Abstract
Background
Studies show that abnormalities in non-coding genes can contribute to carcinogenesis; microRNA levels may modulate cancer growth and metastatic diffusion.
Method
MicroRNA libraries were built and sequenced from two osteosarcoma cell lines (MG-63 and 143B), which differ in proliferation and transmigration. By cloning and transfection, miR-93, expressed in both cell lines, was then investigated for its involvement in osteosarcoma progression.
Results
Six of the 19 miRNA identified were expressed in both cell lines with higher expression levels of miR-93 in 143B and in primary osteosarcoma cultures compared to normal osteoblasts. Interestingly, levels of miR-93 were significantly higher in metastases from osteosarcoma than in paired primary tumours. When 143B and MG-63 were transfected with miR-93, clones appeared to respond differently to microRNA overexpression. Ectopic expression of miR-93 more significantly increased cell proliferation and invasivity in 143B than in MG-63 clones. Furthermore, increased mRNA and protein levels of E2F1, one of the potential miR-93 targets, were seen in osteosarcoma cellular clones and its involvement in 143B cell proliferation was confirmed by E2F1 silencing.
Conclusion
Although further studies are needed to evaluate miRNA involvement in osteosarcoma progression, miR-93 overexpression seems to play an important role in osteosarcoma cell growth and invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Osteosarcoma (OS), the most common bone tumour, is a rare malignant neoplasm [1], for which biologic and pathologic information are still largely incomplete. In recent years surgery combined with chemotherapy has markedly improved patient survival [2]; however the use of anti-cancer drugs is still associated with serious problems, such as frequent acquisitions of drug-resistant phenotypes, highlighting the need for urgent novel treatment approaches [3]. A better understanding of the cellular and molecular mechanisms underlying pre- and post-surgery metastasis formation and the development of chemo-resistance in these tumours are fundamental for prognosis and to unveil novel parameters predicting therapeutic response [4]. Recently, much attention has focused on the impact of microRNA (miRNAs) on tumorigenesis and cancer progression. miRNAs, a class of post-transcriptional regulators, are single strand RNA molecules of about 22-nucleotides in length and regulate gene translation into proteins acting either as tumour suppressor genes or oncogenes, thus affecting mRNA stability and translation [5]. They are transcribed as pri-miRNA precursors and cleaved by Drosha endonuclease in the nucleus, exported into cytoplasm by Exportin-5 as a pre-miRNA, prepared for entry into a large protein complex (RISC) by Dicer endonuclease-removing harping structure [6]. One of the RNA strands, corresponding to mature miRNA, is complementary to mRNA target(s) and 30% of human protein-encoding genes are hypothesized to be regulated by miRNAs. miRNAs were stated as developmental modulators, with an important regulatory role in a broad range of biological processes including development timing, cell differentiation, proliferation, apoptosis, and tumorigenesis [7–9]. Furthermore, there is increasing evidence that abnormalities in certain miRNAs play a role in the molecular mechanism involved in malignant progression, acting as potential regulators of cell motility and/or mediators of cancer spreading [10]. “Oncomir” ectopic expression appears to be associated with transformation, metastatic progression, increased cell viability and proliferation in many solid tumours [11–15]. Other miRNAs, belonging to the tumour suppressor family, have been found down-regulated in a variety of tumours, such as miR-Let-7 in breast cancer [16], miR-335 required for metastasis formation of highly malignant cells [17], and miR-34a, a potent effector of the p53 transcriptional network in uveal melanoma [18]. Many in vitro studies demonstrated that up-regulation of tumour-suppressor miRNAs resulted in a decrease of cell metastatic potential, modulating adhesion, migration, and invasion pathways [19–22]. However, to date very few data are reported in human sarcomas and cell lines [23, 24]. In the present study, microRNA libraries were built and sequenced from two OS cell lines, MG-63(p53−/−) and 143B(p53+/+), with different trans-endothelial migration and proliferation rate, in the effort to identify markers involved in OS progression. We then investigated the role of miR-93, present in both OS cell lines, in primary OS cultures and in clinical samples in relation to cell behaviour.
2 Materials and methods
2.1 Cell line and clinical specimens
Human osteosarcoma cell lines MG-63 (p53−/−), 143B (p53+/+) and Saos-2 (p53−/−), mesenchymal cells and osteoblast cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA; n° CRL-8303, n° CRL-1427 and n° HTB-85, n° CRL-1486 and n° CRL-11372 respectively). Primary OS cell lines IOR/OS10, IOR/OS14, IOR/OS15, IOR/OS18 and IOR/MOS were kindly provided by Dr Massimo Serra (Pharmacogenomics and Pharmacogenetics Research Unit, Rizzoli Institute, Bologna, Italy).
Cells were routinely cultured in humidified atmosphere (5% CO2) using D-MEM medium 10% FBS. Twenty-three primary classic OS samples, 11 paired normal tissues and 10 OS lung metastases were referred to the Rizzoli Orthopaedic Institute. Fresh non-necrotic tissue (≥90% viable tumour cells) was used and diagnosis based on hematoxylin-eosin stained samples according to histopathological criteria.
2.2 FATIMA system for cell migration detection
Cell migration assay was performed as previously described [25]. Cell transmigration kinetic was determined on standard procedure-extracted HUVEC (Human Umbilical Vein Endothelial Cells) previously cultured in M199 Endothelial medium (Cambrex—Rockland, USA) and 20% FBS on 1% gelatine layer (37°C, 5% CO2 humidified atmosphere) by fluorescence plate reader (595 nm, Tecan Spectra Fluor Instrument) at different times.
2.3 Small-RNA library construction
RNA was extracted from MG-63 and 143B osteosarcoma cell lines by Trizol® total RNA isolation reagent (Gibco BRL, Life Technologies, Gaitherburg, MD, USA) according to the manufacturer’s instructions. The two libraries were constructed as previously described by Elbashir et al. [26].
2.4 Sequencing of small-RNA cDNA libraries
Random selected colonies were picked up from ampicillin/kanamicin plate and resuspended in 20 μl of water. Cells were lysed by heating in PCR machine (Gen Amp® PCR System 2700—Applied Biosystem), and DNA was amplified using the following program 95°C/5′, (95°C/30″, 61°C/30″, 72°C/1′) ×30, 72°C/7′, 16°C/∞, Go Taq Polimerase (Promega) and the primers fwd GTT TTC CCA GTC ACG ACG TTG TA and rev CAC AGG AAA CAG CTA TGA CC. The longest amplification products were purified by using Qiaquick Gel Extraction Kit (Qiagen GmbH, Hilden-Germany) and sequenced by DTCS Quick Start Master Mix of Genome Lab Dye Terminator Cycle Sequencing Kit (Beckmann). Termocycling was performed in Gen Amp® PCR System 2700 machine following the program (96°C/20″, 50°C/20″, 60°C/4′) ×30, 16°C/∞. Final products were purified with standard procedure and analyzed in Beckman Coulter CEQTM 2000 DNA Analysis System.
Each sequence identified by Chromas Lite Software was analyzed using megablast software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). After vector and adapter sequence masking and redundancy removing, 20–23 base inserts were compared with the miRNA registry (http://microrna.sanger.ac.uk/sequences) and mapped into human genome by using BLAST human genome (http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html).
2.5 Cloning and transfection of the osteosarcoma cells
miR-93 was inserted in pCDBGW-miR vector by using gateway system. In brief, two complementary strands of 60 nt containing miR-93 were annealed for 4′ at 95°C then ligated overnight in a pENTRY vector. After checking the transformation in DH5α subcloning cells and ligation with PvuII (Promega Corporation, Madison, WI, USA) and BsrgI (New England Biolabs) enzymes, a clonase reaction was performed (LR Clonase—Invitrogen). Final ligation in pCDBGW-miR was tested with MscI restriction enzyme (New England Biolabs). miR-93 vector was transfected in MG-63 wild-type (wt), together with a vector containing G418 resistance in the ratio of 5:1, by using Metafectene reagent (Biontex, Munchen—Germany) and following the manufacturer’s protocol. For insertion in 143B wild-type (wt) cells, pCDBGWmiR-93 retrovirus was prepared by using CaPO4 and HEPES solution following standard procedures and then infection was performed after transfection of a plasmid containing ecotropic receptor. After 2 days, green colonies were picked, cultured and subsequently dispensed in a 96 multi-plate (one cell every three wells) to obtain single cell clones (143Bcl4A, 143Bcl4B, MG-63clB and MG-63clDB). Cells infected with empty vector were used as control.
2.6 miR-93 expression analysis
Reverse transcription and RT-Real Time PCR were carried out following TaqMan MicroRNA Assay Protocol (Applied Biosystems, Foster City, CA) and miR-93 expression was quantified using 2-∆∆CT comparative method (Applied Biosystems, User Bulletin N°2, P/N 4303859) and normalized using RNU6B and RNU44 as endogenous reference (TaqMan miRNA assay n° 001093 and n° 4373384 respectively—PE Applied Biosystems, Foster City, CA). cDNAs from mesenchymal stem cells (MSC) and osteoblasts were used as relative calibrators for OS cell lines and clinical samples. miRNA was considered more expressed where the value of light emission calculated by the method 2-∆∆CT was higher than the value 1.5 SD or less expressed in all those cases in which the value was below 0.5 SD. miRNA average values were obtained from replicates applying the 2-ΔΔCT method.
2.7 mRNA expression analysis of target genes
Reverse transcription of mRNA was carried out in 100 μl final volume from 400 ng total RNA using High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions.
Quantitative RT-PCR was performed using ABI 7900 sequence detection system (Applied Biosystems, Foster City, CA). Expression of target genes ANK2, E2F1 and LATS2 was quantified using TaqMan Expression Assays (Hs00153998_m1, Hs00153451_m1 and Hs00324396_m1 respectively; Applied Biosystems, Foster City, CA) according to manufacturer’s protocol. For calculation of gene expression we used the 2-∆∆CT comparative method. Expression of target genes was normalized to a housekeeping ACTB (Hs99999903_m1 gene; TaqMan Expression Assays-Applied Biosystems, Foster City, CA) and osteoblasts were used as calibrator.
2.8 Immunoblot analysis
According to standard procedures, protein extracts were prepared by mincing and homogenizing fresh samples in extraction buffer (50 mM Tris-HCl at pH 8, 150 mM NaCl, 1 mM DTT, 50 mM NaF, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, 0.1 mM PMFS, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 5 mM pepstatin, 0.1 mM NA3VO4, 1 mM microcystin). 50 microgram of protein extracts were analyzed by 10% SDS-PAGE and Western Blot (WB) was performed by using anti-E2F1 (Santa Cruz Biothech, CA, USA), dil 1:500. The signal was visualized by Supersignal West Pico Chemiluminiscent Substrate (Pierce, Rockford, IL, USA) and quantified by using GS-670 imaging densitometer (BioRad, Hercules, CA, USA). HELA lysate was used as control.
2.9 Proliferation assay
The proliferation assay was performed in a 96-well format using CellTitre 96 One Solution Cell Proliferation Assay (Promega Corporation, Madison, WI, USA). In an individual experiment, proliferation under each condition (0%, 0.1%, 1%, 10% FBS) was studied in duplicate and the whole experiment was repeated at least three times.
2.10 Wound healing assay
2.0 × 104 143B cells and 2.5 × 104 MG-63 cells were seeded in Ibidi Culture Insert in DMEM with 1% of FBS. The plate was incubated overnight at 37°C in humidified atmosphere (5% of CO2). The following day the support was removed, cells were washed with PBS 1× and new DMEM with 1% of FBS was added. To investigate cell migration, the plate was photographed at 0, 8, 24 h with 10× enlargement. The experiment was performed at least three times.
2.11 Migration assay
Cell migration assay was carried out using Transwell Permeable Supports (Corning Incorporated, Corning, NY, USA). Human osteosarcoma cell lines (MG-63 wt, MG-63clB, MG-63clDB, 143B wt, 143Bcl4A, 143Bcl4B) were carefully transferred on the top chamber of each transwell apparatus at a density of 1,000,000 cells/ml (100 μl per chamber). Cells were allowed to migrate for 24 h at 37°C. Cells that had penetrated to the bottom of the membrane were then fixed in methanol, stained using hematoxylin, and countered at microscope. This experiment was performed three times.
2.12 Invasion assay
Cell invasion was analyzed using Cultrex 24 Well BME Cell Invasion Assay (Trevigen, Inc, Gaithersburg, MD) according to standard procedures. Briefly, 103 cells/100 ml serum free media were seeded into the upper well of transwells previously coated with Matrigel basement extract, while 500 μl of media were added in the bottom well. After 24 h of CO2 incubation at 37°C, non-invasive cells on the upper surface were removed and the cells migrated to the lower surface were fixed in 500 μl of cell dissociation solution/Calcein-AM, incubated at 37°C in CO2 for one hour and quantified by fluorimetric analysis (485 nm excitation, 520 nm emission).
2.13 TUNEL assay
10−3 OS cells grown on the slides were washed and fixed with 4% paraformaldehyde in PBS pH 7.4 for 30 min, rinsed with PBS and permeabilized for 2 min on ice (0.1% Triton1 X–100, 0.1% sodium citrate). For the TUNEL technique, all reagents were from in Situ Cell Death Detection Kit (ROCHE Diagnostic, Mannheim, Germany) and procedures followed the manufacturer’s instructions. For negative control, 50 μl Label solution instead of TUNEL reaction mixture was added. Positive control was treated by DNase I recombinant for 10 min at 15–25°C to induce DNA strand breaks, and then 50 μl TUNEL reaction mixture on every sample and on positive controls was added. Samples were incubated for 60 min at 37°C in a dark humidified atmosphere and then rinsed three times with PBS. Finally, we added 50 μl Converter-POD on samples incubated in a humidified chamber for 30 min at 37°C, we rinsed slides three times with PBS, we added 50 μl DAB substrate for 10 min at 15–25°C, we then analyzed samples under light microscope.
2.14 Small interfering RNA pool and transfection
A small interfering RNA (siRNA) pool targeting E2F1 (On-Target Plus SMART Pool, Thermo Scientific Dharmacon, Chicago, USA) was used in 143Bcl4B cell line. For cell growth experiment, cells were seeded in six well plates in triplicate (200.000 cells/well) and transfected 24 h later for 5 h with specific siRNA or control (ctr) siRNA (50 nM) using DharmaFECT (Dharmacon), according to the manufacturer’s protocol. At the end of transfection, fresh medium were placed and 24 h and 48 h later OS cells were recovered with trypsin. As above described RNA and protein extracts were analyzed by RT-PCR and WB to evaluate the efficiency of E2F1 target gene expression down-regulation. The remaining cells were seeded in 96-well flat-bottom plates at 103 cells per well and allowed to attach overnight and proliferation assay was performed.
2.15 Statistical analysis
miRNA expression data are shown as median (m) and 25th–75th percentile for their strong non-Gaussian distribution. Non-parametric two-side Wicoxon’s rank sum tests were performed to compare miRNA expression profile in paired samples; p values were set at <0.05.
3 Results
3.1 Transmigration and proliferation rate of OS cell lines
Cell kinetic was determined for different OS cell lines by FATIMA assay. Differences were found in cell migration. In particular, cells lacking p53, MG-63 and Saos-2, showed a faster trans-endothelial migration with respect to 143B cells expressing wild-type p53. In particular, the kinetic behaviour of MG-63 and Saos-2 was similar, about double per time unit in percentage of migrated cells compared to 143B (≈0.75 vs ≈0.4) (Fig. 1A, B).
On the other hand, OS cell growth rate using different experimental conditions, i.e. different serum concentrations, showed a direct correlation between cell proliferation and serum concentration with a higher proliferation rate of 143B as compared with MG-63 and Saos-2 for up to 72 h at the same conditions (Fig. 2A, B, C).
3.2 Screening of microRNA libraries
A number of miRNAs reported in the miRNA database register were found expressed in MG-63 and 143B OS cell lines (Table 1). Of the 19 miRNAs identified, the majority act as tumour suppressor or oncogenes that control growth and apoptosis. Six out of 19 were expressed in both MG-63 and 143B (Table 1) and included miR-17-5p which regulates cancer cell proliferation, miR-16-1 and −2 that reside within the same cluster of miR-15 controlling anti-apoptotic gene BCL2, miR-130a that regulates expression of homeoboxes GAX and HOXA5, and miR-93, belonging to the miR 17–92 family, that is recognized as an important regulatory factor in various tumours. Other miRNAs were present in either MG-63 or 143B such as miR-21, miR-195, miR-106, miR-20b, miR-24 and miR-323.
A prediction of target genes for known miRNAs was undertaken using software for miRNA targets version as Miranda, PicTar and Target Scan 5.1. In particular, the databases identified 968 candidate targets for miR-16a and -16b including TMEM, SPRED1, KIFB, PLAG1, CCNE1 and BCL2L2. In addition, the most important candidate targets were ATXN1, PFN2, MAP3K2 and MYT1L for miR-17-5p, MI-ERI, MYBL1 and CFL2 genes for miR-130 and LATS2, E2F1, ANK2, ZNFX1, ARID4B, HLF for miR-93.
3.3 miR-93 expression levels in different sarcoma cell lines and clinical specimens
Seeing its importance in different tumour types, we investigated miR-93 expression in sarcoma cell lines using MSC and osteoblasts as calibrators. RT- PCR data showed that 143B cells had respectively a 20- and 40-fold higher miR-93 expression (2-∆∆CT = 21.48 ± 0.90 and 38.05 ± 0.90) when compared with MSC and osteoblasts as reference cells. In MG-63 and SAOS-2, miR-93 levels ranged within normal values, similar to osteoblasts when MSC was taken as calibrator (Fig. 3A). Furthermore, all primary human OS cell lines, specifically IOR/OS14 and IOR/OS15, had higher levels of miR-93 compared to normal human osteoblasts (Fig. 3A).
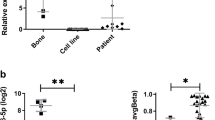
A miR-93 expression in OS cell lines and osteoblasts using MSC as relative calibrator. miRNA average values obtained from replicates applying the 2-∆∆CT method. B Non-Gaussian distribution of miR-93 in OS clinical samples using MSC as relative calibrator. In metastasis specimens miR-93 median values were significantly higher than in primary tumours (Wilcoxon’s text) *p < 0.05
To validate these results we performed RT- PCR in 23 primary OS samples, ten lung metastases and 11 paired normal tissues. Median values were 2.69 (25th–75th = 0.86–4.15), 6.61 (25th–75th = 3.12–23.14) and 1.98 (25th–75th = 0.60–4.01) respectively, and Wilcoxon’s analysis showed a statistically significant miR-93 expression increase in metastases as compared with primary lesions (p = 0.016) (Fig. 3B), while non-significant differences were seen between primary tumours and paired normal tissue.
3.4 miR-93 Target genes expression
mRNA expression of ANK2, E2F1 and LATS2 putative target genes was performed on miR-93 transfected cell clones of both 143B and MG-63 according to 2-∆∆CT comparative method.
The analysis revealed that the E2F1 gene mRNA expression increased in MG-63 and 143B clones (Table 2), the latter showing an evident increase also at protein level as compared to wild-type cell line (Fig. 4).
3.5 Influence of miR-93 on OS cell proliferation, migration and invasive behaviour
Effects of miR-93 on OS cell growth, migration and invasiveness were then tested on infected/transfected cell clones of both 143B and MG-63.
RT-PCR analysis assessed significantly increased miR-93 expression levels in transfected clones when compared with controls (Fig. 5A, B).
miR-93 ectopic expression of significantly increased cell proliferation in 143Bcl4A and 143Bcl4B clones up to 72 h, only slightly affected cell growth of MG-63 clones (Fig. 6A, B).
The wound healing assay performed using serum content restrictions (1%) revealed that ectopic expression of miR-93 induced migration decrease in 143B and MG-63 clones when compared with wild-type and empty vector transfected cells used as controls (Fig. 7A, B). However, migration through polycarbonate membrane and invasion through Matrigel basement membrane showed that miR-93 overexpression reduced 143B cell motility, but significantly increased cell invasiveness in 143B clones 4A and 4B (p = 0.01 and p = 0.04 respectively), without significantly affecting the ability of MG-63 clones to migrate and invade through either membranes (Fig. 8A, B).
3.6 Apoptosis
After miR-93 transfection, 143B clones markedly decreased apoptotic cell fraction measured by TUNEL assay that detects DNA strand breakages in apoptotic cells. No significant variations were seen in MG-63 clones when compared with non-transfected cells (Fig. 9A, B).
3.7 E2F1 siRNA affects 143B cell proliferation
To confirm involvement of E2F1 in the proliferative response of 143Bcl4B to miR-93 overexpression we used siRNA approach to down-regulate E2F1expression. Transfection of siRNA targeting E2F1 markedly reduced its mRNA and protein levels, with a concomitant significant decrease of cell proliferation after 24 h and 48 h from the end of transfection (Fig. 10A, B, C). A slight invasivity decrease (−10%) was also seen. E2F1 expression and proliferative rate were not affected by control siRNA.
Transfection effect of E2F1 siRNA in 143Bcl4B cells incubated for 5 h with siRNA targeting E2F1 or ctr siRNA. A RT-PCR analysis of E2F1. 400 ng total RNA were analyzed 24 h and 48 h from the end of transfection. Osteoblasts were used as calibrator. B Western blot analysis of E2F1. 50 microgram of protein extracts were analyzed 48 h later from the end of transfection. Control loading is shown by actin. HELA lysate was used as control. C Cell growth inhibition after transfection with E2F1 siRNA and control (ctr) siRNA. Each value indicates the average of three independent experiments. **p < 0.01
4 Discussion
miRNA deregulation has been identified in a variety of cancers, where changes in specific miRNA expression may contribute to tumour growth, progression, metastasis, and drug resistance [27]. Deregulation of miRNAs occurs frequently during tumorigenesis [28], making them attractive candidates for molecular detection of malignancy. To date very few studies have been carried out on OS samples and cell lines showing different miRNA expression related to metastatic potential and/or genetic background [29].
In particular, miR-34 and miR-140 affecting target gene expression in a p53 dependent manner induced chemosensitivity in wt-p53 human OS [24, 30].
A very recent study on sarcoma cell lines by Gougelet et al. [31] identified specific miRNAs as discriminating of drug-response using microfluidic PCR technology on TLDA platform preloaded with dehydrated specific primers and probes. In 7 OS samples, using miRNA microarray technology Maire et al. [32] found 38 miRNAs differentially expressed as compared with human osteoblasts.
With a different approach, we identified microRNAs related to a different proliferation and migration behavior by preparing microRNA libraries from OS cell lines which differ in p53 phenotype and in cell growth and migration rate by transwell assay with HUVEC layer. Specifically, MG-63 (p53−/−) and Saos-2 (p53−/−) showed similar transmigration kinetics per time unit, whereas 143B (p53+/+) migrated at about half speed. On the other hand, when proliferation rate was assessed data showed that cell growth of 143B was higher than MG-63 and Saos-2 in all experimental serum conditions. Sequencing analysis identified 19 miRNA, of which 6 were present in both MG-63 and 143B cell lines. Gao et al. [33] identified 25 microRNAs in SOPS-67 osteosarcoma cells. In both studies the same identification methods were used, and the discrepancy of data (19 versus 25) could be due to the loss of less expressed microRNAs during purification processes before cloning reaction.
MiR-21 and miR-106b, considered as the major microRNAs overexpressed in cancers, were expressed in MG-63 but not in 143B. Recent studies showed overexpression of miR-21 in OS tissues and confirmed that miR-21 controls cell adhesion and migration, but not proliferation, through repression of tumour suppressor proteins such as TIMP1, MARCK5 PTEN [34, 35]. Thus, the role of miR-21 appears in conflict with miR-143 that was found to reduce OS viability and metastasis by affecting Bcl2 and matrix metalloproteinases-13 expression [29, 36].
All miRNAs found in the study appear related to cancer development. miR-106b that belongs to the cluster miR-106b-25, is necessary for cell proliferation and anchorage-independent growth [37], miR-17-5p has been characterized as microRNA associated to different cancer types [38], while miR-130a seems to regulate molecule expression inhibiting angiogenesis in vascular endothelial cells [39] and has been found down-regulated in ovarian cancer cell lines resistant to chemotherapy [40]. miR-16-1 and −2 and miR-15 belong to a cluster where deletion or down-regulation appear to be associated with poor prognosis in chronic lymphocytic leukemia [41].
Many expressed miRNAs are “oncomirs” belonging to the 106b-25 microRNA cluster, a polycistronic miRNA highly conserved in all vertebrates, implicated in normal development of the heart, lungs, and immune system and recently found to be highly deregulated in some tumour types [42, 43]. Our attention was fixed on miR-93, recently described as an important molecule during stem cell differentiation [44], involved in different cancer types [45–47], but not yet investigated in sarcoma cells.
RT-PCR specific analysis of miR-93 in the three OS cells showed that MG-63 and Saos-2 cells had miRNA expression similar to that found in osteoblasts when mesenchymal cells were considered as calibrators, whereas a higher expression of miR-93 was detected in 143B, which also showed a higher growth rate. In addition, we assessed miR-93 levels in primary cultures of human OS, and found higher expression levels as compared to osteoblasts. On the basis of these findings, the role of miR-93 in OS cell behaviour was further investigated. In this context, MG-63 and 143B cell lines were transfected/infected by a plasmid containing this miRNA. Two independent cell clones from each OS cell line were obtained with miR-93 expression higher than in wild-type cells, but responding in a different manner to microRNA expression. 143B clones that express p53 oncosuppressor genes increase cell proliferation and viability following miR-93 overexpression more than MG-63, which lack functional p53.
Although there is evidence that hundreds of targets are predicted for each miRNA, recent observations in gastric carcinoma showed that miR-93 cooperate to inhibit p21 post-transcriptional expression through E2F1 regulation, also impairing the TGFb tumour suppressor pathway [45]. It is well known that p21, an inhibitor of cyclin-dependent kinases and proliferating-cell nuclear antigen, is under the transcriptional control of p53 suggesting that p21 might promote p53-dependent cell cycle arrest or apoptosis [48]. Moreover, a recent study demonstrated that knockdown of miR-93 and miR-130b increased p53expression, consistent with the increase of apoptotic rate [46]. Interestingly, we found induction of E2F1 expression following ectopic expression of miR-93 and demonstrated that silencing E2F1 by siRNA significantly decreased cell proliferation as compared to control siRNA transfected cells. Although further studies are needed, these observations suggest an involvement of miR-93 in E2F1 regulation and cell cycle control of osteosarcoma cells.
On the other hand, the role of miR-93 in cell invasion and motility is unclear. Although ectopic miR-93 expression resulted in a slower cell migration through polycarbonate membrane, an increase in cell invasiveness was seen in 143B clones when Matrigel basement coated membrane was used. The observation that in OS metastasis specimens levels of miR-93 were significantly higher than in paired primary tumours might highlight the ability of miR-93 to confer a highly malignant, metastatic phenotype. In conclusion, we demonstrated differences in miRNA profiling of OS cell lines related to a different proliferation and migration behaviour suggesting that up-regulation of miR-93 might be involved in E2F1 transcription factor regulation of p53-positive cells.
Ongoing studies directly addressing the actual targets for miR-93 cluster will be of interest to clarify mechanisms involved in this phenomenon and to better understand how this alteration might contribute to development osteosarcoma.
References
M. Campanacci, Bone and soft tissue tumors, 2nd edn. (Springer, Wien, 1999)
S.S. Bielack, B. Kempf-Bielack, G. Delling, G.U. Exner, S. Flege, K. Helmke, R. Kotz, M. Salzer-Kuntschik, M. Werner, W. Winkelmann, A. Zoubek, H. Jurgens, K. Winkler, Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 20, 776–790 (2002)
M. Wachtel, B.W. Schäfer, Targets for cancer therapy in childhood sarcomas. Cancer Treat. Rev. 36, 318–327 (2010)
J. Toguchida, T. Nakayama, Molecular genetics of sarcomas: applications to diagnoses and therapy. Cancer Sci. 100, 1573–1580 (2009)
R. Schieckel, B. Boyerinas, S.M. Park, M.E. Peter, MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27, 5959–5974 (2008)
S.M. Hammond, Dicing and ilincing: the core machinery of the RNA interference pathway. FEBS Lett. 579, 5822–5829 (2005)
A. Fire, S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, C.C. Mello, Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391, 744–745 (1998)
I. Alvarez-Garcia, E.A. Miska, MicroRNAs functions in animal development and human disease. Development 132, 4653–4662 (2005)
A. Esquela-Kerscher, F.J. Slack, Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 53, 627–635 (2006)
L. Ma, R.A. Weinberg, Micromanagers of malignancy: role of microRNAs in regulating metastasis. Cell 24, 448–456 (2008)
P.M. Voorhoeve, C. le Sage, M. Schrier, A.J. Gillis, H. Stoop, R. Nagel, Y.P. Liu, J. van Duijse, J. Drost, A. Griekspoor, E. Zlotorynski, N. Yabuta, G. De Vita, H. Nojima, L.H. Looijenga, R. Agami, A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumours. Cell 124, 1169–1181 (2006)
Q. Huang, K. Gumireddy, M. Schrier, C. le Sage, R. Nagel, S. Nair, D.A. Egan, A. Li, G. Huang, A.J. Klein-Szanto, P.A. Gimotty, D. Katsaros, G. Coukos, L. Zhang, E. Puré, R. Agami, The microRNAs miR-373 and mir-520c promote tumour invasion and metastasis. Nat. Cell. Biol. 10, 202–210 (2008)
J.A. Chan, A.M. Krichevsky, K.S. Kosik, MicroRNA-21 is an anti-apoptotic factor in human glioblastoma cell. Cancer Res. 65, 6029–6033 (2005)
S.A. Ciafrè, S. Galardi, A. Mangiola, M. Ferracin, C.G. Liu, G. Sabatino, M. Negrini, G. Maira, C.M. Croce, M.G. Farace, Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 334, 1351–1358 (2005)
C. Roldo, E. Missiaglia, J.P. Hagan, M. Falconi, P. Capelli, S. Bersani, G.A. Calin, S. Volinia, C.G. Liu, A. Scarpa, C.M. Croce, MicroRNA expression abnormalities in pancreatic and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 24, 4677–4684 (2006)
F. Yu, H. Yao, P. Zhu, X. Zhang, Q. Pan, C. Gong, Y. Huang, X. Hu, F. Su, J. Lieberman, E. Song, Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007)
S.F. Tavazoie, C. Alarcon, T. Oskarsson, D. Padua, Q. Wang, P.D. Bos, W.L. Gerald, J. Massague, Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008)
D. Yan, X. Zhou, X. Chen, D.N. Hu, X.D. Dong, J. Wang, F. Lu, L. Tu, J. Qu, MicroRNA-34a inhibits uveal melanoma cell proliferation and migration thrugh down-regulation of c-Met. Invest. Ophthalmol. Vis. Sci. 50, 1559–1565 (2009)
P.A. Gregory, A.G. Bert, E.L. Paterson, S.C. Barry, A. Tsykin, G. Farshid, M.A.Y. Vadas, Y. Khew-Goodall, G.J. Goodall, The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008)
M. Korpal, E.S. Lee, G. Hu, Y. Kang, The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of e-cadherin transcriptional repressor ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008)
M. Crawford, E. Brawner, K. Batte, L. Yu, M.G. Hunter, G.A. Otterson, G. Nuovo, C.B. Marsh, S.P. Nana-Sinkam, MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 373, 607–612 (2008)
C. Evangelisti, M.C. Florian, I. Massimi, C. Dominici, G. Giannini, S. Galardi, M.C. Bluè, S. Massalini, H.P. McDowell, E. Messi, A. Gulino, M.G. Farace, S.A. Ciafrè, MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 23, 4276–4287 (2009)
S. Subramanian, W.O. Lui, C.H. Lee, I. Espinosa, T.O. Nielsen, M.C. Heinrich, C.L. Corless, A.Z. Fire, M. van de Rijn, MicroRNA expression signature of human sarcomas. Oncogene 27, 2015–2026 (2008)
C. He, J. Xiong, X. Xu, W. Lu, L. Liu, D. Xiao, D. Wang, Functional elucidation of miR-34 in osteosarcoma cells and primary tumor samples. Biochem. Biophys. Res. Commun. 388, 35–40 (2009)
P. Spessotto, K. Lacrima, P.A. Nicolosi, E. Pivetta, M. Scapolan, R. Perris, Fluorescence-based assays for in vitro analysis of cell adhesion and migration. Methods Mol. Biol. 522, 221–250 (2009)
S.M. Elbashir, W. Lendeckel, T. Tuschl, RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001)
O.A. Kent, J.T. Mendell, A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25, 6188–6196 (2006)
W. Zhang, J.E. Dahlberg, W. Tam, MicroRNAs in tumorigenesis: a primer. Am. J. Pathol. 171, 728–738 (2007)
M. Osaki, F. Takeshita, Y. Sugimoto, N. Kosaka, Y. Yamamoto, Y. Yoshioka, E. Kobayashi, T. Yamada, A. Kawai, T. Inoue, H. Ito, M. Oshimura, T. Ochiya, MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloproteinase-13 expression, Mol. Ther. Mar 22 (2011), [Epub ahead of print], PMID:21427707.
B. Song, Y. Wang, Y. Xi, K. Kudo, S. Bruheim, G.I. Botchkina, E. Gavin, Y. Wan, A. Formentini, M. Kornmann, O. Fodstad, J. Ju, Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28, 4065–4074 (2009)
A. Gougelet, J. Perez, D. Pissaloux, A. Besse, A. Duc, A.V. Decouvelaere, D. Ranchere-Vince, J.Y. Blay, L. Alberti, miRNA Profiling: how to bypass the current difficulties in the diagnosis and treatment of sarcomas, Sarcoma Feb 22(2011), PMID:21437224.
G. Maire, J.W. Martin, M. Yoshimoto, S. Chilton-MacNeill, M. Zielenska, A. Jeremy, Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 204, 138–146 (2011)
J. Gao, T.T. Yang, X.C. Qiu, B. Yu, J.W. Han, Q.Y. Fan, B.A. Ma, Cloning and identification of microRNA from human osteosarcoma cell line SOSP-9607. Ai Zheng 26, 561–565 (2007)
S. Baranwal, S.K. Alahari, MiRNA contro and tumour cell invasion and metastasis. Int. J. Cancer 126, 1283–1290 (2010)
W. Ziyan, W. Shuhua, W. Xiufang, L. Xiaoyun, MicroRNA-p21 is involved in osteosarcoma cell invasion and migration, Med. Oncol. May 18 (2010), [Epub ahead of print] PMID: 20480266.
H. Zhang, X. Cai, Y. Wang, H. Tang, D. Tong, F. Ji, MicroRNA 143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol. Rep. 24, 1363–1369 (2010)
Y. Li, W. Tan, T.W. Neo, M.O. Aung, S. Wasser, S.G. Lim, T.M. Tan, Role of miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 100, 1234–1242 (2009)
C. Volinia, G. Calin, C.G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R.L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C.C. Harris, C.M. Croce, A microRNA expression segnature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103, 2257–2261 (2006)
Y. Chen, D.H. Gorski, Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates anti-angiogenic homeobox genes GAX and HOXA5. Blood 111, 1217–1226 (2008)
M.T. Van Joorsveld, J. Helleman, E.M. Berns, E.A. Wiemer, MicroRNAs in ovarian cancer biology and therapy resistance, Int. J. Biochem. Cell. Biol. (2010) 18 [Epub ahead of print] PMID: 20083225.
G. Calin, A. Cimmino, M. Fabbri, M. Ferracin, S.E. Wojcik, M. Shimizu, C. Taccioli, N. Zanesi, R. Garzon, R.I. Aqeilan, H. Alder, S. Volinia, L. Rassenti, X. Liu, C.G. Liu, T.J. Kipps, M. Negrini, C.M. Croce, Mir-15a e Mir-16-1 clusters function in human leukaemia. Proc. Natl. Acad. Sci. USA 105, 5166–5171 (2008)
A. Ventura, A.G. Young, M.M. Winslow, L. Lintault, A. Meissner, S.J. Erkeland, J. Newman, R.T. Bronson, D. Crowley, J.R. Stone, R. Jaenisch, P.A. Sharp, T. Jacks, Targeted dekletion reveals essential and overlapping functions of the miR-17 trough 92 family of miRNA clusters. Cell 132, 875–886 (2008)
C. Xiao, L. Srinivasan, D.P. Calado, H.C. Patterson, B. Zhang, J. Wang, J.M. Henderson, J.L. Kutok, K. Rajewsky, Lymphoprolferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9, 405–414 (2008)
K.M. Foshay, G.I. Gallicano, Small RNAs, big potential: the role of MicroRNAs in stem cell function. Curr. Stem Cell Res. Ther. 2, 264–271 (2007)
F. Petrocca, R. Visone, M.R. Onelli, M.H. Shah, M.S. Nicoloso, I. de Martino, D. Iliopoulos, E. Pilozzi, C.G. Liu, M. Negrini, L. Cavazzini, S. Volinia, H. Alder, L.P. Ruco, G. Baldassarre, C.M. Croce, A. Vecchione, E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272–286 (2008)
M.L. Yeung, J. Yasunaga, Y. Bennasser, N. Dusetti, D. Harris, N. Ahmad, M. Matsuoka, K.T. Jeang, Roles of MicroRNAs, miR-93 and miR-103b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 68, 8976–8985 (2008)
K.E. Resnick, H. Alder, J.P. Hagan, D.L. Richardson, C.M. Croce, D.E. Cohn, The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 112, 55–59 (2009)
J. Brugarolas, C. Chandrasekaran, J.I. Gordon, D. Beach, T. Jacks, G.J. Hannon, Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377, 552–557 (2002)
Acknowledgements
The authors thank Paola Spessotto for transwell and invasion assays, Arcispedale Santa Maria Nuova for providing umbilical cord, Prof. Paolo Malatesta for technical support, Cristina Ghinelli for the graphic work and Alba Balladelli, B.A. for text revision. This work was supported by grants from the Italian Ministry of Health (MSB, RP), Italian Istituto Superiore della Sanità (MSB, RP), EuroBoNeT Consortium (MSB) and Institutional funds from the University of Parma (RP).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Montanini, L., Lasagna, L., Barili, V. et al. MicroRNA cloning and sequencing in osteosarcoma cell lines: differential role of miR-93. Cell Oncol. 35, 29–41 (2012). https://doi.org/10.1007/s13402-011-0059-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-011-0059-z