-34a can effectively inhibit endogenous MYCN expression in the tumor cells. The results obtained indicate that miR-34a, which was transferred to the T-98G, LN229 and A-172 cells through exosomes, effectively inhibited the expression of MYCN, and this effect was found to be blocked when GW4869 and DMA were added (Fig. 7f). These data indicate that exogenous miR-34a can be transferred from BMSCs to cancer cells via exosomes.
Delivery of miR-34a to GBM cells via BMSC-derived exosomes. a, co-culture of T-98G, LN229 and A-172 cells transfected with pCDNA3.1-GFP and BMSCs transfected with miR-34a-Cy3. b, miR-34a expression in miR-34a-treated BMSCs and BMSC-derived exosomes determined by RT-qPCR. *, p < 0.001 vs. the BMSCs + miR-34a NC group; data between two groups were compared using t test. c, identification of exosomes by transmission electron microscopy (scale bar = 200 nM); d, miR-34a delivery to T-98G, LN229 and A-172 cells by BMSCs observed under a fluorescence microscope (400×). e, expression of miR-34a in T-98G, LN229 and A-172 cells after co-culture determined by RT-qPCR. f, changes in MYCN expression determined by RT-qPCR. Data among multiple groups were compared using one-way analysis of variance. *, p < 0.05 vs. the miR-34a NC + DMSO group in panel e and f; the experiments were repeated 3 times. miR-34a, microRNA-34a; BMSCs, bone marrow mesenchymal stem cells; GBM, glioblastoma; NC, negative control; RT-qPCR, reverse transcription quantitative polymerase chain reaction; DMSO, dimethylsulfoxide
3.8 miR-34a delivered by hBMSC-derived exosomes in combination with TMZ increases the efficacy of GBM treatment
Subsequently, the invasion and migration of the T-98G, LN229 and A-172 cells was investigated after co-culture with BMSCs with or without exogenous miR-34a expression. We found that the invasion and migration capacities of the respective GBM cells were notably suppressed when they were co-cultured with miR-34a-transfected BMSCs (Fig. 8a). In addition, we found that the number of colonies generated by the T-98G, LN229 and A-172 cells co-cultured with miR-34a-transfected BMSCs was remarkably decreased compared to those of the NC group, indicating that miR-34a delivered by BMSCs to these GBM cells may act as a tumor suppressor (Fig. 8b).
MiR-34a released by BMSC-derived exosomes inhibits the proliferation, migration and invasion of GBM cells and enhances their chemosensitivity, thus improving GBM treatment. a, invasion and migration abilities of T-98G, LN229 and A-172 cells detected by Transwell assay (× 200). b, clonality of T-98G, LN229 and A-172 cells detected by colony formation assay. c, viability of T-98G, LN229 and A-172 cells treated with 100 μM TMZ tested by CCK-8 assay. d, apoptosis of T-98G, LN229 and A-172 cells cultured with exosomes derived from BMSC-miR-NC, treated with TMZ or cultured with exosomes derived from BMSCs-miR-34a, detected by TUNEL assay. The t test was used to analyze data between two groups, and repeated measurement variance analysis was used to compare data at different time points. *, p < 0.05 vs. the GBM cells-BMSCs-miR-34a NC group. miR-34a, microRNA-34a; BMSCs, bone marrow mesenchymal stem cells; GBM, glioblastoma; NC, negative control; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling; CCK-8, cell counting kit-8; TMZ, temozolomide
We also evaluated the effect of miR-34a delivered by BMSC-derived exosomes on chemotherapy of T-98G, LN229, and A-172 cells. To this end, the GBM cells were co-cultured with miR-34a-transfected BMSC cells and treated with TMZ. The viability of the T-98G, LN229 and A-172 cells after TMZ treatment was assessed using a CCK-8 assay. The results obtained implied that TMZ treatment alone led to a significant decrease in the viability of the T-98G, LN229 and A-172 cells, while miR-34a delivered by BMSCs exerted a stronger inhibitory effect on the viability of these cells (Fig. 8c). In addition, a TUNEL assay was performed to determine the combined efficacy of BMSC-miR-34a and TMZ on the induction of apoptosis in the T-98G, LN229 and A-172 cells. We found that the combined treatment of BMSC-miR-34a and TMZ evidently induced apoptosis of the GBM cells compared to BMSCs-miR-NC or TMZ alone (Fig. 8d). These results indicate that BMSC-derived exosomes can deliver miR-34a to GBM cells and increase the efficacy of TMZ treatment.
3.9 MiR-34a transferred from hBMSC-derived exosomes to GBM cells suppresses tumor growth in vivo
Nude mice were subcutaneously injected with BMSCs-miR-34a, BMSCs-miR-NC and PBS, respectively, to further evaluate the in vivo effect of BMSC-derived exosomes and the inhibitory effect of miR-34a on GBM tumor growth. Differences in tumor volume and tumor weight were recorded, and the tumor tissues were subjected to immunohistochemical staining. We found that the tumor volumes and tumor weights in the BMSCs-miR-34a group were significantly lower than those in the BMSCs-miR-NC group and the PBS group (Fig. 9a–c), supporting the inhibitory effect of miR-34a on tumor growth in vivo. The immunohistochemical staining results showed that the expression of MYCN in tumor tissues of the mice injected with BMSCs-miR-34a was significantly lower than that in the mice treated with BMSCs-miR-NC and PBS (Fig. 9d). We also found that miR-34a could be transferred from the BMSCs to the cancer cells, thereby reducing the expression of MYCN in the tumor tissues. Subsequently, we tested the in vivo effect of miR-34a on the apoptotic rate of T-98G cells using a TUNEL assay. We found that the apoptotic rate of these cells in the BMSCs-miR-34a group was significantly higher than that in the other two groups (Fig. 9e), which confirms that miR-34a can promote the apoptosis of the T-98G cells in vivo. These results indicate that miR-34a transferred by BMSC-derived exosomes can act as a tumor suppressor in GBM in vivo.
MiR-34a delivered to the GBM cells by BMSC-derived exosomes leads to inhibition of tumor growth in vivo. a, representative tumor sizes in different groups. b, changes in tumor volume after injection with BMSCs-miR-34a, BMSCs-miR-NC and PBS. c, tumor weight after injection with BMSCs-miR-34a, BMSCs-miR-NC and PBS. d, expression of MYCN determined by immunohistochemical staining. e, T-98G cell apoptosis detected by TUNEL assay. n = 6; *, p < 0.05 vs. the PBS group; #, p < 0.05 vs. the BMSCs-miR-NC group; one-way analysis of variance was used for multi-group comparison, and repeated measurement variance analysis was used for data comparison at different time points. miR-34a, microRNA-34a; BMSCs, bone marrow mesenchymal stem cells; GBM, glioblastoma; PBS, phosphate buffered saline; NC, negative control; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling
4 Discussion
Gliomas are brain tumors that can be categorized into oligodendroglioma, astrocytoma, GBM and several other subtypes [30]. GBM is the most aggressive brain tumor in humans [31]. Despite multiple therapeutic strategies available for their treatment, including radiation, chemotherapy and aggressive surgery, patients suffering from GBM usually only have a median survival of 12–15 months [32]. The aim of this present study was to explore the therapeutic effect of hBMSC-secreted exosomal miR-34a on GBM cells. Our data indicate that miR-34a transferred by hBMSC-secreted exosomes results in inhibited expression of MYCN, thereby suppressing GBM cell proliferation, invasion and migration and enhancing the chemosensitivity of GBM cells to TMZ.
Initially, we found that miR-34a was down-regulated while MYCN was up-regulated in primary GBM tissues and GBM-derived cell lines and that miR-34a can target and negatively regulate the expression of MYCN. Rathod et al. have shown that miR-34a is down-regulated in glioma stem cells and primary gliomas compared to normal brain tissues, which is consistent with our findings [21]. Others have shown that miR-34a expression was significantly down-regulated in GBMs compared to normal brain tissues [17]. MYCN expression is closely related to the growth and the invasiveness of neuroblastoma cells, and increased MYCN expression has been correlated with an advanced tumor stage, an accelerated progression and a poor prognosis in patients with neuroblastoma [33]. Moreover, it has been reported that up-regulation of MYCN expression in neural stem cells of the developing mouse forebrain can result in the development of GBM [18]. In addition, it has been found that restoration of miR-34a expression can inhibit the expression of multiple target genes, including MYCN [34], which is in line with the results of our current study.
In addition, we found that miR-34a could be delivered to GBM cells by BMSC-secreted exosomes. A recent study has suggested that BMSCs may exert their therapeutic capacity through exosomes [10]. Accumulating evidence indicates that exosomes may serve as important mediators in various cancers [35], i.e., they can modulate intercellular communication through the transfer of mRNAs, miRNAs and proteins [36]. Even though miRNAs represent only a relatively small proportion of exosomal RNAs, it has been found that miRNAs delivered by exosomes can function as suppressors of their targets in vitro and in vivo [37]. It has e.g. been found that miR-340 transferred by BMSC-derived exosomes can lead to inhibited tube formation and angiogenesis [38]. Likewise, exosomal miR-221 has been found to induce tumor progression and TMZ resistance in glioma [39].
Subsequently, we found that miR-34a transferred by BMSC-derived exosomes resulted in the in vitro inhibition of cell proliferation, invasion and migration, as well as in vivo GBM tumor growth, and enhanced the chemosensitivity of GBM cells to TMZ by silencing MYCN. Shatsberg et al. revealed that human U-87 MG GBM cells transfected with nanogels-miR-34a nano-polyplexes, another strategy of miRNA delivery for GBM therapy, exhibited a significant down-regulation of miR-34a target genes that play major roles in cell cycle arrest and apoptosis, leading to suppression of cell migration and proliferation [23]. These findings are consistent with our results. In addition, data collected by Yin et al. suggest that miR-34a acts as an inhibitor by suppressing the growth of GBM cells in vitro and in vivo, as well as regulating the expression of cell cycle-related proteins and EGFR [40]. MYCN can directly up-regulate the expression of genes that suppress proliferation and promote apoptosis. The ultimate effect results from a complex balance between pro- and anti-oncogenic functions [41]. TMZ is an essential chemotherapeutic drug for GBM treatment [42], and it has been shown that the anti-tumor function of TMZ results from the induction of DNA damage [43]. Moreover, miR-30a has been shown to promote the chemosensitivity of U251 GBM cells to TMZ by binding to beclin 1 [44].
In summary, we provide new data on hBMSC-derived exosomes and the inhibitory effect of exosomal miR-34a on GBM tumor progression. We found that exosomes carrying miR-34a can be effectively internalized by GBM cells. Our study provides evidence that hBMSC-derived exosomes overexpressing miR-34a can suppresses MYCN expression, leading to a decrease in GBM cell proliferation, invasion and migration and an increase in chemo-sensitization towards TMZ (Fig. 10).
Exosomes derived from BMSCs restore cancer-suppressive effect of miR-34a on GBM cells. BMSC-derived exosomes carrying overexpressed miR-34a inhibit GBM cell proliferation, invasion and migration, while increasing their chemo-sensitization towards TMZ by down-regulating MYCN. miR-34a, microRNA-34a; BMSCs, bone marrow mesenchymal stem cells; GBM, glioblastoma; TMZ, temozolomide
Change history
19 August 2021
A Correction to this paper has been published: https://doi.org/10.1007/s13402-021-00627-8
03 December 2023
A Correction to this paper has been published: https://doi.org/10.1007/s13402-023-00900-y
References
I. Vassallo, P. Zinn, M. Lai, P. Rajakannu, M.F. Hamou, M.E. Hegi, WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene 35, 12–21 (2016)
S. Lehrer, Glioblastoma and dementia may share a common cause. Med. Hypotheses 75, 67–68 (2010)
L.M. Nusblat, M.J. Carroll, C.M. Roth, Crosstalk between M2 macrophages and glioma stem cells. Cell. Oncol. 40, 471–482 (2017)
P. Ofek, M. Calderon, F.S. Mehrabadi, A. Krivitsky, S. Ferber, G. Tiram, N. Yerushalmi, S. Kredo-Russo, R. Grossman, Z. Ram, R. Haag, R. Satchi-Fainaro, Restoring the oncosuppressor activity of microRNA-34a in glioblastoma using a polyglycerol-based polyplex. Nanomedicine 12, 2201–2214 (2016)
J. Wang, E. Cazzato, E. Ladewig, V. Frattini, D.I. Rosenbloom, S. Zairis, F. Abate, Z. Liu, O. Elliott, Y.J. Shin, J.K. Lee, I.H. Lee, W.Y. Park, M. Eoli, A.J. Blumberg, A. Lasorella, D.H. Nam, G. Finocchiaro, A. Iavarone, R. Rabadan, Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016)
D. Matias, J. Balca-Silva, L.G. Dubois, B. Pontes, V.P. Ferrer, L. Rosario, A. do Carmo, J. Echevarria-Lima, A.B. Sarmento-Ribeiro, M.C. Lopes, V. Moura-Neto, Dual treatment with shikonin and temozolomide reduces glioblastoma tumor growth, migration and glial-to-mesenchymal transition. Cell. Oncol. 40, 247–261 (2017)
M.K. Nicholas, R.V. Lukas, S. Chmura, B. Yamini, M. Lesniak, P. Pytel, Molecular heterogeneity in glioblastoma: Therapeutic opportunities and challenges. Semin. Oncol. 38, 243–253 (2011)
N. Hata, N. Shinojima, J. Gumin, R. Yong, F. Marini, M. Andreeff, F.F. Lang, Platelet-derived growth factor BB mediates the tropism of human mesenchymal stem cells for malignant gliomas. Neurosurgery 66, 144–156; discussion 156-147 (2010)
T. Birnbaum, J. Hildebrandt, G. Nuebling, P. Sostak, A. Straube, Glioblastoma-dependent differentiation and angiogenic potential of human mesenchymal stem cells in vitro. J. Neuro-Oncol. 105, 57–65 (2011)
L. Chen, F.B. Lu, D.Z. Chen, J.L. Wu, E.D. Hu, L.M. Xu, M.H. Zheng, H. Li, Y. Huang, X.Y. Jin, Y.W. Gong, Z. Lin, X.D. Wang, Y.P. Chen, BMSCs-derived miR-223-containing exosomes contribute to liver protection in experimental autoimmune hepatitis. Mol. Immunol. 93, 38–46 (2018)
M. Ono, N. Kosaka, N. Tominaga, Y. Yoshioka, F. Takeshita, R.U. Takahashi, M. Yoshida, H. Tsuda, K. Tamura, T. Ochiya, Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 7, ra63 (2014)
V. Sundararajan, F.H. Sarkar, T.S. Ramasamy, The versatile role of exosomes in cancer progression: Diagnostic and therapeutic implications. Cell. Oncol. 41, 223–252 (2018)
F. Lan, Q. Qing, Q. Pan, M. Hu, H. Yu, X. Yue, Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell. Oncol. 41, 25–33 (2018)
P. Sumazin, X. Yang, H.S. Chiu, W.J. Chung, A. Iyer, D. Llobet-Navas, P. Rajbhandari, M. Bansal, P. Guarnieri, J. Silva, A. Califano, An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147, 370–381 (2011)
G.S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C. Polytarchou, K.B. Marcu, A.G. Papavassiliou, R. Sandaltzopoulos, E. Kolettas, A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 40, 303–339 (2017)
F. Guessous, Y. Zhang, A. Kofman, A. Catania, Y. Li, D. Schiff, B. Purow, R. Abounader, microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 9, 1031–1036 (2010)
W.B. Li, M.W. Ma, L.J. Dong, F. Wang, L.X. Chen, X.R. Li, MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol. Ther. 12, 477–483 (2011)
L. Bjerke, A. Mackay, M. Nandhabalan, A. Burford, A. Jury, S. Popov, D.A. Bax, D. Carvalho, K.R. Taylor, M. Vinci, I. Bajrami, I.M. McGonnell, C.J. Lord, R.M. Reis, D. Hargrave, A. Ashworth, P. Workman, C. Jones, Histone H3.3. Mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 3, 512–519 (2013)
K. Campbell, J.M. Gastier-Foster, M. Mann, A.H. Naranjo, C. Van Ryn, R. Bagatell, K.K. Matthay, W.B. London, M.S. Irwin, H. Shimada, M.M. Granger, M.D. Hogarty, J.R. Park, S.G. DuBois, Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: A report from the Children's oncology group. Cancer 123, 4224–4235 (2017)
X.Y. He, Z.L. Tan, Q. Mou, F.J. Liu, S. Liu, C.W. Yu, J. Zhu, L.Y. Lv, J. Zhang, S. Wang, L.M. Bao, B. Peng, H. Zhao, L. Zou, microRNA-221 enhances MYCN via targeting nemo-like kinase and functions as an oncogene related to poor prognosis in neuroblastoma. Clin. Cancer Res. 23, 2905–2918 (2017)
S.S. Rathod, S.B. Rani, M. Khan, D. Muzumdar, A. Shiras, Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio. 4, 485–495 (2014)
S. Sarkar, S. Jun, S. Rellick, D.D. Quintana, J.Z. Cavendish, J.W. Simpkins, Expression of microRNA-34a in Alzheimer's disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 1646, 139–151 (2016)
Z. Shatsberg, X. Zhang, P. Ofek, S. Malhotra, A. Krivitsky, A. Scomparin, G. Tiram, M. Calderon, R. Haag, R. Satchi-Fainaro, Functionalized nanogels carrying an anticancer microRNA for glioblastoma therapy. J. Control. Release 239, 159–168 (2016)
E.A. Toraih, N.M. Aly, H.Y. Abdallah, S.A. Al-Qahtani, A.A. Shaalan, M.H. Hussein, M.S. Fawzy, MicroRNA-target cross-talks: Key players in glioblastoma multiforme. Tumour Biol. 39, 1010428317726842 (2017)
J.S. Wei, Y.K. Song, S. Durinck, Q.R. Chen, A.T. Cheuk, P. Tsang, Q. Zhang, C.J. Thiele, A. Slack, J. Shohet, J. Khan, The MYCN oncogene is a direct target of miR-34a. Oncogene 27, 5204–5213 (2008)
T. Lv, K. Song, L. Zhang, W. Li, Y. Chen, Y. Diao, Q. Yao, P. Liu, MiRNA-34a decreases ovarian cancer cell proliferation and chemoresistance by targeting HDAC1. Biochem. Cell Biol. 96, 663–671 (2018)
M. Xu, D. Li, C. Yang, J.S. Ji, MicroRNA-34a inhibition of the TLR signaling pathway via CXCL10 suppresses breast Cancer cell invasion and migration. Cell. Physiol. Biochem. 46, 1286–1304 (2018)
A. Savina, M. Furlan, M. Vidal, M.I. Colombo, Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 278, 20083–20090 (2003)
K. Trajkovic, C. Hsu, S. Chiantia, L. Rajendran, D. Wenzel, F. Wieland, P. Schwille, B. Brugger, M. Simons, Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008)
E. Lages, A. Guttin, M. El Atifi, C. Ramus, H. Ipas, I. Dupre, D. Rolland, C. Salon, C. Godfraind, F. deFraipont, M. Dhobb, L. Pelletier, D. Wion, E. Gay, F. Berger, J.P. Issartel, MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One 6, e20600 (2011)
A.T. Bayrac, K. Sefah, P. Parekh, C. Bayrac, B. Gulbakan, H.A. Oktem, W. Tan, In vitro selection of DNA aptamers to glioblastoma Multiforme. ACS Chem. Neurosci. 2, 175–181 (2011)
C.S. Niu, Y. Yang, C.D. Cheng, MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int. J. Oncol. 42, 1533–1540 (2013)
Y. Chen, Y.H. Tsai, S.H. Tseng, Inhibition of cyclin-dependent kinase 1-induced cell death in neuroblastoma cells through the microRNA-34a-MYCN-survivin pathway. Surgery 153, 4–16 (2013)
T. Thor, A. Kunkele, K.W. Pajtler, A.K. Wefers, H. Stephan, P. Mestdagh, L. Heukamp, W. Hartmann, J. Vandesompele, N. Sadowski, L. Becker, L. Garrett, S.M. Holter, M. Horsch, J. Calzada-Wack, T. Klein-Rodewald, I. Racz, A. Zimmer, J. Beckers, F. Neff, T. Klopstock, P. De Antonellis, M. Zollo, W. Wurst, H. Fuchs, V. Gailus-Durner, U. Schuller, M.H. de Angelis, A. Eggert, A. Schramm, J.H. Schulte, MiR-34a deficiency accelerates medulloblastoma formation in vivo. Int. J. Cancer 136, 2293–2303 (2015)
J.K. Yang, J. Song, H.R. Huo, Y.L. Zhao, G.Y. Zhang, Z.M. Zhao, G.Z. Sun, B.H. Jiao, DNM3, p65 and p53 from exosomes represent potential clinical diagnosis markers for glioblastoma multiforme. Ther. Adv. Med. Oncol. 9, 741–754 (2017)
J. Wang, A. Hendrix, S. Hernot, M. Lemaire, E. De Bruyne, E. Van Valckenborgh, T. Lahoutte, O. De Wever, K. Vanderkerken, E. Menu, Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood 124, 555–566 (2014)
S.R. Baglio, K. Rooijers, D. Koppers-Lalic, F.J. Verweij, M. Perez Lanzon, N. Zini, B. Naaijkens, F. Perut, H.W. Niessen, N. Baldini, D.M. Pegtel, Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 6, 127 (2015)
T. Umezu, S. Imanishi, K. Azuma, C. Kobayashi, S. Yoshizawa, K. Ohyashiki, J.H. Ohyashiki, Replenishing exosomes from older bone marrow stromal cells with miR-340 inhibits myeloma-related angiogenesis. Blood Adv. 1, 812–823 (2017)
J.K. Yang, J.P. Yang, J. Tong, S.Y. Jing, B. Fan, F. Wang, G.Z. Sun, B.H. Jiao, Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro-Oncol. 131, 255–265 (2017)
D. Yin, S. Ogawa, N. Kawamata, A. Leiter, M. Ham, D. Li, N.B. Doan, J.W. Said, K.L. Black, H. Phillip Koeffler, miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene 32, 1155–1163 (2013)
J.M. Shohet, R. Ghosh, C. Coarfa, A. Ludwig, A.L. Benham, Z. Chen, D.M. Patterson, E. Barbieri, P. Mestdagh, D.N. Sikorski, A. Milosavljevic, E.S. Kim, P.H. Gunaratne, A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 71, 3841–3851 (2011)
A. Nadkarni, M. Shrivastav, A.C. Mladek, P.M. Schwingler, P.T. Grogan, J. Chen, J.N. Sarkaria, ATM inhibitor KU-55933 increases the TMZ responsiveness of only inherently TMZ sensitive GBM cells. J. Neuro-Oncol. 110, 349–357 (2012)
N. Kondo, A. Takahashi, E. Mori, T. Noda, M.Z. Zdzienicka, L.H. Thompson, T. Helleday, M. Suzuki, Y. Kinashi, S. Masunaga, K. Ono, M. Hasegawa, T. Ohnishi, FANCD1/BRCA2 plays predominant role in the repair of DNA damage induced by ACNU or TMZ. PLoS One 6, e19659 (2011)
J. Xu, H. Huang, R. Peng, X. Ding, B. Jiang, X. Yuan, J. Xi, MicroRNA-30a increases the chemosensitivity of U251 glioblastoma cells to temozolomide by directly targeting beclin 1 and inhibiting autophagy. Exp. Ther. Med. 15, 4798–4804 (2018)
Acknowledgments
We would like to thank our researchers for their hard work and the reviewers for their valuable advice.
Author information
Authors and Affiliations
Contributions
Bin Wang, Zhong-Hua Wu and Ping-Yang Lou designed the study. Chang Chai and Shuang-Yin Han collated the data, designed and developed the database, carried out data analyses and produced the initial draft of the manuscript. Jian-Fang Ning and Ming Li contributed to drafting the manuscript. All authors have contributed to the revision and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None declared.
Ethical approval and informed consent
This study was carried out and approved by the Institutional Review Board and the Institutional Animal Care and Use Committee of Henan Province People’s Hospital, Zhengzhou University. All patients signed informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s13402-021-00627-8
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Wu, ZH., Lou, PY. et al. RETRACTED ARTICLE: Human bone marrow-derived mesenchymal stem cell-secreted exosomes overexpressing microRNA-34a ameliorate glioblastoma development via down-regulating MYCN. Cell Oncol. 42, 783–799 (2019). https://doi.org/10.1007/s13402-019-00461-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-019-00461-z