Abstract
Background
Tumors contain a functional subpopulation of cells that exhibit stem cell properties. These cells, named cancer stem cells (CSCs), play significant roles in the initiation and progression of cancer. Long non-coding RNAs (lncRNAs) can act at the transcriptional, posttranscriptional and translational level. As such, they may be involved in various biological processes such as DNA damage repair, inflammation, metabolism, cell survival, cell signaling, cell growth and differentiation. Accumulating evidence indicates that lncRNAs are key regulators of the CSC subpopulation, thereby contributing to cancer progression. The aim of this review is to overview current knowledge about the functional role and the mechanisms of action of lncRNAs in the initiation, maintenance and regulation of CSCs derived from different neoplasms. These lncRNAs include CTCF7, ROR, DILC, HOTAIR, H19, HOTTIP, ATB, HIF2PUT, SOX2OT, MALAT-1, CUDR, Lnc34a, Linc00617, DYNC2H1–4, PVT1, SOX4 and ARSR Uc.283-plus. Furthermore, we will illustrate how lncRNAs may regulate asymmetric CSC division and contribute to self-renewal, drug resistance and EMT, thus affecting the metastasis and recurrence of different cancers. In addition, we will highlight the implications of targeting lncRNAs to improve the efficacy of conventional drug therapies and to hamper CSC survival and proliferation.
Conclusions
lncRNAs are valuable tools in the search for new targets to selectively eliminate CSCs and improve clinical outcomes. LncRNAs may serve as excellent therapeutic targets because they are stable, easily detectable and expressed in tissue-specific contexts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Although it has been assumed that the vast majority of the human genome (85%) is transcribed, only ~2% of the transcripts are translated into proteins [1]. Since it has become evident that the remaining transcripts are not translated into proteins they are, concordantly, called non-coding RNAs (ncRNAs) [2]. Long non-coding RNAs (lncRNAs) represent a very interesting subgroup of ncRNAs that have recently come into light as powerful players in various diseases, including cancer. Up to date, the GENCODE database [3], which has the largest compilation of transcripts, reports a total of 7258 small ncRNAs and 15,767 annotated lncRNAs (http://www.gencodegenes.org/stats/current.html).
LncRNAs represent a heterogeneous mix of transcripts, most of them with unknown function. These RNAs have been grouped into 5 categories based on their location relative to the nearest protein-coding genes: 1) sense lncRNAs that overlap with coding mRNAs on the coding strand of a gene, 2) antisense lncRNAs that overlap with coding mRNAs on the non-coding strand of a gene, 3) bidirectional lncRNAs that share its transcription start site with a coding gene on the opposite strand, 4) intronic lncRNAs that are transcribed from an intronic region of a coding gene and 5) intergenic lncRNAs that are located between coding genes [4].
2 Molecular mechanisms involving lncRNAs
In humans, lncRNAs are mainly transcribed by RNA Polymerase (Pol) II or III, but they can also be transcribed by polymerase V in other eukaryotic organisms [5]. It has been found that LncRNAs that are transcribed by different RNA polymerases may exhibit distinct epigenetic marks. LncRNAs transcribed by RNA polymerase II usually exhibit, for instance, histone H3 trimethylation of lysine 4 (H3K4me3) and histone H3 trimethylation of lysine 36 (H3K36me3). RNA polymerase III-transcribed genes on the other hand usually exhibit mono-, bi- or trimethylation of histone H3 lysine 4 (H3K4me1/2/3) or acetylation of histone H3 lysine 4 (H3K4ac) as long as H3K36me3 and bi-methylation of histone H3 lysine 27 (H3K27me2) are absent [6]. Furthermore, Pol II-transcribed lncRNAs are processed as mRNAs, i.e., 5′ caps and 3′ poly-A tails are added, while Pol III-transcribed lncRNAs are not polyadenylated. Mature lncRNAs can interact with an array of diverse molecules, creating supramolecular structures such as RNA:RNA, RNA:DNA (double or triple chains), RNA:Protein, DNA:RNA:Protein or DNA:RNA:RNA complexes [7].
LncRNAs may exhibit distinct subcellular localization patterns including nuclear, cytoplasmic or both. Inside the nucleus, lncRNAs may stay close to the site of transcription anchored to the chromatin or accumulate away from the transcription site. Once transcribed, lncRNAs may act in cis (controlling local gene expression) or in trans (controlling distant gene expression) resulting in the silencing or activation of tissue-specific genes [8]. Cytoplasmic lncRNAs have been found to play essential roles in multiple molecular mechanisms, including mRNA stability and translation regulation, protein modification mediation, serving as microRNA precursors or as competing endogenous RNAs. In general, lncRNAs are thought to act as master regulators of transcription, since they can remodel chromatin and create binding domains for the transcriptional machinery, as well as interact with several repressor complexes to block transcription start sites [9].
LncRNAs have been found to be involved in a broad range of biological processes and to exhibit multiple modes of action [10, 11]. Based on this, LncRNAs can be classified by function as 1) decoy lncRNAs that can bind and sequester proteins to modify their catalytic activity or avoid their interaction with targets, 2) guide lncRNAs that can recruit chromatin modifiers to specific genomic loci, 3) scaffold lncRNAs that can function as adaptors to bring together two or more proteins into a complex, 4) lncRNA sponges that can interact with miRNAs to avoid their effect on mRNA targets, 5) competitive endogenous lncRNAs that can provide stability to mRNAs for correct translation and 6) enhancers that function by stabilizing chromosomal loops between gene enhancers and promoters [12,13,14,15].
LncRNAs are differentially expressed during normal physiological processes such as development, differentiation and imprinting, as well as during pathological processes such as cancer [16]. Interestingly, abnormal modulation of specific lncRNAs has been reported in cancer stem cell (CSC) subpopulations [9, 17, 18]. Here, we will summarize recent advances on the involvement of specific lncRNAs in the regulation of CSCs (Fig. 1).
Role of CSCs in the relapse of solid tumors. Since CSCs are resistant to chemotherapeutic agents, current cancer treatments fail to eradicate them. This failure eventually allows CSCs to self-renew and provoke tumor relapse. Therefore, it is essential to identify specific targets to eliminate the CSC fraction and, thereby, to ensure the eradication of cancer. Recent work has shown that various lncRNAs may play crucial roles in both the nucleus and the cytoplasm of CSCs, thereby allowing them to self-renew and to promote tumor growth
3 Cancer stem cells
Cancer represents a group of diseases with certain commonalities (‘hallmarks’) that behave as dynamic, interrelated and multidimensional evolutionary systems, centered around deregulated genomic and epigenomic processes [19,20,21]. In recent years, it has been suggested that tumors harbor functional cell subpopulations with stem cell features, i.e., the ability to self-renew and to produce a phenotypically diverse progeny [22, 23] . These cells, named cancer stem cells (CSCs), are able to divide either asymmetrically or symmetrically, possess a limitless proliferative capacity, have a high tumorigenic, invasive and metastatic potential and are resistant to commonly used chemotherapeutic agents [24,25,26]. CSCs achieve self-renewal through asymmetric cell division, in which one daughter cell retains the self-renewal capacity and the other undergoes differentiation. The mechanisms that regulate asymmetric versus symmetric division are key to cancer progression, since the deregulation of this process is intrinsically associated with neoplastic transformation and tumor growth.
CSCs can be identified using specific cell surface markers such as CD44, CD24, CD133, EpCAM and CD117 [27, 28]. CSCs express high levels of transcription factors that are associated with pluripotency and epithelial-mesenchymal transition (EMT), and they exhibit an increased potential to form new tumors in e.g. immunodeficient mice or other animal models [25, 29]. Since strong evidence indicates that CSCs are responsible for local and/or distant recurrences (Fig. 1), it is imperative to understand the molecular mechanisms that govern CSCs in order to design specific therapeutic strategies directed against CSCs.
4 The function of lncRNAs in CSCs
Until recently, most research has focused on the role of coding genes in cancer development, providing a basis for most of the knowledge that we have to date. It has, however, become clear that also lncRNAs may participate in cancer development and progression. Since lncRNAs are emerging as master regulators of transcription and as possible oncogenes or tumor suppressors, their role in the establishment of CSC phenotypes has recently been explored. It has for example been shown that the overexpression, deficiency or mutation of lncRNAs may have functional implications for the self-renewal capacity of CSCs (Fig. 1). The role of various lncRNAs in the regulation of CSCs has been studied in several cancers including colon, breast, prostate, esophagus, lung, liver, kidney, stomach, bone and liver cancers. In the next section, we will describe the functions and regulatory roles of lncRNAs in CSCs in different cancers (Table 1).
5 LncRNAs associated with CSCs
LnchPVT1
LnchPVT1 is a nuclear lncRNA that has been found to be significantly upregulated in hepatocellular carcinomas (HCC) and to be associated with hepatitis B virus (HBV) infection [30]. LnchPVT1 has recently been linked to the expansion of CSCs [31, 32]. Notably, lnchPVT1 has been found to be regulated by the TGF-β pathway, which can be activated by HBV in HCC tissues. Gain and loss of function experiments have been used to demonstrate that lnchPVT1 can enhance liver cancer stem cell abilities both in vitro and in vivo. Specifically, it has been found that this lncRNA can mediate the acquisition of stem cell-like properties in HCC cells by stabilizing the nucleolar protein NOP2 [31]. Remarkably, a high lnchPVT1 expression has been found to be associated with a poor clinical outcome.
Linc00617
Linc00617 is a chromosome 14 associated long intergenic non-coding RNA (lincRNA) with a size of 2937 nt. This lincRNA has been found to be highly expressed in advanced breast cancer tissues and its associated lymph node metastases. Gain of function studies have demonstrated that this lincRNA can promote the migration and invasion of breast cancer cells. Notably, overexpression of linc00617 has been found to induce EMT by reducing the level of E-cadherin and increasing that of N-cadherin and Vimentin [33]. This lincRNA is not only associated with EMT, but also with the self-renewal and expansion of breast CSCs. Overexpression of this lincRNA has been found to increase the mammosphere forming and tumorigenic abilities of breast cancer cell populations due to an enrichment of the CSC fraction. Interestingly, in vivo assays have shown that linc00617 deficiency may lead to a dramatic reduction in the number of metastatic nodules.
The molecular mechanism underlying stemness control by linc00617 has recently been elucidated. Linc00617 has been identified as a nuclear RNA that binds to the promoter of the Sox2 gene and activates its transcription through the recruitment of hnRNP-K (Fig. 2a). SOX2 regulation by linc00617 has been verified through loss and gain of function experiments showing a positive strong correlation between the expression levels of linc00617 and SOX2. It was concluded that linc00617 probably exhibits oncogenic activity through the regulation of SOX2, which stimulates EMT and enhances the self-renewal ability of CSCs.
Mechanisms of action of lncRNAs in CSC nuclei. a Lnc00617 may act as a scaffold of hnRNP-K and promote the self-renewal of CSCs. b Lnc34a may act as a scaffold of Dnmt3a, PHB2 and HDAC1 to avoid the transcription of miR34a, thus activating the NOTCH and WNT signaling pathways. c LncTCF7 may act as a scaffold to recruit the chromatin remodeling complex SWI/SNF to the promoter of TCF7, thereby activating the WNT signaling pathway. ci LncTCF7 may promote invasion through the induction of SLUG expression
HIF2PUT
The hypoxia-inducible factor-2α promoter upstream transcript (HIF2PUT) is a novel lncRNA with key regulatory functions in osteosarcoma and colon cancer stem cells [34,35,36]. HIF2PUT is an antisense lncRNA located in the promoter upstream region of the hypoxia-inducible factor-2α (HIF-2α) gene. HIF2PUT regulates the transcriptional activity of its host gene HIF-2α in bone and colon tissues. Overexpression of this lncRNA leads to HIF-2α upregulation whereas HIF2PUT deficiency has been found to result in reduced levels of HIF-2α in both osteosarcoma and colon cancer-derived cell lines. Notably, HIF-2α has been associated with the presence of CSCs in various types of cancer, where it exerts a role in CSC regulation. Interestingly, HIF-2α and HIF2PUT upregulation has been found to be a common feature of aggressive osteosarcomas and a high expression of HIF2PUT has been found to predict a poor prognosis in osteosarcoma patients [35].
HIF2PUT exerts different regulatory roles in osteosarcoma and colon cancer CSCs. In osteosarcoma, HIF2PUT acts as a potent inhibitor of CSC self-renewal. Inhibition of HIF2PUT has been found to enhance the proliferation, migration and self-renewal of CSCs while its overexpression has been found to inhibit these features [36]. Conversely, in colon cancer HIF2PUT expression has been found to be associated with enrichment of the population of cells with a CSC phenotype [34], whereas HIF2PUT deficiency has been found to impair CSC properties, including proliferation, self-renewal, migration and invasion. In addition, HIF2PUT inhibition has been found to result in a reduction in CSC markers such as Oct4, Sox2 and CD44. Together, these data indicate that this lncRNA may exert opposite roles in the regulation of CSCs derived from different tissues types. We believe that functional characterization of lncRNAs in different tissue lineages is crucial for the development of tissue-specific therapies. The communication between lncRNAs and components of the microenvironment, such as stromal cells and extracellular components, may have additional implications for cancer progression and therapy development.
LncSOX2OT
SOX2 overlapping transcript (SOX2OT) is a lncRNA deduced from human chromosome 3q26.3. This lncRNA is transcribed in the same orientation as SOX2, one of the major regulators of pluripotency, which is embedded within the intronic region of SOX2OT. Several studies have shown that there is a positive correlation between SOX2OT and SOX2 expression [37,38,39,40]. SOX2OT has been found to be co-upregulated with SOX2 in embryonic stem cells, in breast CSCs and in esophageal squamous carcinoma cells. SOX2OT and SOX2 have also both been found to be highly expressed in estrogen receptor-positive breast cancer and to be associated with tamoxifen sensitivity [38]. Interestingly, in esophageal squamous cell carcinoma, SOX2OT has been found to be co-upregulated with Oct4, another master regulator of pluripotency [37].
SOX2OT is spliced into at least 8 distinct transcripts, and their expression patterns play an emerging role in stem cell biology and tumorigenesis [37, 41]. It has, for example, been found that SOX2OT variant 7 and 8 are highly expressed in human embryonal carcinoma NT2 cells that exhibit stem cell-like properties. During neuronal differentiation of these cells the SOX2OT-7 variant is dramatically downregulated [41].
HOTAIR
Hox transcript antisense intergenic RNA (HOTAIR) is an oncogenic lncRNA of which the expression has been found to be altered in various types of cancer, including breast, ovary, colon, pancreas and cervix cancer [42, 43]. This lncRNA is able to induce activation or silencing of its target genes. HOTAIR can recruit the MLL1 methyltransferase and induce histone H3 trimethylation of lysine 4 (H3K4me3), thereby relaxing chromatin and allowing binding of the transcription machinery. HOTAIR can also recruit the PRC2 (Polycomb Repressive Complex 2) complex, induce H3K27me3 and, ultimately, provoke gene silencing [12, 13].
Elevated HOTAIR expression has recently been reported in CSCs derived from breast, oral and colon carcinomas, and from gliomas [44,45,46]. The expression of this lncRNA has been associated with the acquisition of stem cell characteristic resulting in an increased tumor growth and metastatic potential [44, 47, 48]. HOTAIR induces stemness mainly through triggering EMT in a TGF-β dependent way [44, 45]. Exogenous expression of HOTAIR has been found to result in upregulation of the EMT inductors Zeb1, SNAIL, TWIST and CTNNA1, as well as in induction of the mesenchymal markers Vimentin and Fibronectin. Accordingly, also epithelial markers such as E-cadherin, BMP7 and ERBB3 were found to be downregulated by HOTAIR. Interestingly, it was found that the genes downregulated during EMT by HOTAIR exhibit increased PRC2 occupancies [45]. The stem cell features induced by HOTAIR have been validated by the occurrence of increased colony formation, migration and self-renewal capacities [46]. It has also been shown that HOTAIR may regulate colony formation through a reduced p53 binding to the p21 promoter. In addition, it has been shown that HOTAIR may promote CSC growth through downregulation of SETD2 [47]. Several studies have established that HOTAIR may also induce the expression of stem cell markers such as SOX1, SOX2, OCT4 and CD44 [45, 46]. Additionally, it has been found that HOTAIR can regulate SOX2 expression through attenuation of the function of miR-34a. HOTAIR has been positively associated with an advanced clinical tumor stage, the occurrence of metastasis and a worse prognosis [49]. The therapeutic significance of this lncRNA has been established using HOTAIR inhibitors that suppress the proliferation, migration, invasion and self-renewal of CSCs. Thus, HOTAIR may serve as a target to attenuate the progression and invasion/metastasis of cancer [48, 50].
LncRNA uc.283-plus
LncRNA uc.283-plus is a RNA of 277 nt deduced from an ultra-conserved region (UCR) on chromosome 10. This lncRNA is expressed only in pluripotent stem cells and in some solid neoplasms such as glioma and prostate adenocarcinoma, but it is absent in normal adult tissues [51]. It has been found that the expression of lncRNA uc.283-plus can discriminate between adult tissues and embryonic stem cells. Although very little is known about this lncRNA, it has been proposed by using bioinformatics tools that it may act as a sponge RNA to recruit miRNAs [52] such as miR-455-5p, miR-640 and miR-1909-3p and, by doing so, allow the expression of target genes such as DICER1, SOX2 and NOTCH1 [51, 53]. Another transcribed RNA called “uc.283-minus” has been found to be deduced from the strand opposite to the uc.283-plus genomic region. The expression of this lncRNA has been found to be regulated by hypermethylation in its CpG islands. Although its function is still unknown, it has been suggested that it may act in the process of tumorigenesis [54].
Lnc34a
Lnc34a is a novel lncRNA that binds to the miR-34a encoding gene and regulates its silencing by recruiting DNA methyltransferase 3a (DNMT3a) and histone deacetylase 1 (HDAC1) to the miR-34a promoter. Previous studies have shown that miR-34a may act as a negative regulator of the Notch and Wnt signaling pathways, which are essential for the self-renewal of CSCs [55,56,57]. Interestingly, Wang et al. [58] showed that lnc34a is highly expressed in colon CSCs where it promotes self-renewal (Fig. 2b). Lnc34a is the first lncRNA identified that exhibits an asymmetric distribution during CSC division, thus producing asymmetric daughter cells with different cell fates (Fig. 2b). Lnc34a suppression leads to CSC differentiation via asymmetric cell division, whereas lnc34a overexpression leads to CSC proliferation via symmetric cell division. We consider that further research is warranted to unravel the mechanisms by which this and other lncRNAs regulate asymmetric CSC division. The discovery of the miR-34a - lnc34a axis highlights the importance of ncRNAs in this process and their potential to orchestrate the CSC self-renewal process.
lncTCF7
LncTCF7 has a length of 3.6 kb and is composed of three exons. This lncRNA can be located in the nucleus as well as in the cytoplasm where it performs different functions depending on the specific cell types or tissues involved. Previous studies have shown that the expression of this lncRNA is regulated through the IL6/STAT3 pathway [59], a key pathway involved in cancer progression [60, 61]. LncTCF7 has been found to interact with three nuclear subunits of the SWI/SNF chromatin remodeling complex (BRG1, BAF170 and SNF5), allowing their recruitment to the TCF7 gene promoter, thus regulating the transcription of TCF7 and activating the Wnt signaling cascade, which is involved in stem cell self-renewal (Fig. 2c) [59, 62] LncTCF7 has been found to regulate 2491 genes, many of which belong to the Wnt signaling pathway (Fig. 2c).
LncTCF7 is highly expressed in nuclei of CSCs derived from hepatocellular carcinomas and it has been reported that inhibition of this lncRNA significantly disrupts the expression of the pluripotency markers Sox2, Nanog and Oct4 and decreases the tumorigenic ability of liver CSCs. Conversely, lncTCF7 overexpression has been found to enhance the tumor-forming ability of liver CSCs [62]. LncTCF7 has also been found to be involved in the regulation of CSCs derived from non-small cell lung cancer (NSCLC) [63]. Together with the SWI/SNF complex, this lncRNA can increase the expression of Slug to promote invasion. Slug is a transcriptional repressor that binds to E-box motifs and represses E-Cadherin transcription (Fig. 2ci). In addition, lncTCF7 increases the expression of the stem cell marker EpCAM to promote self-renewal. This latter effect is not mediated by the SWI/SNF complex, but by competing with EpCAM for binding of the microRNA miR-200c and, thereby, avoiding EpCAM degradation (Fig. 3e) [64]. This sponge effect has been demonstrated in prostate cancer cells as well, thus supporting a more generalized function of this lncRNA. Together, these results show that lncRNAs like lncTCF7 may carry out several molecular functions in the same cells, thereby expanding the possible roles of these RNAs. We consider that this lncRNA depicts a clear example of the multifunctionality of lncRNAs, since some of them may affect the same biological process by acting in different ways according to cellular needs. As we mentioned earlier, lncRNAs can interact directly with DNA, mRNA or proteins to regulate a variety of physiological and pathological processes.
Mechanisms of action of lncRNAs in the cytoplasm of CSCs. a Absence of lncRNA H19 may inhibit the translation of LIN28 through miRLet-7. b LncRNA H19 may act as a sponge RNA for miRLet-7 and allow the translation of LIN28. In addition, H19, let-7 and LIN28 may form a negative feedback loop favoring CSC self-renewal. c Decreases in lncRNA ATB levels may allow miR-200 to inhibit ZEB1/2 mRNAs. In addition, the stability of the IL-11 mRNA may be decreased. d lncRNA ATB may act as a sponge RNA for miR-200 to promote the translation of ZEB1/2 and, thus, to promote EMT. LncRNA ATB may also interact directly with IL-11 mRNA and regulate its stability, thereby contributing to cell colonization. e LncRNA TCF7 may act as a sponge for miR-200c and, thereby, inhibit the repression of EpCAM. f LncDYNC2H1–4 may act as a sponge for miR-145 in the presence of gemcitabine, thereby leading to an increment in stem cell markers
LncH19
H19 is one of the first ncRNAs identified as a cancer-related lncRNA. Hitherto, this lncRNA has been found to be involved in the development and progression of many different cancer types. H19 is an imprinting gene located in the 11p15.5 region and encodes a 2.3 kb lncRNA that is expressed exclusively from the maternal allele [65]. H19 is highly expressed during vertebrate embryonic development, but is downregulated in most tissues after birth. Loss of imprinting and, consequently, strong H19 expression has been extensively documented in several types of cancer. Recent studies have shown that H19 is overexpressed in stem-like cells of breast [66] and prostate [67] cancers and glioblastomas [68].
In breast cancer, it has been found that ectopic overexpression of H19 significantly promotes migration, as well as clone and sphere forming capacities. On the contrary, inhibition of H19 has been found to disrupt the growth and tumor forming capacities of breast cancer cells. H19 is mostly found in the cytoplasm of breast cancer cells where it functions to sponge miRNA let-7, which leads to an increase in the expression of LIN28, a well-known let-7 target (Fig. 3a). H19 can also be repressed by let-7 via a negative feedback loop. Notably, H19 and LIN28 are co-expressed in primary breast carcinomas and both have been found to play a critical role in the maintenance of breast CSCs [66]. LIN28 can also block mature let-7 production, thereby avoiding the repression of H19 and reversing the suppression of breast CSC properties mediated by the loss of H19 [66, 69]. Collectively, these results suggest that H19/let-7/LIN28 forms a double negative feedback loop to promote breast CSC maintenance (Fig. 3b).
In prostate cancer, H19 upregulation has been found to correlate with the expression of stem cell markers such as Sox2, Oct4, Notch1, Klf4, c-Myc and Abcg2. Remarkably, lncH19 level modulations also affect the clonogenic ability of prostate cancer cells [67]. H19 also plays a role in the self-renewal of CSCs in glioblastoma [68]. Interestingly, H19 expression has been found to be mainly restricted to the CSC fraction and exogenous expression has been found to result in an increased migration, as well as neurosphere and tumor formation capacity of this fraction.
Furthermore, H19 is the primary precursor of the proliferation-suppressing miR-675 [70], which has been found to be involved in both neuronal [71, 72] and muscle differentiation [73]. MiR-675 is embedded into the first exon of H19 and its expression is barely detectable in fetal tissues, despite abundant H19 expression [74]. The processing of miR-675 from H19 in embryonic tissues is inhibited by an RNA-binding protein, HuR. MiR-675 is only released in response to cellular stress or oncogenic signals [74]. H19 and miR-675 are both expressed in undifferentiated bone marrow mesenchymal stem cells, but their expression decreases during differentiation of these cells into a neural phenotype. The down-regulation of miR-675 is concomitant with the up-regulation of its target IGF-1R during the differentiation of neural cells [72]. H19 also has an essential function in skeletal muscle differentiation mediated by the microRNAs embedded within it. Strikingly, both miR-675-3p and miR-675-5p promote myogenic differentiation by repressing Smad1, Smad5 and Cdc6. Smad1 and Smad5 are involved in the BMP pathway and Cdc6 is a DNA replication factor that needs to be downregulated during myoblast differentiation [73]. These results support the idea that miR-675 may confer functionality to H19 to control stem cell populations and that disruption of the H19/miR-675 axis may alter the stem cell phenotype.
H19 also functions as a competing endogenous RNA for miR-138, miR-200a and miR-141, which are involved in the regulation of CSCs [75,76,77]. Notably, H19 interferes with miR-138 and miR-200a, thereby avoiding the repression of Vimentin, Zeb1, and Zeb2 and, concomitantly, inducing EMT. Up-regulation of H19 has been found to result in the modulation of multiple genes involved in EMT, which may promote stemness features [78].
LncATB
LncRNA-ATB has been recognized as an onco-lncRNA commonly overexpressed in human neoplasms such as colon, gastric, stomach and liver cancer. This lncRNA is a non-polyadenylated RNA mainly located in the cytoplasm. Recent studies have shown that lncATB is associated with EMT. LncATB modulates TFG-β which, in turn, regulates several master regulators of EMT such as Snail, Slug, Zeb1 and Zeb2 [79]. Yuan et al. [80] have found that lncATB harbors binding sites for miR-200, which prevents EMT through targeting of the Zeb1 and Zeb2 mRNAs. Several studies have reported that lncATB may induce mesenchymal features and promote cell invasion and metastasis. LncATB binds and stabilizes IL-11, leading to activation of STAT3 signaling and the promotion of cell invasion and metastasis (Fig. 3c) [80, 81]. LncATB has been found to be upregulated in HCC metastases and to be correlated with vascular invasion and a poor patient survival. Recent studies using orthotopic xenografts have demonstrated that lncATB may promote HCC cell intravasation and organ colonization [81]. LncATB may affect metastasis by independent transcription-related mechanisms, first by sponging miR-200 and second by stabilizing IL-11 mRNA (Fig. 3a, b). Both mechanisms lead to the induction of EMT [82, 83]. LncATB has also been found to be involved in trastuzumab resistance in patients with breast cancer [84].
Linc-DYNC2H1–4
Linc-DYNC2H1–4 is an intergenic lncRNA, transcribed from the same sense strand as its nearby gene MMP3, which encodes an important protein for the development of pancreatic cancer [85]. Yuran Gao and colleagues [86] established a gemcitabine-resistant pancreatic ductal adenocarcinoma cell line (BxPC-3-Gem) and found that linc-DYNC2H1–4 was upregulated in these cells. Using in vitro and in vivo assays, it was shown that the resistance to gemcitabine resulted in an enrichment in the CSC fraction. The association of this lincRNA with drug resistance and the CSC phenotype has recently been addressed. It was found that linc-DYNC2H1–4 increases the expression of stemness markers such as Lin28, Nanog, Sox2 and Oct4, as well as that of Zeb1, an EMT regulator. The involvement of this lncRNA in regulating the CSC phenotype was further substantiated by in vitro experiments showing that knockdown of linc-DYNC2H1–4 reduced the colony and spheroid forming abilities, as also the invasive behavior of the gemcitabine-resistant cells. In addition, it was found that exogenous expression of this lincRNA promoted the acquisition of EMT and stemness features in the parental gemcitabine sensitive cells.
Linc-DYNC2H1–4 is mainly located in the cytoplasm, where it acts as a sponge for miR-145, thereby upregulating the expression of its targets Oct4, Lin28, Nanog, Sox2, MMP3 and Zeb1, resulting in EMT progression and CSC enrichment in pancreatic cancer cell populations (Fig. 3f) [86, 87].
HOTTIP
LncRNA HOTTIP is transcribed from the 5′ tip of the HOXA locus and acts in cis to regulate the expression of several HOXA genes. HOTTIP binds the adaptor protein WDR5 and targets the WDR5/MLL complex to the HOXA locus, resulting in trimethylation of histone H3 lysine 4. HOXA members play essential roles in the pluripotency, differentiation and self-renewal of stem cells.
HOTTIP is overexpressed in pancreatic ductal adenocarcinoma (PDAC) and promotes its progression, invasion and drug resistance [88]. This lncRNA also promotes EMT and regulates pancreatic CSCs [88]. Zhiqiang Fu et al. [89] found that HOTTIP is highly expressed in the nucleus of pancreatic CSCs and enhances CSC properties through the Wnt/β-catenin pathway. The role of HOTTIP in the regulation of CSCs is based on the induction of HOXA9 and the subsequent activation of the Wnt pathway. The HOTTIP/HOXA9/WNT axis contributes to stemness by controlling CSC maintenance and self-renewal. HOTTIP and HOX9 may serve as potential therapeutic targets and molecular biomarkers for PDAC, since their expression may predict survival and prognosis [88, 90].
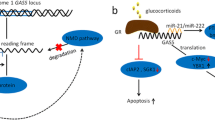
LncRNA-Hh
LncRNA-Hh was first identified in breast CSCs expressing high levels of Twist. This lncRNA is transcriptionally regulated by Twist to enrich the CSC population. LncRNA-Hh directly targets GAS1 to stimulate the Sonic Hedgehog-Patched-Gli pathway and to upregulate the expression of SOX2 and OCT4. Notably, Hedgehog signaling is essential for CSC self-renewal and cell fate determination [91]. LncRNA-Hh overexpression leads to EMT and CSC self-renewal, and promotes tumorigenic abilities, while its silencing reverses these effects [92].
LncARSR
Activated in renal cell carcinoma with sunitinib resistance (ARSR) is a recently identified lncRNA that enhances sunitinib and doxorubicin resistance in renal [93] and hepatocellular [94] carcinomas, respectively. Strikingly, this lncRNA is packaged into exosomes and transmitted to sensitive cells, thereby inducing drug resistance [93]. This lncRNA promotes drug resistance via direct binding to miR-34/miR-449, thereby leading to AXL/c-Met expression and reactivation of STAT3, AKT and ERK signaling [93]. It is worth mentioning that this lncRNA is highly expressed in renal CSCs and is essential for maintenance of their stem cell phenotype. Loss of function analysis of lncARSR has shown that this lncRNA is essential for promoting the self-renewal, tumorigenic and metastatic capacities of renal CSCs. A high lncARSR level has been found to serve as an independent predictor for a poor prognosis of clear cell renal cell carcinoma patients. Mechanistically, this lncRNA binds to Yes-associated protein (YAP) and facilitates its nuclear translocation by blocking the interaction of YAP with the large tumor suppressor kinase-1 (LATS1) [95]. YAP is highly expressed in CSC nuclei where it acts as a transcription co-activator in Hippo signaling, which has been reported to play a critical role in CSC expansion. LncARSR not only plays a role in CSC regulation by inducing Hippo signaling, but also acts on cancer cells with defective Hippo signaling, possibly through other mechanisms [95].
It is well-known that CSC populations do not only emerge from pre-existing CSCs, but also from reprogrammed differentiated cells, possibly through exosomal transfer of lncRNAs. We believe that lncRNA transfer to neighboring cells may play an important role in cancer cell behavior. We consider that the use of extracellular vesicles as delivery vehicles of lncRNAs may serve as an attractive approach to eliminate CSC populations in tumors.
lincROR
The lincRNA regulator of reprogramming (lincROR) was first identified in induced pluripotent stem cells (iPSCs). This lincROR acts as an oncogene and plays essential roles in the maintenance of embryonic stem cells (ESCs) and the reprogramming of differentiated cells into iPSCs [96]. Accumulating evidence indicates that lincROR is associated with EMT and tumorigenesis in many malignancies including breast, liver, lung, pancreatic and colon cancer [97]. LincROR appears to play a role in induction of the EMT program, promoting stem cell-like characteristics, drug resistance and metastasis in ovarian, lung and breast cancer [98,99,100]. Recent studies have revealed that this lncRNA is located in the cytoplasm and acts as a sponge of miR-145 [99, 101], miR-205, miR-34a and let-7 [98]. MiR-205 targets the Zeb1, Zeb2, ErbB3 and VEGF mRNAs and, by doing so, negatively regulates stem cell features [98]. MiR-34a and let-7 also play a role in suppressing breast CSC features [98, 102]. Collectively, these studies indicate that lincROR may be an important regulator of tumor suppressor miRNAs that control stem cell characteristics. LincROR is highly expressed in gastric [103], pancreatic [102] and lung CSCs [101], and its expression has been found to lead to upregulation of several stemness transcription factors such as OCT4, SOX2, NANOG and CD133 [101, 103]. LincROR has been found to acts as a miR-145 sponge, resulting in increased expression of the miR-145 targets OCT4, SOX2 and NANOG, allowing the acquisition of lung and endometrial CSC properties [101, 104]. Upregulation of this lincRNA inhibits the differentiation of endometrial CSCs [104]. Accumulating evidence also indicates that lincROR promotes the proliferation and invasion of CSCs [101], inhibits the apoptosis of CSCs [103] and contributes to the acquisition of stem cell properties by acting as a microRNA sponge to regulate gene transcription [101, 102] .
MALAT-1
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is a highly conserved lncRNA. MALAT-1 has been found to be overexpressed in several human neoplasms and to promote tumor cell invasion and metastasis [105] . Recent studies have shown that MALAT-1 is overexpressed in CSCs derived from pancreas and breast tumors [106, 107]. Through in vitro and in vivo analyses, it has been shown that MALAT-1 enhances CSC phenotypes and regulates their proliferation, colony formation and migration, as well as their self-renewal capacity [106,107,108,109]. This stemness regulating role of MALAT-1 is mainly based on the triggering of EMT through Snail, Slug, E-cadherin, N-cadherin and Vimentin regulation [110]. It has also been shown that downregulation of MALAT-1 may reduce the expression of stem cell markers such as Bmi1, Nanog, SOX2 and Nestin in gliomas and pancreatic cancers [106, 108, 109]. MALAT-1 harbors sites complementary to miR-200c and miR-145 and may, therefore, act as an endogenous sponge for these miRNAs resulting in upregulation of the expression of SOX2 [106, 107].
A more recent study has shown that MALAT-1 can cooperate with lncRNA HULC to increase the expression, phosphorylation and sumoylation of telomere repeat-binding factor 2 (TRF2) and to accelerate liver CSC proliferation, resulting in tumor progression (Fig. 4). Additionally, it has been found that TRF2 depletion may abrogate the oncogenic functions of MALAT-1 and HULC. MALAT-1 combined with HULC may also enhance telomerase activity and promote interactions between TERT and TERC, thereby prolonging the telomere length and, thus, lifespan of liver CSCs (Fig. 4) [109] .
Role of lncRNAs in telomere elongation. Role of different lncRNAs associated with telomeric DNA and proteins that promote telomer elongation. CUDR, MALAT and HULC promote the transcription of TRF2 through different mechanisms. MALAT and HULC may form a complex with TRF2 to demethylate the promoters of TERC and TERT, thereby provoking CST and AAF proteins to be displaced from the telomeres and favoring telomeric elongation. CUDR may also promote the transcription of lncRNA H19 to promote the expression of TERC and TERT
CUDR
Cancer up-regulated drug resistance (CUDR) is a lncRNA that plays a role in cancer progression by affecting cell cycle progression and proliferation. CUDR has been found to be involved in drug resistance of several types of cancer through the induction of WNT expression, which turns CUDR into an attractive target to overcome such resistance [111,112,113]. Since CUDR is an oncofetal gene, its upregulation may be relevant for cancer development. Recent evidence has indicated that CUDR expression may lead to disruption of stem cell populations and induce malignant transformation of normal stem cells. Zheng and colleagues [114] have shown that excessive CUDR cooperates with IL6 to triggering the malignant transformation of human embryonic stem cell-derived hepatocyte-like stem cells through the NF-κB/STAT3 pathway. CUDR and IL6 induce the expression of SUV39h1, a histone methyltransferase that induces tri-methylation of histone H3 at lysine 9 (H3K9me3) to promote the expression and phosphorylation of NF-κB and, subsequently, STAT3 phosphorylation. STAT3 can, in turn, bind to the promoter regions of several miRNAs and lncRNAs such as miR-21, miR-155, miR-17, CUDR, HOTAIR, MALAT-1 and HULC. Abnormal expression of these ncRNAs results in increased telomere length and increased microsatellite instability. Gui et al. [115] have also found that excessive CUDR may trigger the malignant transformation of hepatocyte stem cells. CUDR induces HULC expression by inhibiting its promoter methylation and induces β-catenin expression by promoting promoter-enhancer chromatin loop formation. Both HULC and β-catenin are crucial for the oncogenic activity of CUDR.
Li et al. [116] found that SET1A, a histone methyltransferase complex component, cooperates with CUDR to trigger the malignant transformation of hepatocyte stem cells though TRF2. CUDR enhances the phosphorylation of RB and the interplay between SET1A and pRB, thereby producing an activated pRB-SET1A complex (Fig. 4). This complex induces tri-methylation of histone H3 (H3K4me3) and loads onto the TRF2 promoter region provoking TRF2 overexpression (Fig. 4). TRF2 is a component of the shelterin complex that protects the ends of chromosomes. Excessive TRF2 binds to telomeric repeats, thereby prolonging their lengths and accelerating the malignant transformation of hepatocyte stem cells and their subsequent cancerous growth [116].
It has been reported that CUDR expression also plays an important role in the occurrence of hepatocellular carcinomas and is associated with their TNM stage and metastatic potential, as well as with postoperative patient survival [117]. Recent evidence shows that CUDR can also act on liver CSCs and promote CSC growth through TERT and c-Myc upregulation. CUDR binds to Cyclin D1, forming a complex that loads onto the H19 lncRNA promoter region to reduce its methylation and to provoke its overexpression (Fig. 4). Excessive H19 increases the binding of TERT to TERC and reduces the interplay between TERT and TERRA, thus enhancing telomerase activity and telomere length (Fig. 4c). CUDR may also mediate c-Myc overexpression through the CUDR-CyclinD1-CTCF complex (Fig. 4). Both TERT and c-Myc expression can lead to liver CSC proliferation [118].
Taken together, these findings indicate that CUDR can interact with other lncRNAs (H19, HULC and MALAT-1) and with proteins (Cyclin D1, TRF2 and SETA1) to enhance telomere elongation, disrupt genomic stability and induce malignant transformation and proliferation of (hepatic) stem cells (Fig. 4).
Lnc-DILC
LncRNA downregulated in liver cancer (Lnc-DILC) was recently identified through high-throughput screening in liver tissues. Lnc-DILC was found to be downregulated in liver cancer stem cells (LCSC) and to be restored during their differentiation. The lnc-DILC expression level was found to be related to the proportion of CSCs, since the number of LCSC increased after lnc-DILC silencing and decreased after lnc-DILC overexpression. Loss and gain function analyses revealed that lnc-DILC can modulate the clonogenic and tumorigenic abilities of LCSCs. Lack of expression of this lncRNA facilitates the expansion of LCSCs and favors the progression of hepatocellular cancer. Conversely, it has been found that lnc-DILC expression exhibits a suppressive role in LCSC expansion. Lnc-DILC binds to the IL6 promoter and inhibits its transcription, thereby blocking the IL-6/STAT3 signaling cascade. These findings indicate that lnc-DILC may mediate crosstalk between TNF/NF-κB signaling and autocrine IL6/STAT3 signaling to regulate LCSC expansion. Additional clinical data revealed that a low lnc-DILC expression predicts early recurrence and a short survival of hepatocellular carcinoma patients, thereby highlighting its clinical significance [119].
6 Therapeutic implications of CSC-associated lncRNAs
CSCs are key players in tumor initiation, maintenance, progression, metastasis and recurrence [120]. In addition, CSCs have intrinsic mechanisms allowing them to resist conventional drug treatments [121, 122]. Since CSCs are molecularly and functionally distinct from the bulk of tumor cells, therapeutic alternatives may be developed to eliminate CSCs that otherwise may cause disease progression and/or recurrence. The putative advantage of such selective therapies is that they may have fewer side effects and exhibit less toxicity to non-cancer stem cells. This approach requires a clear understanding of the molecular mechanisms that directly regulate stem cell features. It has amply been shown that lncRNAs exhibit important capacities for inducing the self-renewal, migration, invasion, drug resistance and differentiation of CSCs [123,124,125,126]. LncRNAs are attractive treatment targets since they commonly exhibit restricted tissue-specific expression patterns. Their levels are frequently indicative for the severity of the disease [127]. LncRNAs are also ideal candidates for cancer screening, since they are readily detectable in body fluids such as blood, plasma, saliva and urine [16, 128, 129]. These features render lncRNAs into ideal targets to noninvasively detect or predict cancer behavior before, during and after therapy. An additional advantage of using lncRNAs as therapeutic targets is the feasibility to induce their degradation, modulate their transcription and/or block their interaction with other regulatory factors. Although a wide range of lncRNAs has been found to be deregulated in CSCs, its consequences are known for only a few of them. Accumulating data now provide insight into the functional implications of lncRNAs in controlling cell division, determining cell fate, conferring differential drug-resistance, protecting telomere ends, maintaining genomic architecture, interacting with key signaling pathways and regulating the transcription or translation of stem cell-related genes [58, 109, 123, 127, 130,131,132].
Several lines of evidence suggest that CSCs of advanced stage tumors utilize a symmetric division strategy to give rise daughter cells able to self-renew and, thus, to rapidly increase the pool of CSCs within a tumor [133] . This finding suggests that controlling or avoiding symmetrical CSC divisions in advanced tumors could be clinically beneficial. Since lncRNAs may play unique roles in regulating the balance between asymmetry and symmetry in CSCs [58, 134], altering their function may perturb the division machinery that establishes the unequal partitioning of cell fate-determining factors between daughter cells. As such, lncRNAs that control the manner in which CSCs divide could be considered as putative targets for the treatment of advanced cancers. It has been shown, for instance, that asymmetric distribution of lnc34a during cell division leads to asymmetric daughter cell fate, but that high lnc34a levels lead to CSC expansion via symmetric self-renewal [58]. Interestingly, this lncRNA is commonly upregulated in late-stage colorectal cancers, so its suppression could lead to asymmetric cell division and differentiation. It has been reported that lnc34a directly targets miR-34a and balances the cell division mode by a differential Notch1 distribution mediated by this miRNA [58]. Recently, a miR-34a mimic (MRX34) has been proposed to restore miR-34a function and, thus, to prevent CSC proliferation and expansion. Currently, phase II clinical trials are being conducted to test the efficacy of MRX34 in advanced solid tumors [135], although the delivery efficiency into the tumors still remains a challenge. Also lnc34a could be considered as a target for therapeutic intervention of advanced tumors since it is feasible to develop small molecule inhibitors that block its function or disrupt its structure.
It is well documented that lncRNAs may also participate in the acquisition and maintenance of drug-resistance in CSCs. These lncRNAs may activate several mechanisms to promote this drug resistance, including modulation of drug transporter expression levels, regulation of survival signaling pathways, avoidance of apoptosis and induction of DNA repair. LncPVT1 and H19 are known to promote drug resistance in some types of cancer by modulating the expression of drug transporters such as MDR1 and MRP1 [130, 136, 137], whereas HOTAIR is known to modulate the DNA damage response pathway through suppression of p21 and p53 [46] . LncRNA CUDR has been found to increase drug-mediated resistance by regulating the Wnt signaling pathway [111] or by inducing Bcl2 expression mediated by miR-204-5p inhibition [138]. LncATB and lncROR have been found to reduce chemotherapy-induced cell death by modulating the expression levels of TGFβ [80, 139]. Since these lncRNAs play crucial roles in drug resistance, they might be used as therapeutic targets to overcome this resistance. Recent work has shown that lncRNAs that induce drug resistance can be disseminated to sensitive cells by vesicles (exosomes) that transmit regulatory lncRNAs. Once inside these cells, they have the ability to induce drug resistance and stem cell phenotypes [130]. LncARSR can, for example, be encapsulated by exosomes that are released into the extracellular environment and transferred to neighboring cells, thereby conferring drug resistance to sensitive cells [93]. LncROR is another lncRNA involved in the modulation of cellular responses to chemotherapy that can be transferred to other cells via extracellular vesicles [139]. The targeting lncRNAs transmitted by vesicles may be a useful approach to improve the responses to conventional therapeutic agents used for the treatment of cancer.
LncRNAs modulating other stem cell features may also be considered for therapeutic intervention. HOTAIR and MALAT-1 are, for example, well-known lncRNAs playing a role in maintaining stem cell features through the modulation of pluripotency stem cell factors [46, 47, 106]. Interestingly, these lncRNAs have been found in the plasma of cancer patients and are considered useful tools for primary cancer and metastasis detection [140]. Mohamed-Moustafa et al. [141] set out to find clinically relevant cancer-associated lncRNA by characterizing temporally expressed lncRNAs during S-phase. They found that most of these lncRNAs appeared to be strongly associated with cancer development. In addition, they found that a large proportion of these lncRNAs may acts as independent prognostic indicators in some types of cancer. Subsequent modulation of these lncRNAs resulted in alterations in cell cycle progression, proliferation, apoptosis, migration and senescence [141]. Hua-Sheng et al. [142] inferred lncRNAs with relevant functions in cancer by predicting their targets using molecular profiles of TCGA primary tumors. They linked lncRNAs with the deregulation of cancer genes and pathways known to influence tumor biology and found that a large number of these lncRNAs regulates hundreds of genes, many of which modulate cancer pathways across multiple tumor types [142]. These latter findings indicate that large-scale screening may result in the identification of potential oncogenic drivers and, thus, potential therapeutic targets.
7 Conclusions and perspectives
The large orchestra of molecular and cellular processes in which lncRNAs participate leads to the activation and/or disruption of a plethora of normal and abnormal biological processes such as cancer. CSCs have been extensively studied since they can drive tumor initiation and progression. CSCs are of particular significance due to their metastatic capacity and their ability to escape from common therapeutic agents, which renders them into the primary cause of tumor relapse. Despite their essential role in tumor development, the regulation of this subpopulation of cells is not completely deciphered yet and, therfore, more in-depth studies are required. Here, we have provided an overview of lncRNAs involved in the self-renewal, maintenance and differentiation of CSCs, as well as some of the underlying molecular mechanisms. LncRNAs can exert their functions at different levels and in different ways. In the nucleus they can act as scaffold, guide or capture, as exemplified by SOX4, HOTAIR and HOTTIP. In the cytoplasm they can function as endogenous competitors or sponges of miRNAs, as exemplified by ATB or DYNC2H1. Some lncRNAs, such as CTCF7 and H19, may act in both cellular compartments in order to maintain the self-renewal of the CSCs. There are also many lncRNAs, such as MALAT-1, H19 and CUDR, that work together to prolong telomeres and, by doing so, expand CSC subpopulations. As yet, only a few lncRNAs have been identified to be involved in CSC stemness, warranting further studies on the mechanisms involved in this feature.
LncRNAs are highly abundant within the genomes of our cells where they regulate multiple processes at the transcriptional, translational and post-translational level. We consider that it is essential to understand the molecular mechanisms whereby lncRNAs exert their functions and to uncover the roles that lncRNAs play in specific tissues. Such information will provide a basis for considering lncRNAs as diagnostic or prognostic markers, or even as targets for future therapies. Further studies are needed to establish which lncRNAs may serve as the best candidates for future therapeutic strategies through hampering the self-renewal and proliferation capacities of CSCs. Since CSCs exhibit intrinsic mechanisms allowing them to overcome conventional therapeutic treatment regimens, specific methods are needed to ensure the eradication of CSCs and, thus, to avoid tumor recurrence. Extensive efforts have yielded ample information on the functional implications of specific lncRNAs in conferring drug resistance, controlling cell division, determining cell-fate and regulating the transcription or translation of stem cell-related genes. Blockage of these lncRNAs may be used to treat specific types of cancer. The fact that lncRNAs can readily be detected in serum, saliva, urine, blood or tissue biopsies, renders them highly attractive for clinical (diagnostic/prognostic) purposes. In addition, different approaches to target lncRNAs for therapeutic purposes can be considered, such as the use of siRNAs or locked nucleic acid molecules to induce lncRNA degradation, and CRISPR/Cas9 mediated gene editing or exosome delivery to modulate their expression. Currently, the major challenge of these approaches is to specifically deliver the respective molecules into their pre-selected tissues/cells. Without any doubt, lncRNAs are emerging as valuable tools for future clinical applications. In fact, some of them are already being tested in clinical trials to determine their putative therapeutic efficacy.
References
S. Djebali, C.A. Davis, A. Merkel, A. Dobin, T. Lassmann, A. Mortazavi, A. Tanzer, J. Lagarde, W. Lin, F. Schlesinger, C. Xue, G.K. Marinov, J. Khatun, B.A. Williams, C. Zaleski, J. Rozowsky, M. Roder, F. Kokocinski, R.F. Abdelhamid, T. Alioto, I. Antoshechkin, M.T. Baer, N.S. Bar, P. Batut, K. Bell, I. Bell, S. Chakrabortty, X. Chen, J. Chrast, J. Curado, T. Derrien, J. Drenkow, E. Dumais, J. Dumais, R. Duttagupta, E. Falconnet, M. Fastuca, K. Fejes-Toth, P. Ferreira, S. Foissac, M.J. Fullwood, H. Gao, D. Gonzalez, A. Gordon, H. Gunawardena, C. Howald, S. Jha, R. Johnson, P. Kapranov, B. King, C. Kingswood, O.J. Luo, E. Park, K. Persaud, J.B. Preall, P. Ribeca, B. Risk, D. Robyr, M. Sammeth, L. Schaffer, L.H. See, A. Shahab, J. Skancke, A.M. Suzuki, H. Takahashi, H. Tilgner, D. Trout, N. Walters, H. Wang, J. Wrobel, Y. Yu, X. Ruan, Y. Hayashizaki, J. Harrow, M. Gerstein, T. Hubbard, A. Reymond, S.E. Antonarakis, G. Hannon, M.C. Giddings, Y. Ruan, B. Wold, P. Carninci, R. Guigo, T.R. Gingeras, Landscape of transcription in human cells. Nature 489, 101–108 (2012)
V. Taucher, H. Mangge, J. Haybaeck, Non-coding RNAs in pancreatic cancer: Challenges and opportunities for clinical application. Cell Oncol 39, 295–318 (2016)
S. Jalali, S. Gandhi, V. Scaria, Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Hum Genomics 10, 35 (2016)
J.L. Rinn, H.Y. Chang, Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166 (2012)
G. Bohmdorfer, S. Sethuraman, M.J. Rowley, M. Krzyszton, M.H. Rothi, L. Bouzit, A.T. Wierzbicki, Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. elife 5, e19092 (2016)
A. Barski, I. Chepelev, D. Liko, S. Cuddapah, A.B. Fleming, J. Birch, K. Cui, R.J. White, K. Zhao, Pol II and its associated epigenetic marks are present at pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 17, 629–634 (2010)
K.W. Vance, C.P. Ponting, Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet 30, 348–355 (2014)
L.L. Chen, Linking long noncoding RNA localization and function. Trends Biochem Sci 41, 761–772 (2016)
J. Cao, The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 16, 11 (2014)
T.R. Mercer, J.S. Mattick, Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20, 300–307 (2013)
M. Guttman, J.L. Rinn, Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012)
X. Wang, S. Arai, X. Song, D. Reichart, K. Du, G. Pascual, P. Tempst, M.G. Rosenfeld, C.K. Glass, R. Kurokawa, Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 (2008)
K.C. Wang, H.Y. Chang, Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914 (2011)
P.O. Angrand, C. Vennin, X. Le Bourhis, E. Adriaenssens, The role of long non-coding RNAs in genome formatting and expression. Front Genet 6, 165 (2015)
A. Ferraro, Altered primary chromatin structures and their implications in cancer development. Cell Oncol 39, 195–210 (2016)
J.R. Prensner, A.M. Chinnaiyan, The emergence of lncRNAs in cancer biology. Cancer Discov 1, 391–407 (2011)
R.B. Perry, I. Ulitsky, The functions of long noncoding RNAs in development and stem cells. Development 143, 3882–3894 (2016)
G. Eades, Y.S. Zhang, Q.L. Li, J.X. Xia, Y. Yao, Q. Zhou, Long non-coding RNAs in stem cells and cancer. World J Clin Oncol 5, 134–141 (2014)
D. Hanahan, R.A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011)
S.L. Floor, J.E. Dumont, C. Maenhaut, E. Raspe, Hallmarks of cancer: Of all cancer cells, all the time? Trends Mol Med 18, 509–515 (2012)
D. Hanahan, L.M. Coussens, Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012)
A. Cicalese, G. Bonizzi, C.E. Pasi, M. Faretta, S. Ronzoni, B. Giulini, C. Brisken, S. Minucci, P.P. Di Fiore, P.G. Pelicci, The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009)
M.F. Clarke, M. Fuller, Stem cells and cancer: Two faces of eve. Cell 124, 1111–1115 (2006)
Y. Welte, J. Adjaye, H.R. Lehrach, C.R. Regenbrecht, Cancer stem cells in solid tumors: Elusive or illusive? Cell Commun Signal 8, 6 (2010)
S. Bugide, V.K. Gonugunta, V. Penugurti, V.L. Malisetty, R.K. Vadlamudi, B. Manavathi, HPIP promotes epithelial-mesenchymal transition and cisplatin resistance in ovarian cancer cells through PI3K/AKT pathway activation. Cell Oncol 40, 133–144 (2017)
M.R. Sam, P. Ahangar, V. Nejati, R. Habibian, Treatment of LS174T colorectal cancer stem-like cells with n-3 PUFAs induces growth suppression through inhibition of survivin expression and induction of caspase-3 activation. Cell Oncol 39, 69–77 (2016)
M. Munz, P.A. Baeuerle, O. Gires, The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res 69, 5627–5629 (2009)
K. Vazquez-Santillan, J. Melendez-Zajgla, L. Jimenez-Hernandez, G. Martinez-Ruiz, V. Maldonado, NF-kappaB signaling in cancer stem cells: A promising therapeutic target? Cell Oncol 38, 327–339 (2015)
A. Sathyanarayanan, K.S. Chandrasekaran, D. Karunagaran, microRNA-145 modulates epithelial-mesenchymal transition and suppresses proliferation, migration and invasion by targeting SIP1 in human cervical cancer cells. Cell Oncol 40, 119–131 (2017)
Q. Zhang, K. Matsuura, D.E. Kleiner, F. Zamboni, H.J. Alter, P. Farci, Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med 14, 328 (2016)
F. Wang, J.H. Yuan, S.B. Wang, F. Yang, S.X. Yuan, C. Ye, N. Yang, W.P. Zhou, W.L. Li, W. Li, S.H. Sun, Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 60, 1278–1290 (2014)
M.A. Parasramka, T. Patel, Long non-coding RNA regulation of liver cancer stem cell self-renewal offers new therapeutic targeting opportunities. Stem Cell Investig 3, 1 (2016)
H. Li, L. Zhu, L. Xu, K. Qin, C. Liu, Y. Yu, D. Su, K. Wu, Y. Sheng, Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol Carcinog 56, 3–17 (2017)
J. Yao, J. Li, P. Geng, Y. Li, H. Chen, Y. Zhu, Knockdown of a HIF-2alpha promoter upstream long noncoding RNA impairs colorectal cancer stem cell properties in vitro through HIF-2alpha downregulation. Onco Targets Ther 8, 3467–3474 (2015)
W. Li, X. He, R. Xue, Y. Zhang, X. Zhang, J. Lu, Z. Zhang, L. Xue, Combined over-expression of the hypoxia-inducible factor 2alpha gene and its long non-coding RNA predicts unfavorable prognosis of patients with osteosarcoma. Pathol Res Pract 212, 861–866 (2016)
Y. Wang, J. Yao, H. Meng, Z. Yu, Z. Wang, X. Yuan, H. Chen, A. Wang, A novel long non-coding RNA, hypoxia-inducible factor-2alpha promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep 11, 2534–2540 (2015)
A. Shahryari, M.R. Rafiee, Y. Fouani, N.A. Oliae, N.M. Samaei, M. Shafiee, S. Semnani, M. Vasei, S.J. Mowla, Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells 32, 126–134 (2014)
M.E. Askarian-Amiri, V. Seyfoddin, C.E. Smart, J. Wang, J.E. Kim, H. Hansji, B.C. Baguley, G.J. Finlay, E.Y. Leung, Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One 9, e102140 (2014)
Z. Hou, W. Zhao, J. Zhou, L. Shen, P. Zhan, C. Xu, C. Chang, H. Bi, J. Zou, X. Yao, R. Huang, L. Yu, J. Yan, A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol 53, 380–388 (2014)
P.P. Amaral, C. Neyt, S.J. Wilkins, M.E. Askarian-Amiri, S.M. Sunkin, A.C. Perkins, J.S. Mattick, Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15, 2013–2027 (2009)
M. Saghaeian Jazi, N.M. Samaei, M. Ghanei, M.B. Shadmehr, S.J. Mowla, Identification of new SOX2OT transcript variants highly expressed in human cancer cell lines and down regulated in stem cell differentiation. Mol Biol Rep 43, 65–72 (2016)
S.S. Saha, R. Roy Chowdhury, N.R. Mondal, B. Chakravarty, T. Chatterjee, S. Roy, S. Sengupta., Identification of genetic variation in the lncRNA HOTAIR associated with HPV16-related cervical cancer pathogenesis. Cell Oncol 39, 559–572 (2016)
S. Sharma Saha, R. Roy Chowdhury, N.R. Mondal, B. Chakravarty, T. Chatterjee, S. Roy, S. Sengupta, Identification of genetic variation in the lncRNA HOTAIR associated with HPV16-related cervical cancer pathogenesis. Cell Oncol 39, 559–572 (2016)
Y.N. Jun Dou, X. He, M.L. Di Wu, S. Wu, R. Zhang, M. Guo, Fengsu, l. Zhao, Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res 8, 98–108 (2016)
C. Padua Alves, A.S. Fonseca, B.R. Muys, E.L.B.R. de Barros, M.C. Burger, J.E. de Souza, V. Valente, M.A. Zago, W.A. Silva Jr., Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 31, 2827–2832 (2013)
J. Deng, M. Yang, R. Jiang, N. An, X. Wang, B. Liu, Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the Cancer stem-like cell (CSC) subpopulation enriched from breast Cancer cells. PLoS One 12, e0170860 (2017)
J.A. Haiyan Li, M. Wu, Q. Zheng, X. Gui, T. Li, P. Hu, D. Lu, LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 6, 27847–27864 (2015)
K. Fang, P. Liu, S. Dong, Y. Guo, X. Cui, X. Zhu, X. Li, L. Jiang, T. Liu, Y. Wu, Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int J Oncol 49, 509–518 (2016)
S.N. Min, T. Wei, X.T. Wang, L.L. Wu, G.Y. Yu, Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: A meta-analysis. Medicine 96, e7084 (2017)
Y.-W.L. Ming-Yi Lu, P.-Y. Chen, P.-L. Hsieh, C.-Y. Fang, C.-Y. Wu, M.-L. Yen, B.-Y. Peng, D.P. Wang, H.-C. Cheng, C.-Z. Wu, Y.-H. Shih, D.-J. Wang, C.-c. Yu, L.-L. Tsai, Targeting LncRNA HOTAIR suppresses cancer stemness and metastasis in oral carcinomas stem cells through modulation of EMT. Oncotarget 8, 98542–98552 (2017)
P.D. Marco Galasso, M. Previati, S. Sandhu, J. Palatini, V. Coppola, S. Warner, M.E. Sana, R. Zanella, R. Abujarour, C. Desponts, M.A. Teitell, R. Garzon, G. Calin, C.M. Croce, S. Volinia, A large scale expression study associates uc.283-plus lncRNA with pluripotent stem cells and human glioma. Genome Medicine 6, 76 (2014)
G.S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C. Polytarchou, K.B. Marcu, A.G. Papavassiliou, R. Sandaltzopoulos, E. Kolettas, A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol 40, 303–339 (2017)
Y. Li, T. Mine, C.G. Ioannides, Short GC-rich RNA similar to miR 1909 and 1915 folds in silico with the 5'-UTR and ORF of notch and responders: Potential for the elimination of cancer stem cells. Oncol Rep 24, 1443–1453 (2010)
A. Lujambio, A. Portela, J. Liz, S.A. Melo, S. Rossi, R. Spizzo, C.M. Croce, G.A. Calin, M. Esteller, CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 29, 6390–6401 (2010)
S.-Y.L. Wei-Yu Chen, Y.-S. Chang, J.J. Yin, Hsiu-lien, T.H. Yeh, O.H. Mouhieddine, W. Abou-Kheir, Y.-N. Liu, MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras activated prostate cancer. Oncotarget 6, 441–457 (2014)
P. Bu, K.Y. Chen, J.H. Chen, L. Wang, J. Walters, Y.J. Shin, J.P. Goerger, J. Sun, M. Witherspoon, N. Rakhilin, J. Li, H. Yang, J. Milsom, S. Lee, W. Zipfel, M.M. Jin, Z.H. Gumus, S.M. Lipkin, X. Shen, A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 12, 602–615 (2013)
U.S.B. Sumithra, A.B. Das, Alternative splicing within the Wnt signaling pathway: Role in cancer development. Cell Oncol 39, 1–13 (2016)
L. Wang, P. Bu, Y. Ai, T. Srinivasan, H.J. Chen, K. Xiang, S.M. Lipkin, X. Shen, A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. elife 5, e14620 (2016)
J. Wu, J. Zhang, B. Shen, K. Yin, J. Xu, W. Gao, L. Zhang, Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res 34, 116 (2015)
S. Wan, E. Zhao, I. Kryczek, L. Vatan, A. Sadovskaya, G. Ludema, D.M. Simeone, W. Zou, T.H. Welling, Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology 147, 1393–1404 (2014)
R. Bharti, G. Dey, M. Mandal, Cancer development, chemoresistance, epithelial to mesenchymal transition and stem cells: A snapshot of IL-6 mediated involvement. Cancer Lett 375, 51–61 (2016)
Y. Wang, L. He, Y. Du, P. Zhu, G. Huang, J. Luo, X. Yan, B. Ye, C. Li, P. Xia, G. Zhang, Y. Tian, R. Chen, Z. Fan, The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413–425 (2015)
J. Wu, D. Wang, Long noncoding RNA TCF7 promotes invasiveness and self-renewal of human non-small cell lung cancer cells. Hum Cell 30, 23–29 (2017)
P. Massoner, T. Thomm, B. Mack, G. Untergasser, A. Martowicz, K. Bobowski, H. Klocker, O. Gires, M. Puhr, EpCAM is overexpressed in local and metastatic prostate cancer, suppressed by chemotherapy and modulated by MET-associated miRNA-200c/205. Br J Cancer 111, 955–964 (2014)
C.C. Poirier F, P.M. Timmons, E.J. Robertson, M.J. Evans, P.W. Rigby, The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 113, 1105–1114 (1991)
F. Peng, T.T. Li, K.L. Wang, G.Q. Xiao, J.H. Wang, H.D. Zhao, Z.J. Kang, W.J. Fan, L.L. Zhu, M. Li, B. Cui, F.M. Zheng, H.J. Wang, E.W. Lam, B. Wang, J. Xu, Q. Liu, H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 8, e2569 (2017)
H. Bauderlique-Le Roy, C. Vennin, G. Brocqueville, N. Spruyt, E. Adriaenssens, R.P. Bourette, Enrichment of human stem-like prostate cells with s-SHIP promoter activity uncovers a role in Stemness for the long noncoding RNA H19. Stem Cells Dev 24, 1252–1262 (2015)
X. Jiang, Y. Yan, M. Hu, X. Chen, Y. Wang, Y. Dai, D. Wu, Y. Wang, Z. Zhuang, H. Xia, Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg 124, 129–136 (2016)
S.R. Viswanathan, G.Q. Daley, Lin28: A microRNA regulator with a macro role. Cell 140, 445–449 (2010)
X. Cai, B.R. Cullen, The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13, 313–316 (2007)
Y. Zheng, X. Lu, L. Xu, Z. Chen, Q. Li, J. Yuan, MicroRNA-675 promotes glioma cell proliferation and motility by negatively regulating retinoblastoma 1. Hum Pathol 69, 63–71 (2017)
A. Farzi-Molan, S. Babashah, B. Bakhshinejad, A. Atashi, M. Fakhr Taha, Down-regulation of the non-coding RNA H19 and its derived miR-675 is concomitant with up-regulation of insulin-like growth factor receptor type 1 during neural-like differentiation of human bone marrow mesenchymal stem cells. Cell Biol Int 42, 940–448 (2018)
B.K. Dey, K. Pfeifer, A. Dutta, The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28, 491–501 (2014)
A. Keniry, D. Oxley, P. Monnier, M. Kyba, L. Dandolo, G. Smits, W. Reik, The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14, 659–665 (2012)
N. Liu, L. Zhong, J. Zeng, X. Zhang, Q. Yang, D. Liao, Y. Wang, G. Chen, Y. Wang, Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer. Neoplasma 62, 550–559 (2015)
C. Liu, R. Liu, D. Zhang, Q. Deng, B. Liu, H.P. Chao, K. Rycaj, Y. Takata, K. Lin, Y. Lu, Y. Zhong, J. Krolewski, J. Shen, D.G. Tang, MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun 8, 14270 (2017)
Q. Yang, X. Wang, C. Tang, X. Chen, J. He, H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol 50, 1801–1809 (2017)
W.-M.F. Wei-Cheng Liang, C.-W. Wong, Y. Wang, G.-X.H.W. Wei-Mao, L. Zhang, L.-J. Xiao, D.C.-C. Wan, Jin-Fang, M.M.-Y.W. Zhang, The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525 (2015)
S. Cufi, A. Vazquez-Martin, C. Oliveras-Ferraros, B. Martin-Castillo, J. Joven, J.A. Menendez, Metformin against TGFbeta-induced epithelial-to-mesenchymal transition (EMT): From cancer stem cells to aging-associated fibrosis. Cell Cycle 9, 4461–4468 (2010)
J.H. Yuan, F. Yang, F. Wang, J.Z. Ma, Y.J. Guo, Q.F. Tao, F. Liu, W. Pan, T.T. Wang, C.C. Zhou, S.B. Wang, Y.Z. Wang, Y. Yang, N. Yang, W.P. Zhou, G.S. Yang, S.H. Sun, A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014)
W. Li, Y. Kang, A new Lnc in metastasis: Long noncoding RNA mediates the prometastatic functions of TGF-beta. Cancer Cell 25, 557–559 (2014)
E. Karamitopoulou, Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front Oncol 2, 209 (2012)
U. Wellner, J. Schubert, U.C. Burk, O. Schmalhofer, F. Zhu, A. Sonntag, B. Waldvogel, C. Vannier, D. Darling, A. zur Hausen, V.G. Brunton, J. Morton, O. Sansom, J. Schuler, M.P. Stemmler, C. Herzberger, U. Hopt, T. Keck, S. Brabletz, T. Brabletz, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11, 1487–1495 (2009)
L.-J.W. Sheng-Jia Shi, B. Yu, Y.-H. Li, Y. Jin, X.-Z. Bai, LncRNA-ATB promotes trastuzumab resistance and invasion metastasis cascade in breast cancer. Oncotarget 6, 11652–11663 (2015)
C. Mehner, E. Miller, D. Khauv, A. Nassar, A.L. Oberg, W.R. Bamlet, L. Zhang, J. Waldmann, E.S. Radisky, H.C. Crawford, D.C. Radisky, Tumor cell-derived MMP3 orchestrates Rac1b and tissue alterations that promote pancreatic adenocarcinoma. Mol Cancer Res 12, 1430–1439 (2014)
Y. Gao, Z. Zhang, K. Li, L. Gong, Q. Yang, X. Huang, C. Hong, M. Ding, H. Yang, Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis 8, e2924 (2017)
T. Han, X.P. Yi, B. Liu, M.J. Ke, Y.X. Li, MicroRNA-145 suppresses cell proliferation, invasion and migration in pancreatic cancer cells by targeting NEDD9. Mol Med Rep 11, 4115–4120 (2015)
Z. Li, X. Zhao, Y. Zhou, Y. Liu, Q. Zhou, H. Ye, Y. Wang, J. Zeng, Y. Song, W. Gao, S. Zheng, B. Zhuang, H. Chen, W. Li, H. Li, H. Li, Z. Fu, R. Chen, The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med 13, 84 (2015)
Z. Fu, C. Chen, Q. Zhou, Y. Wang, Y. Zhao, X. Zhao, W. Li, S. Zheng, H. Ye, L. Wang, Z. He, Q. Lin, Z. Li, R. Chen, LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett 410, 68–81 (2017)
L. Quagliata, M.S. Matter, S. Piscuoglio, L. Arabi, C. Ruiz, A. Procino, M. Kovac, F. Moretti, Z. Makowska, T. Boldanova, J.B. Andersen, M. Hammerle, L. Tornillo, M.H. Heim, S. Diederichs, C. Cillo, L.M. Terracciano, Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 59, 911–923 (2014)
C.R. Cochrane, A. Szczepny, D.N. Watkins, J.E. Cain, Hedgehog signaling in the maintenance of Cancer stem cells. Cancers (Basel) 7, 1554–1585 (2015)
M. Zhou, Y. Hou, G. Yang, H. Zhang, G. Tu, Y.E. Du, S. Wen, L. Xu, X. Tang, S. Tang, L. Yang, X. Cui, M. Liu, LncRNA-Hh strengthen Cancer stem cells generation in Twist-positive breast Cancer via activation of hedgehog signaling pathway. Stem Cells 34, 55–66 (2016)
L. Qu, J. Ding, C. Chen, Z. J. Wu, B. Liu, Y. Gao, W. Chen, F. Liu, W. Sun, X.F. Li, X. Wang, Y. Wang, Z.Y. Xu, L. Gao, Q. Yang, B. Xu, Y.M. Li, Z.Y. Fang, Z.P. Xu, Y. Bao, D.S. Wu, X. Miao, H.Y. Sun, Y.H. Sun, H.Y. Wang and L.H. Wang, Exosome-transmitted lncARSR promotes Sunitinib resistance in renal Cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016)
Y. Li, Y. Ye, B. Feng, Y. Qi, Long Noncoding RNA lncARSR promotes doxorubicin resistance in hepatocellular carcinoma via modulating PTEN-PI3K/Akt pathway. J Cell Biochem 118, 4498–4507 (2017)
L. Qu, Z. Wu, Y. Li, Z. Xu, B. Liu, F. Liu, Y. Bao, D. Wu, J. Liu, A. Wang, X. Chu, Y. Sun, C. Chen, Z. Zhang, L. Wang, A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat Commun 7, 12692 (2016)
C.M. Loewer, M. Guttman, Y.H. Loh, K. Thomas, I.H. Park, M. Garber, M. Curran, T. Onder, S. Agarwal, P.D. Manos, S. Datta, E.S. Lander, T.M. Schlaeger, G.Q. Daley, J.L. Rinn, Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42, 1113–1117 (2010)
Y. Pan, C. Li, J. Chen, K. Zhang, X. Chu, R. Wang, L. Chen, The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem 40, 219–229 (2016)
P. Hou, Y. Zhao, Z. Li, R. Yao, M. Ma, Y. Gao, L. Zhao, Y. Zhang, B. Huang, J. Lu, LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5, e1287 (2014)
Y. Pan, et al., Long noncoding RNA ROR regulates chemoresistance in docetaxelresistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget 20, 33144–33158 (2017)
Y. Lou, H. Jiang, Z. Cui, L. Wang, X. Wang, T. Tian, Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/β-catenin signaling. Oncotarget 8, 69983–69994 (2017)
F. Xia, Y. Xiong, Q. Li, Interaction of lincRNA ROR and p53/miR-145 correlates with lung cancer stem cell signatures. J Cell Biochem 1 (2017)
Z. Fu, G. Li, Z. Li, Y. Wang, Y. Zhao, S. Zheng, H. Ye, Y. Luo, X. Zhao, L. Wei, Y. Liu, Q. Lin, Q. Zhou, R. Chen, Endogenous miRNA sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov 3, 17004 (2017)
S. Wang, F. Liu, J. Deng, X. Cai, J. Han, Q. Liu, Long noncoding RNA ROR regulates proliferation, invasion, and Stemness of gastric Cancer stem cell. Cell Reprogram 18, 319–326 (2016)
X. Zhou, Q. Gao, J. Wang, X. Zhang, K. Liu, Z. Duan, Linc-RNA-RoR acts as "sponge" against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol Oncol 133, 333–339 (2014)
S. Chen, S. Nagel, B. Schneider, H. Dai, R. Geffers, M. Kaufmann, C. Meyer, C. Pommerenke, K.S. Thress, J. Li, H. Quentmeier, H.G. Drexler, R.A.F. MacLeod, A new ETV6-NTRK3 cell line model reveals MALAT1 as a novel therapeutic target - a short report. Cell Oncol 41, 93–101 (2018)
F. Jiao, H. Hu, T. Han, C. Yuan, L. Wang, Z. Jin, Z. Guo, L. Wang, Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci 16, 6677–6693 (2015)
L. Zeng, Y. Cen, J. Chen, Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol Lett 15, 2117–2122 (2017)
Y. Han, L. Zhou, T. Wu, Y. Huang, Z. Cheng, X. Li, T. Sun, Y. Zhou, Z. Du, Downregulation of lncRNA-MALAT1 affects proliferation and the expression of Stemness markers in glioma stem cell line SHG139S. Cell Mol Neurobiol 36, 1097–1107 (2016)
M. Wu, Z. Lin, X. Li, X. Xin, J. An, Q. Zheng, Y. Yang, D. Lu, HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci Rep 6, 36045 (2016)
F. Jiao, H. Hu, C. Yuan, L. Wang, W. Jiang, Z. Jin, Z. Guo, L. Wang, Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep 32, 2485–2492 (2014)
Y. Fan, B. Shen, M. Tan, X. Mu, Y. Qin, F. Zhang, Y. Liu, Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 281, 1750–1758 (2014)
M. Jiang, O. Huang, Z. Xie, S. Wu, X. Zhang, A. Shen, H. Liu, X. Chen, J. Wu, Y. Lou, Y. Mao, K. Sun, S. Hu, M. Geng, K. Shen, A novel long non-coding RNA-ARA: Adriamycin resistance-associated. Biochem Pharmacol 87, 254–283 (2014)
W.P. Tsang, T.W. Wong, A.H. Cheung, C.N. Co, T.T. Kwok, Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 13, 890–898 (2007)
Q. Zheng, Z. Lin, X. Li, X. Xin, M. Wu, J. An, X. Gui, T. Li, H. Pu, H. Li, D. Lu, Inflammatory cytokine IL6 cooperates with CUDR to aggravate hepatocyte-like stem cells malignant transformation through NF-kappaB signaling. Sci Rep 6, 36843 (2016)
X. Gui, H. Li, T. Li, H. Pu, D. Lu, Long noncoding RNA CUDR regulates HULC and beta-catenin to govern human liver stem cell malignant differentiation. Mol Ther 23, 1843–1853 (2015)
T. Li, Q. Zheng, J. An, M. Wu, H. Li, X. Gui, H. Pu, D. Lu, SET1A cooperates with CUDR to promote liver Cancer growth and hepatocyte-like stem cell malignant transformation epigenetically. Mol Ther 24, 261–275 (2016)
F. Wang, H.-Q. Ying, B.-S. He, Y.-Q. Pan, Q.-W. Deng, H.-L. Sun, J. Chen, X. Liu, S.-K. Wang, Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 6, 7899–7917 (2015)
Q.Z.H. Pu, H. Li, M. Wu, J. An, X. Gui, T. Li, D. Lu, CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget 6, 40775–40798 (2015)
X. Wang, W. Sun, W. Shen, M. Xia, C. Chen, D. Xiang, B. Ning, X. Cui, H. Li, X. Li, J. Ding, H. Wang, Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol 64, 1283–1294 (2016)
V. Plaks, N. Kong, Z. Werb, The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16, 225–238 (2015)
M. Prieto-Vila, R.U. Takahashi, W. Usuba, I. Kohama, T. Ochiya, Drug resistance driven by Cancer stem cells and their niche. Int J Mol Sci 18, e2574 (2017)
L.T.H. Phi, I.N. Sari, Y.G. Yang, S.H. Lee, N. Jun, K.S. Kim, Y.K. Lee, H.Y. Kwon, Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in Cancer treatment. Stem Cells Int 5416923, 2018 (2018)
S. Lee, H.H. Seo, C.Y. Lee, J. Lee, S. Shin, S.W. Kim, S. Lim, K.C. Hwang, Human long noncoding RNA regulation of stem cell potency and differentiation. Stem Cells Int 6374504, 2017 (2017)
A.M. Schmitt, H.Y. Chang, Long noncoding RNAs in Cancer pathways. Cancer Cell 29, 452–463 (2016)
A.C.P. Ioannis Grammatikakis, K. Abdelmohsen, M. Gorospe, Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging 6, 992–1009 (2014)
Y.T. Liting Yang, F. Xiong, H. Yi, F. Wei, S. Zhang, B.X. Can Guo, M. Zhou, N. Xie, X. Li, Y. Li, G. Li, W.X.Z. Zeng, LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget 9, 1426–1443 (2018)
A. Bhan, M. Soleimani, S.S. Mandal, Long noncoding RNA and Cancer: A new paradigm. Cancer Res 77, 3965–3981 (2017)
A. Sahu, U. Singhal, A.M. Chinnaiyan, Long noncoding RNAs in cancer: From function to translation. Trends Cancer 1, 93–109 (2015)
T. Shi, G. Gao, Y. Cao, Long noncoding RNAs as novel biomarkers have a promising future in Cancer diagnostics. Dis Markers 2016, 9085195 (2016)
e.a. Qin-nan Chen, Long non-coding RNAs in anti-cancer drug resistance. Oncotarget 8, 1925–1936 (2017)
J.R. Evans, F.Y. Feng, A.M. Chinnaiyan, The bright side of dark matter: lncRNAs in cancer. J Clin Invest 126, 2775–2782 (2016)
L. Cheng, H. Ming, M. Zhu, B. Wen, Long noncoding RNAs as organizers of nuclear architecture. Sci China Life Sci 59, 236–244 (2016)
B.M. Boman, M.S. Wicha, J.Z. Fields, O.A. Runquist, Symmetric division of cancer stem cells--a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther 81, 893–898 (2007)
L. Landskron, V. Steinmann, F. Bonnay, T.R. Burkard, J. Steinmann, I. Reichardt, H. Harzer, A.S. Laurenson, H. Reichert, J.A. Knoblich, The asymmetrically segregating lncRNA cherub is required for transforming stem cells into malignant cells. elife 7 (2018)
M.S. Beg, A.J. Brenner, J. Sachdev, M. Borad, Y.K. Kang, J. Stoudemire, S. Smith, A.G. Bader, S. Kim, D.S. Hong, Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig New Drugs 35, 180–188 (2017)
M. Al-Hajj, M.S. Wicha, A. Benito-Hernandez, S.J. Morrison, M.F. Clarke, Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100, 3983–3988 (2003)
H. Fan, J.H. Zhu, X.Q. Yao, Knockdown of long noncoding RNA PVT1 reverses multidrug resistance in colorectal cancer cells. Mol Med Rep 17, 8309–8315 (2018)
Z. Bian, L. Jin, J. Zhang, Y. Yin, C. Quan, Y. Hu, Y. Feng, H. Liu, B. Fei, Y. Mao, L. Zhou, X. Qi, S. Huang, D. Hua, C. Xing, Z. Huang, LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep 6, 23892 (2016)
K. Takahashi, I.K. Yan, T. Kogure, H. Haga, T. Patel, Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 4, 458–467 (2014)
M. Rasool, A. Malik, S. Zahid, M.A. Basit Ashraf, M.H. Qazi, M. Asif, A. Zaheer, M. Arshad, A. Raza, M.S. Jamal, Non-coding RNAs in cancer diagnosis and therapy. Non-coding RNA Research 1, 69–76 (2016)
M.M. Ali, V.S. Akhade, S.T. Kosalai, S. Subhash, L. Statello, M. Meryet-Figuiere, J. Abrahamsson, T. Mondal, C. Kanduri, PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun 9, 883 (2018)
H.S. Chiu, S. Somvanshi, E. Patel, T.W. Chen, V.P. Singh, B. Zorman, S.L. Patil, Y. Pan, S.S. Chatterjee, N. Cancer Genome Atlas Research, A.K. Sood, P.H. Gunaratne, P. Sumazin, Pan-Cancer analysis of lncRNA regulation supports their targeting of Cancer genes in each tumor context. Cell Rep e212, 297–312 (2018)
Acknowledgments
We thank graphic designer Cristina Oropeza Ramírez for her excellent contribution.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Castro-Oropeza, R., Melendez-Zajgla, J., Maldonado, V. et al. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 41, 585–603 (2018). https://doi.org/10.1007/s13402-018-0406-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-018-0406-4