Abstract
Purpose
Previously, compounds containing a piperidone structure have been shown to be highly cytotoxic to cancer cells. Recently, we found that the piperidone compound P2 exhibits a potent anti-neoplastic activity against human breast cancer-derived cells. Here, we aimed to evaluate two piperidone compounds, P1 and P2, for their potential anti-neoplastic activity against human leukemia/lymphoma-derived cells.
Methods
Cytotoxicity and apoptosis induction were evaluated using MTS, annexin V-FITC/PI and mitochondrial membrane potential polychromatic assays to confirm the mode of action of the piperidone compounds. The effects of compound P1 and P2 treatment on gene expression were assessed using AmpliSeq analysis and, subsequently, confirmed by RT-qPCR and Western blotting.
Results
We found that the two related piperidone compounds P1 and P2 selectively killed the leukemia/lymphoma cells tested at nanomolar concentrations through induction of the intrinsic apoptotic pathway, as demonstrated by mitochondrial depolarization and caspase-3 activation. AmpliSeq-based transcriptome analyses of the effects of compounds P1 and P2 on HL-60 acute leukemia cells revealed a differential expression of hundreds of genes, 358 of which were found to be affected by both. Additional pathway analyses revealed that a significant number of the common genes were related to the unfolded protein response, implying a possible role of the two compounds in the induction of proteotoxic stress. Subsequent analyses of the transcriptome data revealed that P1 and P2 induced similar gene expression alterations as other well-known proteasome inhibitors. Finally, we found that Noxa, an important mediator of the activity of proteasome inhibitors, was significantly upregulated at both the mRNA and protein levels, indicating a possible role in the cytotoxic mechanism induced by P1 and P2.
Conclusions
Our data indicate that the cytotoxic activity of P1 and P2 on leukemia/lymphoma cells is mediated by proteasome inhibition, leading to activation of pro-apoptotic pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The World Health Organization (WHO) states that cancer is the second leading cause of death worldwide next to heart disease [1, 2]. In the USA, it was estimated that in 2016 there were approximately 1.7 million new cancer cases with 595,690 of those leading to mortality [3]. These numbers are expected to increase as factors that contribute to cancer incidence (i.e., smoking, occupational exposure and diet/physical inactivity) are spreading to developing countries [4]. There are several hallmarks that provide cancer cells with their notable characteristics [5]. It is also well known that multiple cellular signaling pathways and (extra) cellular components may govern the development of this disease [5, 6], and it has amply been shown that exogenous and endogenous carcinogens may cause mutations in the genome, driving the conversion of normal cells into cancerous cells [6, 7]. Cancer may also present itself as a genetic disease, since cancer-causing mutations in tumor suppressor genes and/or DNA repair genes may be transferred from one generation to the next [8]. Currently, several therapeutic strategies are being employed to combat cancer, including immune therapy and the use of specific (targeted) drugs and hormones [9]. In addition, combination drug therapy is being used but, although successful in certain cases [10], drug resistance is a recurring problem [11,12,13]. Also, the targeting of one signal transduction pathway may be effective [10] but, again, resistance may occur [14]. Thus, there is a clear need for the development of new therapies that may reduce drug evasion by cancer cells.
Previously, proteasome inhibition has been found to lead to the induction of proteotoxic stress and apoptosis [15]. Bortezomib (Velcade®) is the first proteasome inhibitor that was FDA approved for the treatment of multiple myeloma [16]. Proteasome inhibitors have been thoroughly investigated due to their selective toxicity towards cancer cells [16]. This selective toxicity is based on the fact that cancer cells exhibit higher endogenous stress levels than normal cells and heavily rely on proteasome activity [17, 18]. Inhibition of the proteasome affects the ubiquitin-proteasome pathway (UPP) leading to the accumulation of misfolded and unfolded proteins [19, 20]. This, in turn, causes proteotoxic stress within the endoplasmic reticulum (ER), which leads to activation of the unfolded protein response (UPR) [20, 21]. Thus, when the proteasome is inhibited, cancer cells become overloaded with UPR stress, which ultimately results in apoptosis [20,21,22]. Recently, it has been reported that proteasome inhibitors such as Bortezomib may induce apoptosis via Noxa, also known as PMAIP1 (Phorbol-12-Myristate-13-Acetate Induced Protein 1) [23]. Noxa is a BH3-only pro-apoptotic protein belonging to the BCL2 family of proteins, which are essential initiators of apoptosis [24, 25]. Although Noxa is typically not over-expressed during tumor development, it has been found to be up-regulated after treatment with proteasome inhibitors [16, 23, 26]. It has also been found that siRNA-mediated silencing of Noxa may lead to inhibition of apoptosis activation after treatment of chronic lymphocytic leukemia [26], melanoma [23], myeloma [23] and squamous cell carcinoma [27] cells with proteasome inhibitors. Therefore, Noxa appears to be a key mediator in the induction of apoptosis by proteasome inhibitors [26, 28].
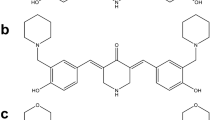
Previously, we reported the development of several new cancer drugs and screening methods [29, 30]. We found, among others, that compounds containing the piperidone structure are highly cytotoxic, and that the P2 piperidone compound exhibits a potent anti-neoplastic activity against breast cancer cells [30, 31]. Next to P2, we generated additional analogs with the intention of increasing the anti-cancer potency of P2. One of these compounds, P1, was deemed most cytotoxic. Here, P1 and P2 (see Fig. 1) were assessed for their cytotoxicity and their capacity to induce cell death pathways. Both compounds were found to activate the intrinsic apoptotic pathway in human leukemia/lymphoma cells and to induce proteotoxic stress via proteasome inhibition.
2 Materials and methods
2.1 Cell lines and culture conditions
The panel of hematological cancer cell lines used in this study consisted of BJAB (Burkitt’s Lymphoma), CCRF-CEM (Acute Lymphoblastic Leukemia, ATCC: 62848170), HL-60 (Acute Myelocytic Leukemia, ATCC: 62690063), Jurkat (Acute Lymphocytic Leukemia, ATCC: 62729601), Nalm-6 (Acute Lymphoblastic Leukemia) and Ramos (Burkitt’s Lymphoma, ATCC: 62927031). Five of the cell lines were cultured in RPMI-1640 medium (Corning 10–040-CV) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals Cat No. S11150) and a mixture of 25 μg/ml amphotericin B, 1000 U/ml penicillin, and 1000 μg/ml streptomycin (Quality Biological 120–096-711). The HL-60 cells were supplemented with 20% FBS in RPMI-1640 medium and the antibiotics mentioned above. The non-cancerous cell lines used were Hs27 (human foreskin fibroblast, ATCC: 3681588) cultured in DMEM medium supplemented with 10% heat-inactivated FBS and a mixture of 25 μg/ml Amphotericin B, 1000 U/ml Penicillin and 1000 μg/ml Streptomycin, and MCF-10A (human breast epithelium, ATCC: CRL-10317) cultured in DMEM/F12 medium supplemented with FBS and antibiotics as mentioned above, as well as 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone (Sigma-Aldrich, H0888) and 10 μg/ ml insulin (Sigma-Aldrich, I1882). All cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
2.2 Compound synthesis
For the synthesis of (3E,5E)-3,5-bis(3,4,5-trimethoxybenzylidene)piperidin-4-one (named P1), we used a well-established 2 component coupling reaction between a 3,4,5-trimethoxybenzaldehyde and piperidin-4-one in acidic buffer at room temperature [31]. The desired HCl salt product (P1) was collected by filtration. The purity of P1 was confirmed by 1H and 13C NMR and the mass was confirmed by LC/MS (see Supplementary Material). 1H NMR (400 MHz, DMSO) δ 9.93 (s, 1H), 7.84 (s, 1H), 6.85 (s, 2H), 4.58 (s, 2H), 3.84 (s, 6H), 3.73 (s, 3H); 13C NMR (101 MHz, DMSO) δ 182.61, 153.37, 139.97, 139.65, 129.68, 127.49, 108.83, 60.66, 56.60, 44.37; LC/MS (MW + 1 = 456.17). In the same manner, compound P2 was synthesized according to previously published synthetic routes, and its purity was established by 1H NMR spectroscopy and elemental analysis [31].
2.3 MTS assay
A colorimetric MTS assay was used for IC50 assessment according to the manufacturer’s instructions (Promega; G1111). Briefly, cells were seeded in a clear flat bottom 96-well plate according to the cell optimization methods provided by the protocol. Lymphoma and leukemia-derived cells were seeded at a density of 25,000 cells/well in 100 μl culture media, whereas Hs27 and MCF-10A were seeded at a density of 10,000 cells/well in 100 μl culture media. Next, the cells were treated at different compound concentrations to determine the 50% inhibitory concentration (IC50). After a 22 h incubation, MTS ([3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]) and PMS (phenazine methosulfate) were added and incubated for an additional 2 h after which colorimetric changes were analyzed using a spectrophotometer. The IC50 values were calculated using a linear interpolation method (https://www.johndcook.com/interpolator.html). The following controls were used: 1 mM H2O2 as a positive control, 1% v/v DMSO as a vehicle control, and untreated cells as a negative control. The selective cytotoxicity index (SCI) was calculated using the following equation: IC50 of non-cancerous cells / IC50 of cancer cells [30]. The SCI indicates the selective profile that a drug exhibits towards cancer cells. Values above 1 indicate a higher selectivity towards cancer cells and vice versa. A chemical compound with a high SCI can be deemed a potential anti-cancer drug candidate [30].
2.4 Annexin V-FITC/PI assay
The HL-60 and Ramos cells were seeded at a density of 200,000 cells/well in a clear flat-bottom 24-well plate in 1 ml medium. Next, the cells were treated with 2 μM P1 and P2 for 24 h, after which the cells were collected and simultaneously stained with propidium iodide and annexin V-FITC according to the manufacturer’s instructions (Beckman Coulter; IM3546). Finally, the samples were analyzed using flow cytometry (Cytomics FC 500; Beckman Coulter). The following controls were used: 1 mM H2O2 as a positive control, 1% v/v DMSO as a vehicle control, and untreated cells as a negative control. Approximately 10,000 events (cells) were acquired per sample and analyzed using the CXP software tool (Beckman Coulter).
2.5 Mitochondrial membrane potential (ΔΨm) polychromatic assay
HL-60 or Ramos cells were seeded at a density of 200,000 cells/well in a clear 24-well plate. HL-60 cells were treated with 2 μM while Ramos cells were treated with 4 μM P1 and P2 for 5 h. Subsequently, the cells were stained with the cationic polychromatic JC-1 (5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide) reagent at a final concentration of 2 μM (MitoProbe; Life Technologies; M34152). In cells with an intact mitochondrial membrane, the JC-1 dye aggregates within the inner mitochondrial membrane causing a shift from green (~529 nm) to red emission (~590 nm). Once mitochondria are depolarized, JC-1 is unable to form aggregates and remains as a monomer emitting a green fluorescence signal [32]. After JC-1 staining, the samples were analyzed using flow cytometry (Cytomics FC 500; Beckman Coulter). The same controls were used as in the other apoptosis assays (see above).
2.6 Caspase-3/7 activation assay
HL-60 cells or Ramos cells (200,000 cells/well) were seeded in a 24-well plate in 1 ml complete RPMI-1640 medium. Next, the cells were treated for 7 h with 2 μM (HL-60 cells) or 4 μM (Ramos cells) P1 and P2 after which caspase-3/7 activation was detected using the fluorogenic NucView 488 caspase-3/7 substrate, designed to identify active caspase-3/7 within live cells (Biotium; 30,029). After flow cytometry (Cytomics FC 500; Beckman Coulter) cells emitting a green fluorescence signal were counted as apoptotic cells with activated caspase 3/7. The same positive and negative controls as in the other apoptosis assays were also used in this series of experiments.
2.7 Transcriptome analysis by AmpliSeq
HL-60 cells (2,000,000 cells/1 ml/well in 24 well plate) were treated with 2 μM P1, P2 or solvent control (0.3% v/v PEG-400) for 6 h. After treatment, the cells were collected in 15 ml conical tubes, centrifuged at 262 g for 5 min, transferred to 1.5 ml centrifuge tubes, washed once with 1 ml ice-cold PBS and spun down at 150 g for 5 min. Next, the supernatants were removed and the pellets were stored at −80 °C. The next day, RNA was extracted using a RNeasy Mini Kit (Qiagen; 74,104) after which the samples were incubated with 10 μl (20 units) DNase I for 15 min at room temperature (25 °C) as indicated in the optional instructions for the kit and total RNA was quantified using the Qubit Fluorometer (Thermo Fisher; Q33216). Total RNA (10 ng) from each independent replicate was reverse transcribed by poly-A priming followed by PCR pre-amplification (12 cycles) according to the protocol supplied with the Ion AmpliSeq RNA Library Kit (Life Technologies; 4,482,335). After primer digestion, adapters and molecular barcodes were ligated to the amplicons followed by magnetic bead purification. This library was amplified, purified and stored at −20 °C. Sequencing was performed using an Ion Proton on a P1v3 Chip (Life Technologies) in the Analytical and Translational Genomics Shared Resource unit at the University of New Mexico Comprehensive Cancer Center. A total of three biological replicates were done for each treatment.
2.8 Connectivity map (CMAP) analysis
The Connectivity Map 02 (CMAP) is a database containing gene expression profiles of human cancer-derived cell lines exposed to known perturbagens (Broad Institute, http://www.broad.mit.edu/cmap/). The cancer-derived cell lines listed in this database are MCF7 (breast cancer), PC3 (prostate cancer), HL-60 (leukemia) and SKMEL5 (melanoma). For our studies, we conducted a transcriptome analysis (AmpliSeq analysis) as mentioned above in the HL-60 cell line. The molecular signatures obtained for P1 and P2 were compared to those in the CMAP database. These analyses were performed as previously reported [33].
2.9 Gene ontology analysis
The PANTHER classification system (pantherdb.org) is an online tool that allows users to classify genes based on biological processes, cellular pathways and/or metabolic processes. Gene ontology analyses were conducted using the PANTHER system. To functionally classify genes of which the expression is altered by P1 and P2, we placed the ID list of the genes in common between P1 and P2 into the PANTHER system. This strategy allowed the identification of biological processes affected in cells treated with P1 and P2.
2.10 Quantitative reverse transcriptase real-time PCR
HL-60 cells (1,000,000) were seeded in 6-well plates and Ramos cells (1,500,000) were seeded in 60 mm cell culture dishes, both at a density of 200,000 cells per ml. Next, the cells were treated for 6 h with 2 μM P1, 2 μM P2 or 0.3% v/v PEG-400. After treatment, the cells were collected and washed once with PBS and spun down at 262 g for 5 min. Dry pellets were used immediately for RNA extraction using a RNeasy Mini kit followed by a QIA-shredder used in the initial steps and an optional DNase I digestion as described above. A total of three biological replicates were used. After storage at −80 °C overnight, the RNA was employed to synthesize cDNA using a RT2 HT First Strand Kit (Qiagen; 330,411). To this end, the RNA was quantified using a NanoDrop N-1000 system after which 500 ng RNA was used for cDNA synthesis per sample. The RNA was diluted with RNase free water to obtain the desired concentration in 9 μl. As per manufacturer’s instructions, samples (9 μl) were incubated with 6 μl GE2 (gDNA elimination buffer) for 5 min at 37 °C, followed by incubation in 6 μl BC4 Reverse Transcriptase Mix for 15 min at 42 °C, 5 min at 95 °C and, finally, a 4 °C hold. Following synthesis, cDNA was stored at −20 °C and the remaining RNA was stored at −80 °C. Quantitative real-time PCR (RT-qPCR) was carried using an iCycler Thermal Cycler (Bio-Rad; 582BR). A 25 μl reaction mixture containing the following components was placed in 8-strip PCR tubes: 12.5 μl RT2 SYBR Green (Qiagen; 330,512), 3.5 μl forward (sense) primer (Bioneer), 3.5 μl reverse (antisense) primer (Bioneer), 1.5 μl nuclease-free water (Promega, P119A) and 4 μl cDNA template. Each biological replicate was run using three technical replicates and a negative control of nuclease-free water. The following PCR cycles were used for the reaction: 1 cycle for 10 min at 95 °C, 40 cycles for 10 s at 95 °C, 10 s at annealing temperature (Supplementary Table 4), 10 s at 72 °C and, finally, 1 cycle for 10 min at 72 °C. A final cycle was added to create a melt curve of the primers’ specificity consisting of 80 cycles of 30 s, beginning at the melting temperature of the corresponding primer pair and increasing by 0.5 °C each cycle. The primers used in this study are listed in Supplementary Table 4 (all primers were obtained from Bioneer). Annealing temperatures were calculated for each individual primer using an online Tm calculator (New England Biolabs). Extension times were calculated using a reference of 1 kb/min.
2.11 Western blotting
A total of 3,000,000 HL-60 cells was seeded in a T-25 flask in 15 ml complete RPMI-1640 medium. Next, the cells were treated with 2 μM P1, 2 μM P2, 2 μM MG-132 or vehicle control (0.3% v/v PEG-400) during different time periods. After incubation, the cells were collected in 15 ml conical tubes, centrifuged at 262 g for 5 min, transferred to 1.5 ml tubes, washed once with 1 ml PBS and spun down at 1000 g for 6 min. The supernatants were removed and the pellets were re-suspended in 70 μl 2× Laemmli buffer (120 mM Tris-HCl, 0.1% β-mercaptoethanol, 4% SDS, 20% glycerol and 0.02% (v/v) bromophenol blue) and boiled at 100 °C for 10 min to lyse the cells. The protein content per sample was quantified using a NanoDrop N-1000 system (Thermo Fischer) after which the samples were used immediately or stored at −80 °C. A total of 100 μg protein in a final volume of 25 μl was loaded per lane on a 10% SDS gel (Noxa) or a 7% SDS gel (Ubiquitin) and separated at 100 V for 1 h. Next, the gel-separated proteins were transferred to PVDF membranes using a wet transfer at 100 V for 1 h. The resulting membranes were blocked overnight at 4 °C in a 5% powder milk solution containing 0.02% sodium azide, and next incubated with primary antibodies for 1 h at room temperature. The following antibodies were used: mouse monoclonal anti-Noxa (Thermo Fischer; MA1–41000) at a dilution of 1:1000 in 5% BSA/TBS-T (0.001% Tween) solution, mouse monoclonal anti-Ubiquitin (Santa Cruz Biotech; sc-8017) at a dilution of 1:1000 in 5% BSA/TBS-T solution, and mouse monoclonal anti-GAPDH (Fitzgerald; 10R-2932) at a dilution of 1:2000 in 5% BSA/TBS-T solution. Unbound antibodies were washed away with TBS-T 3 times for 15 min each, after which a secondary antibody (polyclonal goat anti-mouse conjugated to horseradish peroxidase; Thermo Scientific; 31,430) at a dilution of 1:10,000 in TBS-T was added for 1 h at room temperature. Unbound antibodies were again washed away with TBS-T 3 times for 15 min. After washing, an ECL substrate (Thermo Scientific; 32,106) was added and the membranes were exposed to an X-ray film in a dark room. Densitometry analysis of each protein band of interest was conducted using the Image Studio™ Lite (LI-COR) Software tool.
2.12 Statistical analysis
All experiments were conducted in triplicate. Average values are reported and standard deviations (SD) were calculated for each average. The Student’s t-test was used to determine p-values between groups and values < 0.05 were deemed significant. Tests for variance were conducted between groups as well as to determine equal or unequal variances.
3 Results
3.1 P1 and P2 exert a selective cytotoxicity towards leukemia/lymphoma cells
A MTS assay was used to determine the cytotoxic effects P1 and P2 on a panel of six leukemia/lymphoma-derived cell lines (see materials and methods). Relatively lower IC50 values in the nano- and micromolar range were observed in the leukemia/lymphoma-derived cell lines compared to those in the non-cancerous cell lines (Hs27 and MCF-10A) tested (Table 1). As previously reported, a compound must have a selectivity index value above 1 in order to exhibit selectivity towards cancer cells [30]. This parameter allowed us to more precisely define the selectivity of the compounds. We found that the Ramos cell line was most sensitive to both P1 (0.23 μM ± 0.01) and P2 (0.67 μM ± 0.01). Overall, we found that P1 and P2 exerted cytotoxic effects at a nanomolar range in a selective manner towards a panel of hematological cancer-derived cell lines (Table 1).
3.2 P1 and P2 activate the intrinsic apoptotic pathway
Phosphatidylserine (PS) is typically found in the inner leaflet of the membrane, but when the membrane is compromised during apoptosis, it becomes exposed [34]. Annexin-V-FITC is commonly used to detect apoptosis because of its ability to bind to PS. We found that treatment of HL-60 cells with P1 and P2 caused significant PS externalization, indicative of a strong induction of apoptosis, compared to treatment with a vehicle control (1% v/v DMSO) (Fig. 2a). The effects of P1 and P2 were similar as they both yielded ~ 40% annexin-V-FITC positive cells compared to controls. Similar to HL-60 cells, we found that treatment of Ramos cells with 2 μM P1 or P2 resulted in a PS externalization of ~60% (P1) and ~75% (P2) (Fig. 2d). Using propidium iodide (PI), necrosis can be detected if cells are stained only with this dye, but not with annexin-V-FITC. By doing so, we found that only a few cells stained PI positive after P1 or P2 treatment (results not shown), indicating that the cell death observed was primarily brought about by apoptosis. As a positive control for apoptosis induction hydrogen peroxide (H2O2) at 1 mM final concentration was used and, as expected, this cytotoxic agent induced a strong PS externalization in the majority of cells (≥ 90%).
P1 and P2 induce cell death via apoptosis. HL-60 cells were treated with 2 μM of either compound (a, b, c), whereas Ramos cells were treated with 2 μM (d) and 4 μM (e, f) of each compound, respectively. (a, d) Annexin V-FITC/PI assay to detect phosphatidylserine exposure after treatment of the cells with P1 and P2 for 24 h. (b, e) Mitochondrial depolarization detected by JC-1 assay after treatment of the cells with P1 and P2 for 5 h. (c, f) Caspase-3/7 activation detected after treatment of the cells with P1 and P2 for 7 h. Time-points were determined based on the occurrence of each event during the time course of apoptosis. The following controls were included in each experiment: DMSO as a vehicle control, H2O2 as a positive (apoptosis-inducing) control and untreated (Unt.) cells as a negative control. The experiments were conducted in triplicate for determination of statistical significance and standard deviation from the mean. Student’s t-test was used to calculate p-values (*indicates a p-value < 0.05)
Disruption of mitochondrial transmembrane potential is a common hallmark of the intrinsic apoptotic pathway [35]. In order to determine whether mitochondrial depolarization was induced by P1 and P2, HL-60 and Ramos cells were treated for 6 h with 2 μM P1, P2, DMSO (1% v/v) or H2O2 (1 mM). We found that the cell lines treated with P1 or P2 exhibited a green fluorescent signal in ≥ 25% of the cells as measured by flow cytometry. The treated samples were compared to vehicle (1% v/v DMSO) treated control cells and showed a significant difference (p < 0.05; Fig. 2b). Interestingly, we found that both cancer cell lines exhibited higher levels of mitochondrial depolarization when treated with P2 than with P1 (Fig. 2b, e).
To further confirm that P1 and P2 induce apoptosis, caspase-3/7 activation was measured after treatment of the cells for 7 h with the respective compounds [35]. To detect cells with active caspase-3 and -7, a membrane-permeable fluorogenic NucView 488 caspase-3/7 enzyme substrate (Biotium, Fremont, CA) was used, which allows the detection of active caspase-3/7 in live cells. The cells emitting a green fluorescent signal were counted as positives using flow cytometry. By doing so, a significant increase (p < 0.05) in caspase-3 cleavage was observed in both HL-60 and Ramos cells after treatment with P1 and P2 (Fig. 2c, f) compared to vehicle (1% v/v DMSO) control treated cells. More than 50% of the cells were found to contain active caspase-3 after compound treatment. Taken together, our data indicate that P1 and P2 can induce the intrinsic apoptotic pathway in both HL-60 and Ramos cells.
3.3 Transcriptome analysis of HL-60 cells treated with P1 and P2 reveals 358 common differentially regulated genes
The effect of P1 and P2 on the transcriptome of HL-60 cells was investigated for each compound. Based to the structural similarity and similar apoptotic activities of P1 and P2, we reasoned that the compounds might affect the expression of a similar set of genes. Concordantly, we found that 358 genes were similarly affected by P1 and P2 treatment (Fig. 3a). A subset of these genes is shown in Fig. 3b, c, whereas a larger set is listed in Supplementary Tables 2 and 3. It is particularly interesting to note that the top 8 up-regulated genes in both transcriptome data are the same, i.e., HMOX1, HSPA6, HSPA1B, HSPA1A, BAG3, DUSP1, ATF3 and DNAJA4. Further analysis of the potential roles of these genes revealed that the majority of them is known to be induced as a result of an unfolded protein response (UPR), leading to proteotoxic stress [36]. In addition, we found that the pro-apoptotic genes PMAIP1 (Noxa) [37] and CHAC1 [38] were up-regulated by both P1 (20.7-fold and 51.3-fold, respectively) and P2 (14.5-fold and 31.1-fold, respectively). The induction of these two pro-apoptotic genes is predicted to result in the induction of apoptosis [37, 38]. In addition, we found that the oncogene MYC was down-regulated −5.1-fold by P1 and − 3.1-fold by P2. MYC up-regulation is known to result in cell immortalization and increased proliferation [39, 40], whereas its down-regulation is known to result in a reduced proliferation [41]. Together, these data indicate that the induction of apoptosis by P1 and P2 is likely mediated by the induction of proteotoxic stress, the up-regulation of pro-apoptotic genes and the down-regulation of proliferation-associated genes.
Transcriptome analysis after treatment of HL-60 cells with P1 and P2 for 6 h reveals many affected genes. (a) 358 genes similarly affected (genes in common in the overlapping circles; shaded middle portion) by P1 and P2 treatment. P1 and P2 treatment resulted in the up- and down-regulation of 246 and 98 genes, respectively. Genes chosen for analysis were differentially regulated by > 2-fold compared to solvent (0.3% v/v PEG-400) treated cells. (b) Genes up-regulated after both P1 and P2 treatment. The top eight up-regulated genes by both compounds are known to play a role in the unfolded protein response (UPR). The pro-apoptotic genes CHAC1 and PMAIP1 are also important contributors to the UPR. (c) Ten most down-regulated genes in the transcriptome data of P1 treated cells and corresponding values of P2 treated cells. The fold change (FC) differences and adjusted p-values (Adj. P-Val) are deduced from vehicle control and compound treated samples
3.4 CMAP analysis reveals similarities with known proteasome inhibitors
The expression signatures of the genes induced by both P1 and P2 were compared to those in the Connectivity Map (CMAP) of drug treated cells. CMAP is a database that contains the gene profiles of five different human cancer-derived cell lines exposed to 1309 chemicals [42]. Since one of the cell lines used in the CMAP study was HL-60, this cell line was selected for transcriptome analyses after treatment with P1 and P2 for 6 h. The genes found to be common through the transcriptome analysis of P1 and P2 treated cells (Fig. 3a) were used to query the CMAP database. Comparison to the CMAP expression profiles revealed that P1 and P2 appear to behave as proteasome inhibitors with gene expression patterns highly similar to those induced by the known proteasome inhibitors MG-132 and MG-262 [43]. The top 12 hits are shown in Table 2 along with their p-values and false-discovery-rates (FDR). In both cases the p-values were < 0.001. The p-values were obtained by comparing the expression profiles of P1 and P2 treated cells to those within the CMAP database.
3.5 Gene ontology analysis corroborates that P1 and P2 induce apoptosis via the unfolded protein response
Next, the 358 differentially regulated genes that were found to be in common between the P1 and P2 treated cells were subjected to gene ontology (GO) analyses using the PANTHER classification system [44]. Such GO analyses allow the identification of biological processes that are most likely involved in the responses to P1 and P2. We found that the respective treatments resulted in a positive regulation of programmed cell death and a response to unfolded protein (UPR; see highlighted rows of Fig. 4). The observation that P1 and P2 induce apoptosis (Fig. 2) was thus corroborated by this GO analysis. Also, the response to unfolded protein was observed above in the transcriptome analyses through up-regulation of critical genes to the UPR after treatment with P1 and P2 (Fig. 3). Based on these results, we next set out to confirm the role of the UPR in the induction of apoptosis by P1 and P2.
Gene ontology (GO) analysis reveals pathways that may be affected by P1 and P2 treatment. The PANTHER classification system was used for GO analysis to classify significantly activated genes by biological process. The 358 genes found in common (Fig. 3a) between the P1 and P2 treatments were analyzed. The graph displays the subsets of genes that belong to each corresponding biological process. Of note are the positive regulation of programmed cell death and the unfolded protein response (UPR), which are boxed in bold lettering
3.6 Confirmation of differentially regulated genes by RT-qPCR
The genes that were found to be significantly deregulated in HL-60 cells through AmpliSeq analyses (Fig. 3), and believed to be important in the induction of cell death and proliferation, were further examined by RT-qPCR in HL-60 and Ramos cells. Of these genes, ATF3 and HMOX1 are involved in the unfolded protein response [36], PMAIP1 and CHAC1 in the induction of the intrinsic apoptosis pathway [38, 45] and MYC in cell proliferation and immortalization [39] (Fig. 5a, b). In HL-60 cells P1 treatment caused the following fold changes (FC) of the aforementioned genes: ATF3 (25.1), HMOX1 (1155.8), PMAIP1 (14.9), CHAC1 (10.6) and MYC (−22.1), whereas P2 treatment caused the following FC changes: ATF3 (47.9), HMOX1 (31.7), PMAIP1 (4.7), CHAC1 (26.8) and MYC (−6.1). In Ramos cells (Fig. 5c, d) P1 treatment caused the following fold changes (FC): ATF3 (151.9), HMOX1 (217.9), PMAIP1 (3.8), CHAC1 (21.2) and MYC (−3.6), whereas P2 treatment caused the following FCs: ATF3 (95.4), HMOX1 (154.7), PMAIP1 (4.1), CHAC1 (24.5) and MYC (−5.2). Although the FC values obtained by AmpliSeq analyses appeared to be slightly higher than those obtained by RT-qPCR, the overall trend was similar between both methods in both P1 and P2 treated samples. Furthermore, we found that the results obtained with the HL-60 and Ramos cells revealed similar trends of up- or down-regulation of genes that were identified in the AmpliSeq study.
Validation of fold-change differences observed in transcriptome analyses of genes involved in cell death induction. RT-qPCR was performed to detect the mRNA levels of phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1), proto-oncogene MYC, activating transcription factor 3 (ATF3), ChaC glutathione-specific gamma-glutamylcyclotransferase 1 (CHAC1) and Heme Oxygenase 1 (HMOX1) in P1 or P2 treated HL-60 (a, b) and Ramos (c, d) cells, respectively. RT-qPCR values are depicted as fold changes (Y-axis) relative to 0.3% v/v PEG-400 treated cells (solvent control). Gene expression levels were normalized to the expression level of the β-actin (ACTB) gene. Each bar denotes the average results (± S.D.) from biological triplicates, with three technical replicates each. For HMOX1 in Ramos cells two biological replicates were used. The comparative Ct method was used for the RT-qPCR analyses
3.7 P1 and P2 treatment causes an accumulation of high molecular weight poly-ubiquitinated proteins
Impairment of proteasome activity has previously been shown to result in an accumulation of poly-ubiquitinated proteins [15, 16]. b-AP15, a compound that has a similar structure as P1 and P2, has been found to inhibit the proteasome by specifically interacting with UCHL5 and USP14, resulting in an accumulation of high molecular weight poly-ubiquitinated proteins [15]. Based on this structural similarity, we set out to examine the accumulation of high molecular-weight (above 100 kDa) poly-ubiquitinated proteins after P1 or P2 treatment (Fig. 6a). We found that the accumulation of poly-ubiquitinated proteins was stronger after 1 h treatment than after treatment for longer periods of time as detected by anti-ubiquitin antibodies. Densitometric analysis of the bands after Western blot analyses revealed a ~4 to 5.5-fold increase in poly-ubiquitinated proteins after P1 and P2 treatment compared to vehicle control treatment (Supplementary Fig. 1). MG-132, which is a known proteasome inhibitor, was used as a positive control and, by doing so, we found a similar pattern of poly-ubiquitinated proteins as in cells treated with P1 or P2.
Western blot analysis reveals poly-ubiquitination and increased Noxa protein expression in cells treated with P1 and P2. (a) HL-60 cells treated with 2 μM P2, 2 μM MG-132 and 2 μM P1 exhibit accumulation of high molecular weight poly-ubiquitinated proteins. (b, c) Cells treated with P1 exhibit a slight increase in Noxa expression, which is validated by densitometric analysis. (d) HL-60 cells treated with 2 μM P2, 2 μM MG-132 and 0.3% v/v PEG-400 (Vehicle) at different time points. Densitometric analysis and Western blotting reveal increased Noxa induction in samples treated with P2, P1 and MG-132 compared to the vehicle control. Image Studio™ Lite (LI-COR) Software was used for the densitometry analyses. Fold change differences were calculated by comparing signal values from the protein of interest (Noxa) to the loading control (GAPDH). Note that lanes (vertical black lines) are spliced between vehicle and treatment samples as indicated
3.8 Noxa, an important regulator of the intrinsic apoptotic pathway, is up-regulated in HL-60 cells treated with P1 and P2
Noxa, which is encoded by the PMAIP1 gene, was found in the AmpliSeq analyses to be up-regulated by both P1 (20.7 FC) and P2 (14.5 FC). Further analysis by RT-qPCR confirmed the up-regulation of this gene in HL-60 cells treated with P1 (14.9) and P2 (4.7). Noxa is a BH3-only protein that has been shown to act as an important factor in the activation of the intrinsic apoptotic pathway [35]. Based on our previous results (Fig. 2) indicating that P1 and P2 induce the expression of the PMAIP1 gene, we set out to assess the Noxa protein levels after P1 and P2 treatment using Western blot analysis and the proteasome inhibitor MG-132 [46]. We found a clear difference between samples treated with the vehicle control (0.3% v/v PEG-400) or P2 (Fig. 6e). Densitometric analysis revealed a 20-fold change in cells treated with P2 for 6 h compared to the vehicle control. Increasing the incubation time to 9 and 12 h resulted in 45.6-fold and 46.6-fold changes in Noxa expression, respectively, compared to the vehicle control (Fig. 6d). Treatment with MG-132 also resulted in an increase in Noxa expression that was similar to that seen with P2 (Fig. 6e). Also P1 treated cells showed an increase in Noxa expression, but to a lesser extent than P2 or MG-132 (Fig. 6b, c). Our results indicate that PMAIP1 gene up-regulation correlates with up-regulated Noxa protein levels. These findings suggest that P1 and P2 induce the expression of the pro-apoptotic protein Noxa and that this protein is the most likely mediator of cell death induction.
4 Discussion
We previously reported that several piperidone compounds, including P2, may exhibit selective cytotoxicity against triple-negative breast cancer cells at low micromolar concentrations [30]. In an attempt to identify additional potent cytotoxic compounds, several piperidone compounds were synthesized. Of these newly synthesized compounds, one (P1) was found to exhibit a stronger cytotoxic activity than P2, with an improved selectivity compared to the non-cancerous cell lines Hs27 and MCF-10A. Both P1 and P2 were also tested on triple-negative breast cancer MDA-MB-231 cells for 72 h and, again, P1 exhibited lower IC50 values (0.47 ± 0.13 μM) than P2 (1.28 ± 0.14 μM; data not shown). To elucidate the mode of action of these compounds, we assessed their role in apoptosis. Induction of apoptosis by anti-cancer drugs is preferred since those that cause necrosis can elicit excessive non-specific cellular/tissue destruction [47]. After we found that P1 and P2 induce apoptosis, additional assays were performed to determine whether P1 and P2 may induce cell death via the intrinsic apoptotic pathway [48] and, indeed, we found that treatment with P1 and P2 resulted in impairment of mitochondrial membrane potential. A final step in apoptosis is activation of the executioner caspase-3. Our results also confirmed activation of caspase-3 after both P1 and P2 treatment. These results were first observed in HL-60 cells, which was the primary focus of our studies [42], and validated in Ramos cells, which were found to be highly sensitive to the piperidone compounds. The Ramos cells also showed a similar mitochondrial depolarization and caspase-3 induction, characteristic of the intrinsic apoptotic pathway. It is interesting to note that the expression of HMOX1 was highly up-regulated in both HL-60 and Ramos cells after P1 treatment, but more consistently in the Ramos cells treated with either P1 or P2. Taken together, we conclude that the mode of cell death induction by P1 and P2 is similar in these two distinct leukemia/lymphoma cell lines. Signaling through cell surface death receptors results in activation of the extrinsic apoptotic pathway, which results in activation of caspase-8 [35, 49]. We found, however, that neither P1 nor P2 induced caspase-8 cleavage (data not shown), which underscores the notion that both compounds act through the intrinsic apoptotic pathway.
In order to further delineate what occurs within cells after treatment with P1 or P2, we conducted targeted whole transcriptome analyses using the Ampliseq technology [50]. Ampliseq whole transcriptome sequencing has an advantage over RNA sequencing (RNAseq) in that it requires smaller quantities of RNA (fewer treated cells) for analysis and compares favorably with RNAseq results [50]. After generating cDNA from HL-60 cells treated with either P1, P2 or 0.3% v/v PEG-400 control for 6 h, the transcriptome data of triplicate samples were compared to each other. The comparison between P1 and P2 revealed 358 genes in common. Since both compounds have a similar structure and induce cell death in a similar fashion, we compared the genes in common to the gene expression profiles within the Broad Institutes’ Connectivity Map (CMAP) that contains more than 7000 microarray-based expression profiles representing the effects of 1309 compounds on several cancer-derived cell lines, including HL-60 [42]. This analysis revealed that P1 and P2 induce expression profiles similar to those induced by known proteasome inhibitors. Poly-ubiquitination of proteins is a hallmark of proteasome inhibition [51] and this effect was subsequently investigated in further detail. We indeed observed accumulation of high molecular weight poly-ubiquitinated proteins in response to P1 or P2 treatment. In addition, Noxa is believed to be a sensor of proteasome integrity. This protein is usually not expressed under normal conditions but when induced, it is rapidly ubiquitinated for degradation [24, 28]. If the proteasome is inhibited, ubiquitin-tagged Noxa cannot be degraded leading to its accumulation [24]. We found that the interplay between proteasomal inhibition and the accumulation of pro-apoptotic proteins like Noxa are likely facilitating apoptosis after P1 and P2 treatment. It has been shown that Noxa is important for the function of proteasome inhibitors [23], and proteasome inhibitors such as Bortezomib have been found to rely on Noxa for selectivity [16]. Also, Noxa has been shown to be regulated through MYC binding sites on its promoter [52] and the selectivity of Bortezomib towards cancer cells has been found to be dependent on these MYC binding sites [16]. However, one of the problems with Bortezomib and other proteasome inhibitors is that, eventually, tumors become resistant to these drugs [16]. Here, we observed up-regulated Noxa expression and down-regulated MYC expression after P1/P2 treatment. This observation is in contrast with previously published results which state that up-regulation of Noxa is correlated with MYC up-regulation [52]. Therefore, we assume that PI and P2 act through different mechanisms that do not rely on MYC. This hypothesis is underscored by the fact that we did observe selectivity towards cancer cells. Down-regulation of MYC expression would be predicted to reduce cancer cell proliferation/survival and, thus, the role of MYC gene down-regulation by P1/P2 treatment would be expected to enhance the cytotoxic effects of these compounds. Additional studies are needed to elucidate the exact role of MYC in the induction of cell death by P1/P2.
Other factors are known, including proteotoxic stress and the UPR, that may contribute to the activity of proteasome inhibitors. To evaluate this possibility, we examined differentially expressed genes (DEGs) from our transcriptome analysis. Interestingly, we found that the same top genes (HMOX1, HSPA6, HSPA1B, HSPA1A, BAG3, DUSP1, ATF3 and DNAJA4) were up-regulated by P1 and P2. These genes are involved in the heat shock response, which is used by cancer cells to alleviate stress [18]. HMOX1, HSPA6 and HSPA1A have been shown to be induced after proteotoxic stress [36]. Due to the relationship between proteotoxic stress and the UPR, we further investigated differentially regulated genes involved in the UPR. The UPR is used in normal cells to maintain protein homeostasis, but it has also been reported that some members of the UPR may serve as anti-cancer targets [53]. Through this analysis we identified PMAIP1 (Noxa) and CHAC1, which are important pro-apoptotic factors involved in the UPR [28, 38], to be differentially expressed. ATF3 is an important transcription factor of the UPR [54]. Protein phosphatase 1 regulatory subunit 15A (PPP1R15A/GADD34), which is a critical component of the UPR, was found to be up-regulated by P1 (29.9 FC) and P2 (14.7 FC). Others have reported that its activation decreases cell survival [22]. Also, cyclin-dependent kinase inhibitor 1 (CDKN1A) was found to be up-regulated 30.9-fold and 16.5-fold by P1 and P2, respectively, and this inhibitor is also known to be involved in the UPR [23]. It is believed that p21 (encoded by CDKN1A) plays an important role in the shift from a pro-survival to a pro-apoptotic role of the UPR [24]. Furthermore, through our gene ontology (GO) analysis, we noted a response to unfolded protein, which supports the induction of the UPR by P1 and P2.
Other DEGs in the transcriptome analysis of P1 and P2 may also play a role in the induction of cytotoxicity. As mentioned before, the MYC proto-oncogene plays an important role in the proliferation of cancer cells [39], and this gene was found to be down-regulated by −5.1 FC and − 3.1 FC after P1 and P2 treatment, respectively. Down-regulation of MYC indicates a role of these compounds in affecting critical genes regulating the lifespan of cancer cells [39]. As mentioned above, we also observed up-regulation of CDKN1A, whose encoded protein p21 has a well-known tumor suppressor function [55]. Therefore, P1 and P2 may be targeting typical hallmarks of cancer through up-regulating tumor suppressors and down-regulating oncogenes [56].
Further DEG analysis led to the identification of two similar compounds, b-AP15 and VLX5710, that induce proteotoxic stress [15, 57]. These compounds act by inhibiting proteasome-related de-ubiquitinases (DUBs). b-AP15 has been found to inhibit the proteasome by inhibiting the DUBs USP14 and UCHL5 [15]. VLX1570, which is an analog of b-AP15, has been found to inhibit the proteasome by preferentially binding to USP14 [57]. The structural similarities between P1 and P2 and the aforementioned compounds support the argument that P1 and P2 may interact with these DUBs. The α,β-unsaturated keto structure is believed to target the cysteine residues within the active sites of DUBs [58]. Several other compounds, including 15-Deoxy-Δ12,14-prostaglandin J2, curcumin, and Gambogic acid, contain a α,β-unsaturated keto group and all have been found to act as proteasome inhibitors through DUB inhibition [58]. Given the fact that also P1 and P2 have a α,β-unsaturated keto structure, we believe that P1 and P2 are also interacting with UCHL5 and USP14. Our in silico docking experiments revealed that P1 and P2 exhibit similar binding scores to these DUBs as b-AP15 and VLX1570 (Supplementary Fig. 2). Future work should reveal whether P1 and P2 actually inhibit the activity of UCHL5 and USP14.
An attractive feature of any anti-cancer drug candidate is that it should possess a high selective cytotoxicity index (SCI) towards cancer cells compared to non-cancerous cells [30]. We found that the SCIs for P1 ranged from 20.83 to 32.6, and those of P2 from 4.52 to 16.88, for all the cancer-derived cell lines included in this study. Thus, P1 possess a superior SCI compared to P2 and it, therefore, appears to be a more attractive and potent anti-cancer drug candidate. It has previously been noted that cancer cells may react differently to anti-cancer compounds based on their gender-derived origin [59]. For this reason, the IC50 values obtained for each cell line and for each piperidone were grouped by gender origin of the cells, as previously recommended [60,61,62]. An average IC50 was calculated for each gender group, which included 3 cancerous cell types. Female-derived cells were BJAB, CEM and HL-60, whereas male-derived cells were Jurkat, Nalm-6 and Ramos. The IC50 average for P1 on the male-derived cells was 0.287 μM, whereas for female-derived cells this average was 0.380 μM (Supplementary Fig. 3). The IC50 average for P2 for male-derived cells was 0.923 μM, whereas for female-derived cells this average was 2.617 μM (Supplementary Fig. 3). Although P1 exhibited lower IC50 average values than P2 in both gender groups, it did not show a statistically significant increase in the killing of either gender cell type (p = 0.33). However, when P2 was tested on male and female-derived cancer cells, the difference was found to be significant (p = 0.04). These observations are in conformity with previously reported findings, supporting the existence of gender dissimilarities in cell sensitivities to cytotoxic compounds [59,60,61,62,63].
Taken together, we conclude that P1 and P2 treatment of cancer cells results in inhibition of proteasome activity, leading to proteotoxic stress and activation of the UPR. The over-abundance of stress (heat shock proteins and UPR-related proteins) within cancer cells and the over-expression of pro-apoptotic proteins, such as Noxa, may eventually result in activation of the intrinsic apoptotic pathway. Further work is required to fully elucidate the mechanism of action and effectiveness of P1 and P2 as proteasome inhibitors.
References
WHO | Cancer WHO Available at: http://www.who.int/mediacentre/factsheets/fs297/en/. (Accessed: 7th November 2017)
WHO Global status report on noncommunicable diseases 2014. WHO Available at: http://www.who.int/nmh/publications/ncd-status-report-2014/en/. (Accessed: 9th November 2017)
American Cancer Society Cancer Facts & Figures 2016. (American Cancer Society, 2016)
X. Ma, H. Yu, Global burden of Cancer. Yale J Biol Med 79, 85–94 (2006)
D. Hanahan, R.A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011)
G. M. Cooper, The development and causes of Cancer. in The Cell: A Molecular Approach (Sinauer Associates, 2000)
G. S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C, Polytarchou, K. B. Marcu, A. G. Papavassiliou, R. Sandaltzopoulos, E. Kolettas, a step-by-step microRNA guide to cancer development and metastasis. Cell Oncol 40, 303–339 (2017)
G. M. Cooper, Applications of molecular biology to Cancer prevention and treatment. In The Cell: A Molecular Approach (Sinauer Associates, 2000)
K.L. Nastiuk, J.J. Krolewski, Opportunities and challenges in combination gene cancer therapy. Adv Drug Deliv Rev 98, 35–40 (2016)
S. Kummar, H.X. Chen, J. Wright, S. Holbeck, M.D. Millin, J. Tomaszewski, J. Zweibel, J. Collins, J.H. Doroshow, Utilizing targeted cancer therapeutic agents in combination: Novel approaches and urgent requirements. Nat Rev Drug Discov 9, 843–856 (2010)
C. Holohan, S.V. Schaeybroeck, D.B. Longley, P.G. Johnston, Cancer drug resistance: An evolving paradigm. Nature Rev Cancer 13, 714–726 (2013)
A. Chavez-Gonzalez, B. Bakhshinejad, K. Pakravan, M.L. Guzman, S. Babashah, Novel strategies for targeting leukemia stem cells: Sounding the death knell for blood cancer. Cell Oncol 40, 1–20 (2017)
Z. Mousavian, A. Nowzari-Dalini, R.W. Stam, Y. Rahmatallah, A. Masoudi-Nejad, Network-based expression analysis reveals key genes related to glucocorticoid resistance in infant acute lymphoblastic leukemia. Cell Oncol 40, 33–45 (2017)
Z. Liu, B. Delavan, R. Roberts, W. Tong, Lessons learned from two decades of anticancer drugs. Trends Pharmacol Sciences 38, 852–872 (2017)
P. D’Arcy, S. Brnjic, M.H. Olofsson, M. Fryknäs, K. Lindsten, M. De Cesare, P. Perego, B. Sadeghi, M. Hassan, R. Larsson, S. Linder, Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med 17, 1636–1640 (2011)
T. Mujtaba, Q.P. Dou, Advances in the understanding of mechanisms and therapeutic use of Bortezomib. Discov Med 12, 471–480 (2011)
P. Moreau, P.G. Richardson, M. Cavo, R.Z. Orlowski, J.F.S. Miguel, A. Palumbo, J.-L. Harousseau, Proteasome inhibitors in multiple myeloma: 10 years later. Blood 120, 947–959 (2012)
C. Dai, S. Dai, J. Cao, Proteotoxic stress of cancer: Implication of the heat-shock response in oncogenesis. J Cell Physiol 227, 2982–2987 (2012)
J. Adams, The proteasome: Structure, function, and role in the cell. Cancer Treatment Rev 29. Supplement 1, 3–9 (2003)
T. Hideshima, P.G. Richardson, K.C. Anderson, Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol Cancer Ther 10, 2034–2042 (2011)
H.-W. Chiu, Y.-C. Tseng, Y.-H. Hsu, Y.-F. Lin, N.-P. Foo, H.-R. Guo, Y.-J. Wang, Arsenic trioxide induces programmed cell death through stimulation of ER stress and inhibition of the ubiquitin–proteasome system in human sarcoma cells. Cancer Lett 356, 762–772 (2015)
E.A. Obeng, L.M. Carlson, D.M. Gutman, W.J. Harrington, K.P. Lee, L.H. Boise, Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916 (2006)
J.-Z. Qin, J. Ziffra, L. Stennett, B. Bodner, B.K. Bonish, V. Chaturvedi, F. Bennett, P.M. Pollock, J.M. Trent, M.J.C. Hendrix, P. Rizzo, L. Miele, B.J. Nickoloff, Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res 65, 6282–6293 (2005)
M. Baou, S.L. Kohlhaas, M. Butterworth, M. Vogler, D. Dinsdale, R. Walewska, A. Majid, E. Eldering, M.J.S. Dyer, G.M. Cohen, Role of NOXA and its ubiquitination in proteasome inhibitor-induced apoptosis in chronic lymphocytic leukemia cells. Haematologica 95, 1510–1518 (2010)
D.C.S. Huang, A. Strasser, BH3-only proteins -essential initiators of apoptotic cell death. Cell 103, 839–842 (2000)
J.E. Guikema, M. Amiot, E. Eldering, Exploiting the pro-apoptotic function of NOXA as a therapeutic modality in cancer. Expert Opin Ther Targets 21, 767–779 (2017)
A.M. Fribley, B. Evenchik, Q. Zeng, B.K. Park, J.Y. Guan, H. Zhang, T.J. Hale, M.S. Soengas, R.J. Kaufman, C.-Y. Wang, Proteasome inhibitor PS-341 induces apoptosis in cisplatin-resistant squamous cell carcinoma cells by induction of Noxa. J Biol Chem 281, 31440–31447 (2006)
A. Craxton, M. Butterworth, N. Harper, L. Fairall, J. Schwabe, A. Ciechanover, G.M. Cohen, NOXA, a sensor of proteasome integrity, is degraded by 26S proteasomes by an ubiquitin-independent pathway that is blocked by MCL-1. Cell Death Differ 19, 1424–1434 (2012)
L.M. Nunes, M. Hossain, A. Varela-Ramirez, U. Das, Y.M. Ayala-Marin, J.R. Dimmock, R.J. Aguilera, A novel class of piperidones exhibit potent, selective and pro-apoptotic anti-leukemia properties. Oncol Lett 11, 3842–3848 (2016)
E. Robles-Escajeda, U. Das, N.M. Ortega, K. Parra, G. Francia, J.R. Dimmock, A. Varela-Ramirez, R.J. Aguilera, A novel curcumin-like dienone induces apoptosis in triple-negative breast cancer cells. Cell Oncol 39, 265–277 (2016)
U. Das, J. Alcorn, A. Shrivastav, R.K. Sharma, E. De Clercq, J. Balzarini, J.R. Dimmock, Design, synthesis and cytotoxic properties of novel 1-[4-(2-alkylaminoethoxy)phenylcarbonyl]-3,5-bis(arylidene)-4-piperidones and related compounds. Eur J Med Chem 42, 71–80 (2007)
E. Robles-Escajeda, D. Lerma, A.M. Nyakeriga, J.A. Ross, R.A. Kirken, R.J. Aguilera, A. Varela-Ramirez, Searching in mother nature for anti-cancer activity: Anti-proliferative and pro-apoptotic effect elicited by green barley on leukemia/lymphoma cells. PLoS One 8, e73508 (2013)
Y. Quan, B. Li, Y.-M. Sun, H.-Y. Zhang, Elucidating pharmacological mechanisms of natural medicines by biclustering analysis of the gene expression profile: A case study on curcumin and si-wu-tang. Int J Mol Sci 16, 510–520 (2014)
K. Segawa, S. Nagata, An apoptotic ‘eat me’ signal: Phosphatidylserine exposure. Trends Cell Biol 25, 639–650 (2015)
S.W.G. Tait, D.R. Green, Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11, 621–632 (2010)
A.L. Davis, S. Qiao, J.L. Lesson, M. Rojo de la Vega, S.L. Park, C.M. Seanez, V. Gokhale, C.M. Cabello, G.T. Wondrak, The quinone methide Aurin is a heat shock response inducer that causes proteotoxic stress and Noxa-dependent apoptosis in malignant melanoma cells. J Biol Chem 290, 1623–1638 (2015)
X.H. Lowman, M.A. McDonnell, A. Kosloske, O.A. Odumade, C. Jenness, C.B. Karim, R. Jemmerson, A. Kelekar, The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell 40, 823–833 (2010)
I.N. Mungrue, J. Pagnon, O. Kohannim, P.S. Gargalovic, A.J. Lusis, CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182, 466–476 (2009)
C.V. Dang, C-Myc target genes involved in cell growth, apoptosis. and metabolism Mol Cell Biol 19, 1–11 (1999)
J. Wang, L.Y. Xie, S. Allan, D. Beach, G.J. Hannon, Myc activates telomerase. Genes Dev 12, 1769–1774 (1998)
B.-J. Chen, Y.-L. Wu, Y. Tanaka, W. Zhang, Small molecules targeting c-Myc oncogene: Promising anti-cancer therapeutics. Int J Biol Sci 10, 1084–1096 (2014)
J. Lamb, E.D. Crawford, D. Peck, J.W. Modell, I.C. Blat, M.J. Wrobel, J. Lerner, J.-P. Brunet, A. Subramanian, K.N. Ross, M. Reich, H. Hieronymus, G. Wei, S.A. Armstrong, P.A. Clemons, R. Wei, S.A. Carr, E.S. Lander, T.R. Golub, The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006)
G. de Bettignies, O. Coux, Proteasome inhibitors: Dozens of molecules and still counting. Biochimie 92, 1530–1545 (2010)
H. Mi, X. Huang, A. Muruganujan, H. Tang, C. Mills, D. Kang, P.D. Thomas, PANTHER version 11: Expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189 (2017)
C. Ploner, R. Kofler, A. Villunger, Noxa: At the tip of the balance between life and death. Oncogene 27, S84–S92 (2009)
N. Guo, Z. Peng, MG132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia-Pacific J Clin Oncol 9, 6–11 (2013)
L. Galluzzi, M.C. Maiuri, I. Vitale, H. Zischka, M. Castedo, L. Zitvogel, G. Kroemer, Cell death modalities: Classification and pathophysiological implications. Cell Death Differ 14, 1237–1243 (2007)
N. Mohana-Kumaran, D.S. Hill, J.D. Allen, N.K. Haass, Targeting the intrinsic apoptosis pathway as a strategy for melanoma therapy. Pigment Cell Melanoma Res 27, 525–539 (2014)
S. Elmore, Apoptosis: A review of programmed cell death. Toxicol Pathol 35, 495–516 (2007)
W. Li, A. Turner, P. Aggarwal, A. Matter, E. Storvick, D.K. Arnett, U. Broeckel, Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genomics 16, 1069 (2015)
S.L. Downey, B.I. Florea, H.S. Overkleeft, A.F. Kisselev, Use of proteasome inhibitors. Curr Protoc Immunol 109, 9.10.1–9.10.8 (2015)
M.A. Nikiforov, M. Riblett, W.-H. Tang, V. Gratchouck, D. Zhuang, Y. Fernandez, M. Verhaegen, S. Varambally, A.M. Chinnaiyan, A.J. Jakubowiak, M.S. Soengas, Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci U S A 104, 19488–19493 (2007)
C. Hetz, E. Chevet, H.P. Harding, Targeting the unfolded protein response in disease. Nat Rev Drug Discov 12, 703–719 (2013)
M. Edagawa, J. Kawauchi, M. Hirata, H. Goshima, M. Inoue, T. Okamoto, A. Murakami, Y. Maehara, S. Kitajima, Role of activating transcription factor 3 (atf3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J Biol Chem 289, 21544–21561 (2014)
I.B. Roninson, Oncogenic functions of tumour suppressor p21Waf1/Cip1/Sdi1: Association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett 179, 1–14 (2002)
X.E. Guo, B. Ngo, A.S. Modrek, W.-H. Lee, Targeting tumor suppressor networks for Cancer therapeutics. Curr Drug Targets 15, 2–16 (2014)
X. Wang, M. Mazurkiewicz, E.-K. Hillert, M.H. Olofsson, S. Pierrou, P. Hillertz, J. Gullbo, K. Selvaraju, A. Paulus, S. Akhtar, F. Bossler, A.C. Khan, S. Linder, P.D. Arcy, The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci Rep 6, srep26979 (2016)
P. D’Arcy, X. Wang, S. Linder, Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther 147, 32–54 (2015)
A. Varela-Ramirez, Female versus male cells in anti-Cancer drug discovery: The winner is …. AAPS Blog (2014) Available at: https://aapsblog.aaps.org/2014/06/18/female-versus-male-cells-in-anti-cancer-drug-discovery-the-winner-is/
L.M. Nunes, E. Robles-Escajeda, Y. Santiago-Vazquez, N.M. Ortega, C. Lema, A. Muro, G. Almodovar, U. Das, S. Das, J.R. Dimmock, R.J. Aguilera, A. Varela-Ramirez, The gender of cell lines matters when screening for novel anti-cancer drugs. AAPS J 16, 872–874 (2014)
Y. Santiago-Vázquez, U. Das, A. Varela-Ramirez, S.T. Baca, Y. Ayala-Marin, C. Lema, S. Das, A. Baryyan, J.R. Dimmock, R.J. Aguilera, Tumor-selective cytotoxicity of a novel pentadiene analogue on human leukemia/ lymphoma cells. Clin Cancer Drugs 3, 138–146 (2016)
J.A. Clayton, F.S. Collins, NIH to balance sex in cell and animal studies. Nature 509, 282–283 (2014)
E. Pollitzer, Cell sex matters. Nature 500, 23–24 (2013)
Acknowledgments
Funding for this work was provided by the National Institute of General Medical Sciences-Support of Competitive Research (SCORE) grant 1SC3GM103713 to RJA, as well as a Canadian Institutes of Health Research-Regional Partnerships Program Saskatchewan grant to JRD. We thank Madan Balal for the compound docking experiments presented in the supplementary material. The synthesis and docking work performed by the Dr. Rachid Skouta research laboratory was partially supported by Lung Cancer Research Foundation and Green Fund grants. The authors also thank the Genomic Analysis and Cytometry, Screening and Imaging Core Facilities at the University of Texas at El Paso (UTEP), which were supported by a Research Centers in Minority Institutions (RCMI) program grant 5G12MD007592 to the Border Biomedical Research Center (BBRC) in UTEP from the National Institute on Minority Health and Health Disparities, a component of the National Institutes of Health. The authors thank Gladys Almodovar (with UTEP) for cell culture expertise. LC was supported by NIGMS RISE training grant R25 GM069621-15.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 6709 kb)
Rights and permissions
About this article
Cite this article
Contreras, L., Calderon, R.I., Varela-Ramirez, A. et al. Induction of apoptosis via proteasome inhibition in leukemia/lymphoma cells by two potent piperidones. Cell Oncol. 41, 623–636 (2018). https://doi.org/10.1007/s13402-018-0397-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-018-0397-1