Abstract
Currently, it is clear that the luxS gene has an impact on the process of biofilm formation in Campylobacter jejuni. However, even within the species, naturally occurring strains of Campylobacter lacking the luxS gene exist, which can form biofilms. In order to better understand the genetic determinants and the role of quorum sensing through the LuxS/AI-2 pathway in biofilm formation, a set of mutant/complemented strains of C. jejuni 81–176 were prepared. Additionally, the impact of the mutagenic strategy used against the luxS gene was investigated. Biofilm formation was affected by both the presence and absence of the luxS gene, and by the mutagenic strategy used. Analysis by CLSM showed that all mutant strains formed significantly less biofilm mass when compared to the wild-type. Interestingly, the deletion mutant (∆luxS) showed a larger decrease in biofilm mass than the substitution (∙luxS) and insertional inactivated (\(\colon\colon\)luxS) mutants, even though all the mutant strains lost the ability to produce autoinducer-2 molecules. Moreover, the biofilm of the ∆luxS mutant lacked the characteristic microcolonies observed in all other strains. The complementation of all mutant strains resulted in restored ability to produce AI-2, to form a complex biofilm, and to develop microcolonies at the level of the wild-type.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Campylobacter jejuni is a microaerophilic food-borne pathogen capable of entering the human gastrointestinal tract (GIT), particularly after consumption of undercooked meat. After reaching the target area, usually the lower GIT, Campylobacter is able to induce the gastroenteritis known as campylobacteriosis, found world-wide (European Food Safety Authority 2019; Bolton 2015; Vondrakova et al. 2015). During transfer along the food chain, Campylobacter cells have to overcome the hostile conditions of the external environment, including temperature changes and the presence of reactive oxygen species (ROS) formed in aerobic conditions (Gambino and Cappitelli 2016). Additionally, in the host, bacteria must survive the impact of acidic pH in the stomach, which ranges between 1.5 and 5.5, depending on many factors including food intake (Askoura et al. 2016; Varsaki et al. 2015). It is therefore surprising that such fastidious and sensitive bacterium like Campylobacter can endure this spectrum of harsh conditions. However, in its small genome of approximately 1.7 Mbp, a wide range of mechanisms are encoded, allowing Campylobacter to survive, persist, and ultimately induce the disease within a host (Bronowski et al. 2014). One of the survival mechanisms is the ability to form a biofilm, where cells are enclosed and protected in an extracellular matrix (Turonova et al. 2016). The extracellular matrix consists mostly of proteins, polysaccharides, and extracellular DNA (Bae et al. 2014; Brown et al. 2015). Biofilms can be attached on different biotic and abiotic surfaces and represent a reservoir of viable cells, which may be released during the dispersion process and serve as a source of contamination of food products (Reuter et al. 2010). In the structure of the biofilm, cells behave as a multicellular organism via coordinated changes in gene expression. This ability is mediated by the presence of signalling molecules used in the cell-to-cell communication system also known as quorum sensing (QSEmerenini et al. 2015; Li and Tian 2012).
Quorum sensing is based on the production, release, and detection of signalling molecules known as autoinducers (AIs). The production and accumulation of AIs allow the population of bacteria to regulate their behaviour and thus affect important processes, including biofilm formation, motility, or expression of virulence factors (Plummer 2012). In gram-negative bacteria, two predominant types of AIs have been previously described. The first type consists of highly species-specific acyl-homoserine lactones (AHLs), also known as autoinducers-1 (AI-1), produced by AHL synthases (Rickard et al. 2010). The second type of signalling molecule, autoinducer-2 (AI-2), contains a characteristic furanone ring and is produced by S-ribosylhomocysteine lyase (LuxS) within the activated methyl cycle (AMC). This molecule is recognised by both gram-positive and gram-negative bacteria (Xavier and Bassler 2003).
Currently, only the LuxS/AI-2 pathway has been identified in C. jejuni (Elvers and Park 2002). However, unlike Escherichia coli and Vibrio harveyi, C. jejuni is not able to internalize a synthetic AI-2 (Adler et al. 2015). It is therefore possible that C. jejuni may produce and recognise a chemically distinct form of AI-2, or it possesses a specific receptor protein for AI-2 recognition (Adler et al. 2015; Miller et al. 2004).
It is well known that both LuxS and AI-2 have a critical impact on the biofilm formation of C. jejuni (Reeser et al. 2007). However, their exact role in this process has not yet been fully investigated. Therefore, a set of C. jejuni 81–176 luxS mutant strains (deletion (∆luxS), substitution (∙luxS), and insertion (\(\colon\colon\)luxS)), and their complements, were created using cassette mutagenesis. The disruption of AI-2 production was confirmed by bioluminescence assays, and the impact of the LuxS/AI-2 QS pathway and mutagenic strategy used on the process of biofilm formation was investigated.
Material and methods
Bacterial strains and cultivation
All strains used in this work were stored at − 80 °C in 20% glycerol stocks. During genetic manipulations, C. jejuni 81–176 (ATCC® BAA-2151™) was routinely cultivated at 42 °C under microaerobic conditions (5% O2, 10% CO2, 85% N2) in a multi-gas incubator (MCO-18 M, Schoeller, Germany) on the Mueller–Hinton (MH) agar or broth (Hi-Media, India), eventually supplemented either with chloramphenicol (20 µg/mL, Merck, USA) for the cultivation of luxS deletion, substitution and insertion inactivated mutant strains, or kanamycin (50 µg/mL, Merck, USA) for the cultivation of luxS complemented strains. For biofilm formation, C. jejuni was pre-cultivated on Karmali agar (Oxoid, UK). Escherichia coli DH5α (Q5® Site-Direct Mutagenesis Kit, NEB, USA) was cultivated aerobically at 37 °C on the Lysogeny agar (LB-A) or broth (LB; Hi-Media, India), eventually supplemented either with kanamycin (50 µg/mL; bacteria carrying a plasmid with the complementation cassette), chloramphenicol (20 µg/mL; bacteria carrying plasmids containing the cassettes for deletion, substitution or insertion inactivation) or ampicillin (100 µg/mL; bacteria carrying plasmids with partially assembled cassettes before the insertion of chloramphenicol or kanamycin selection marker. For the bioluminescence assay, Vibrio campbellii ATCC® BAA-1117™, designated as Vibrio harveyi BB170 (AI-2 sensitive reporter strain), and V. campbellii ATCC® BAA-1119™ designated as Vibrio harveyi BB152 (AI-2 positive control) were cultivated on the Autoinducer bioassay (AB) agar or broth (ATCC Medium: 2746) aerobically at 30 °C.
Genetic engineering of Escherichia coli DHα and Campylobacter jejuni 81–176
For all cloning experiments, amplification parameters of the specific regions of C. jejuni 81–176 were designed using the FastPCR software (Kalendar et al. 2017). Primers for site-specific mutagenesis with substitution and insertion inactivation cassettes were designed in the NEBaseChanger® (NEB, USA). Genomic DNA of C. jejuni 81–176 was isolated according to the protocol described by He (2011). Competent E. coli DH5α cells were prepared and transformed by routine heat-shock transformation according to the protocol described by Sambrook and Russell (2006). The electro-competent cells of C. jejuni were prepared according to the protocol described by Wassenaar et al. (1993) with the modification of replacing the saponin agar plates with MH agar without supplement. All plasmids used in this study were isolated by the GenElute HP Plasmid Miniprep Kit (Merck, USA). The restriction enzymes used in this study (XbaI, BamHI, NcoI, EcoRI and SacI) were purchased from NEB, USA. Ligase T4 DNA was used for ligations, and Q5® High-Fidelity DNA Polymerase was used for all PCRs (both NEB, USA). The accuracy of assembled cassettes in both E. coli and transformed C. jejuni strains was confirmed by PCR and Sanger sequencing. All PCR programs used in this study are available in Online Resource 1.
Construction of the luxS deletion cassette
To prepare the deletion cassette (Fig. 1; Fig. S1a, Online Resource 1), border sequences of approx. 600 bp in front of and behind the luxS gene were amplified by two primer sets containing specific restriction sites (Set 1 and Set 2; Table 1). The fragments from PCR reactions were purified by Wizard® SV Gel and PCR Clean-Up System (Promega, USA). Subsequently, approximately 1 µg of each product was digested with 10 units (U) of appropriate restriction enzymes, together with 1 µg of plasmid pGEM®-T easy vector (Promega, USA). Digested fragments and plasmids were electrophoretically separated in 1% agarose gel, cut out, and purified using the Wizard kit. The concentration of all purified fragments was measured using Nanophotometer™ (Implen, Germany) and the appropriate ligation ratio (F1:F2:plasmid = 3:3:1) was calculated in silico using NEBioCalculator® (NEB, USA). Ligation was performed according to a protocol provided by the manufacturer of T4 ligase. After an overnight ligation at 16 °C, 5 µl of the ligation mixture was introduced into competent E. coli DH5α by heat shock as described by Sambrook and Russell (2006). Transformed cells were cultivated 24 h at 37 °C on LB agar containing 100 µg/mL of ampicillin. Resulting plasmids were isolated from 10 randomly selected colonies. The assembly of plasmids was verified by restriction analysis with BamHI, as a unique BamHI site was created between the F1 and F2 fragments (Fig. S1a, Online Resource 1). Finally, chloramphenicol acetyltransferase (cat, GenBank: M35190.1) was amplified from plasmid pRY111 (Yao et al. 1993) by primer Set 3 (Table 1). After its purification and BamHI treatment, the cassette was ligated into the created BamHI site, replacing the luxS gene. The final plasmid was transformed into E. coli DH5α as mentioned above. Colonies containing the deletion cassette were selected on the LB-A plate with chloramphenicol (20 µg/mL). The sequence of the deletion cassette was verified by Sanger sequencing using primer Set 4 (Table 1). Data from sequencing are available in Online Resource 1 (Fig. S1b and Fig. S1c).
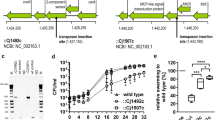
Design of the mutagenic and complementation cassettes of C. jejuni 81–176 and the electropherogram of the confirmation PCR with primer Set 11. (A) wild-type (wt); (B) substitution (∙luxS 256A); (C) insertional inactivation (::luxS); (D) deletion (ΔluxS); (E) complementation (∙luxSc,::luxSc, ΔluxSc). The putative regulation promoter sequence (TATA), the Shine-Dalgarno (SD) sequence, and + 1 as start codon were left intact in all mutants, except for the ΔluxS where the whole luxS gene was removed including the start codon
Construction of the luxS substitution cassette
Substitution cassette (Fig. 1; Fig. S2a, Online Resource 1) was created as follows. A fragment of 1.2 kbp containing the luxS gene with approx. 600 bp in front of and 100 bp behind the gene was amplified by primer Set 5 containing modified regions with restriction sites (Table 1). The PCR product was purified and 1 µg was digested with 10 U of SacI and BamHI, together with plasmid pUC19 (NEB, USA). Products of digestion were then treated as described in the previous chapter, and they were subsequently ligated and transformed into competent E. coli DH5α by heat shock. Transformed cells were cultivated 24 h at 37 °C on LB agar containing 100 µg/mL of ampicillin, 50 µg/mL of IPTG, and 40 µg/mL of X-Gal, which allows the blue-white screening as described by Juers et al. (2012). After cultivation, 10 random white colonies were selected to confirm the presence of the fragment of interest by colony PCR using primer Set 5 (Table 1). A point substitution 256A previously identified by Plummer et al. (2011) was incorporated into the luxS gene using specifically designed primer Set 6 (Table 1) and Site-Direct Mutagenesis Kit (NEB, USA). To confirm the successful substitution, a small region containing the luxS gene was amplified with modified primer Set 7 (Shagieva et al. 2020) and subsequently cloned between the NcoI and EcoRI restriction sites of the pGEM-T easy vector. The substitution was confirmed by Sanger sequencing using the commercially available M13/forward primer (Fig. S2b, Online Resource 1). Finally, the complementary border sequence (approx. 500 bp) located behind the luxS gene was amplified (primer Set 8, Table 1), purified, and digested by BamHI and XbaI. The products of digestion were ligated at a standard ratio of 3:1, incorporating the border sequence behind the first fragment containing the point mutation 256A (Fig. S2a, Online Resource 1). After transformation into E. coli, positive colonies were confirmed by restriction analysis with BamHI and XbaI enzymes. Finally, the cat cassette was cloned between both fragments, into the BamHI site, and positive colonies were selected on the LB-A plates with chloramphenicol (20 µg/mL).
Construction of the cassette for insertion inactivation of luxS
The modified primer Set 9 containing restriction sites for NcoI and EcoRI (Table 1) was used for amplification of the fragment of approx. 1.5 kbp containing the luxS gene with border sequences on both sides. This fragment was purified from the PCR reaction and subsequently digested with NcoI and EcoRI, together with the plasmid pGEM-T easy vector. The products of digestion were then electrophoretically separated, purified, ligated, and transformed into the E. coli DH5α as described in the sections above. Transformed cells were cultivated for 24 h at 37 °C on LB agar containing 100 µg/mL of ampicillin, 50 µg/mL of IPTG, and 40 µg/mL of X-Gal, which allows the blue-white screening. Positive (white) colonies were confirmed by the colony PCR using primer set 9 (Table 1). In silico analysis of the luxS gene revealed a nucleotide sequence similar to the BamHI restriction site, suitably situated close to the middle of the luxS, requiring substitution of three nucleotides. Using the primer Set 10 (Table 1), where the forward primer carried the required substitution, and Q5® Site-Direct Mutagenesis Kit (NEB, USA), the substitution was introduced and the BamHI site was created and confirmed by the restriction analysis. Into the BamHI unique restriction site in the middle of luxS, the amplified cat cassette was cloned as described above, disrupting the luxS gene (Fig. S3a, Online Resource 1). Positive colonies were selected on the LB-A plates with chloramphenicol (20 µg/mL). All modifications were confirmed using Sanger sequencing (Fig. S3b and Fig. S3c, Online Resource 1).
Construction of the luxS complementation cassette
A fragment containing a functional copy of the luxS gene (approx. 1.2 kbp) was amplified (primer Set 11, Table 1), purified, and digested with NcoI and BamHI. Plasmid pGEM-T easy vector containing the complete deletion cassette was digested in the same way as the fragment. After electrophoretic separation in 1% agarose gel, the characteristic band of the pGEM-T easy vector containing the fragment F2 was cut out and purified (Fig. S4b, Online Resource 1), and was subsequently ligated with the functional copy of the luxS gene. Positive colonies were confirmed by colony PCR using primer Set 11 (Table 1). For complementation, the kanamycin resistance cassette from plasmid pRY107 was amplified using the modified primer Set 12 (Table 1; Shang et al. 2016) and was cloned between both fragments into the BamHI site (Fig. S4a, Online Resource 1). Positive colonies were selected on the LB-A plate with kanamycin (50 µg/mL). The correct assembly of the complementation cassette was confirmed through the Sanger sequencing (Fig. S4c, Fig. S4d, and Fig. S4e, Online Resource 1) using commercially available primers M13 and primer SubF (Table 1) to assure a precise sequencing of the long construct.
Transformation of Campylobacter jejuni 81–176 by cassette mutagenesis
The transformation was performed as described by Wassenaar et al. (1993). Briefly, the PCR with primer set 13 was performed for amplification of deletion, substitution, insertion inactivation, and complementation cassettes (1 µg of DNA), which were subsequently electroporated into freshly prepared electro-competent cells of C. jejuni 81–176 using MicroPulser™ (Bio-Rad, Hercules, California, USA) and pre-cooled 0.2 cm cuvettes (Bio-Rad, Hercules, California, USA). For each electroporation experiment, exactly one pulse of 2500 V was used. After electroporation, the cuvette was rinsed two times with 100 µL of pre-warmed MH broth, and each suspension was transferred (without spreading) on a surface of pre-warmed MH agar (without antibiotics) and incubated microaerobically at 42 °C overnight. Subsequently, the culture was harvested with 1 mL of the pre-warmed MH broth and 100 µL aliquots were spread on MH agar plates containing 20 µg/mL of chloramphenicol and incubated for 3–7 days. After the incubation, colonies grown on the selective agar plate were individually inoculated on the surface of fresh antibiotic-supplemented plates and were incubated microaerobically at 42 °C overnight. To confirm the recombination, a part of each grown overnight culture was harvested with a sterile inoculation loop, and its DNA was isolated by thermal lysis. The presence of mutagenic cassettes within the genome was confirmed by PCR (primer Set 13, Table 1). After visualization of products in 1% agarose gel, the remaining cells of positive cultures were collected with pre-warmed MHB containing 20% glycerol and stored at − 80 °C.
To complement the mutant strains, the complementation cassettes were electroporated into electro-competent cells of the respective luxS mutant strains. Positive colonies were selected on the MH agar plates with kanamycin (50 µg/mL).
The presence/absence of the luxS gene was confirmed by PCR (primer Set 14, Table 1). For final sequencing of the incorporated cassettes (mutagenic/complementation), these were amplified from DNA of positive colonies (primer Set 9, Table 1) and cloned into the pGEM®-T easy vector as described above. All sequences were deposited to the GenBank under the following numbers: deletion MN640707; substitution MN738698; insertional inactivation MN738697; and complementation MN711445.
Bioluminescence assay for AI-2
Bioluminescence assay was performed according to the protocol described by Rickard et al. (2006) with some modifications. To prepare the cell-free supernatants (CFSs), one colony of an overnight culture of tested strain was inoculated on a fresh pre-warmed Karmali with appropriate antibiotics (C. jejuni strains) or AB agar (V. harveyi BB152, positive control), and was incubated for 24 h either microaerobically at 42 °C for C. jejuni strains, or aerobically at 30 °C for V. harveyi strain. The cultures were then harvested from plates with 1 mL of pre-warmed MH or AB broth. All cultures were diluted to the OD600 0.1 ± 0.005 and were used to inoculate the MH/AB broth in a ratio of 1:30. The resulting suspensions were then incubated for 24 h either microaerobically at 42 °C for C. jejuni strains, or aerobically at 30 °C for V. harveyi strain. After the incubation, grown cultures were transferred into 50 mL plastic centrifugation tubes and centrifuged at 9000 × g for 10 min in a pre-cooled centrifuge (4 °C). The supernatant was subsequently filtered through a 0.22 µm PES filter (P-Lab, Czech Republic).
The overnight culture of the V. harveyi BB170 (AI-2 sensitive reporter strain) was harvested with 2 mL of AB medium and diluted to the OD600 1.0 ± 0.005. The diluted culture was mixed with the sterile AB medium in a ratio 1:500. Subsequently, 900 µL of the suspension was mixed with 100 µL (10% (v/v)) of the CFS. A parallel with a sterile MH/AB broth instead of CFS served as a negative control, which was used for the normalisation of the background signal. The suspension was briefly homogenised and 100 µL of each sample was transferred to the Corning® Costar 96 Well White Plate (Merck, USA) in triplicate. The bioluminescence was measured in 30-min intervals, 10 s per well, by SpectraMax® i3x Multi-Mode Detection Platform (Molecular Devices, UK) in the kinetic mode for 16 h. The resulting bioluminescence of one biological replicate was interpreted as an average of the relative light units (RLU) of the three technical replicates normalised by subtracting the negative control with sterile MH/AB broth. To avoid the possibility, that the resulted bioluminescence could be influenced by a different growth rates of the tested C. jejuni strains and V. harveyi BB152, colonies were counted according to a protocol described by Miles et al. (1938). Only the data obtained from cultures with comparable growth rates were used for the interpretation of the bioluminescence assay results. The experiment was performed in three independent biological replicates. The statistical significance of the generated numerical data was determined by paired T-test. Results with a p-value lower than 0.05 were considered statistically significant.
Biofilm formation
Biofilms were prepared as described by Turonova et al. (2015) with some modifications. Campylobacters were resuscitated from stock on Karmali agar (with chloramphenicol 20 µg/mL for mutant and kanamycin 50 µg/mL for complemented strains) and incubated microaerobically for 24 h at 42 °C. After cultivation, a single colony of each strain was selected and inoculated on a fresh Karmali plate with the appropriate antibiotics and incubated again for 24 h at 42 °C. The culture was then harvested with 1 mL of pre-warmed MH broth and diluted to the OD600 0.8 ± 0.005. Each well of a 96-well clear-bottom plate (thickness of 190 ± 5 µm; Greiner Bio-one, Germany)) was inoculated with 250 µL of suspension in triplicates and incubated for 5 h at 42 °C. Then, the suspension was carefully replaced with 250 µL of a sterile pre-warmed MH broth and cultivated for another 24 h at 42 °C. After the cultivation, the wells were manually washed by gently removing 150 µL of the suspension and replacing it with 150 µL of sterile and filtrated pre-warmed physiological solution. The washing was repeated 5 times for each well. After the final aspiration of the well content, 50 µL of a physiological solution containing Syto-9 (Thermo Scientific, USA) in a concentration of 1 µL/mL was added to each well. Biofilms were stained for 15 min and subsequently scanned using Olympus IX81F- ZDC2 (Olympus, Japan) confocal scanning laser microscope with a spinning disc (CLSM), equipped with Andor IQ software (Andor, Belfast, UK). The signal was recorded in the green channel (excitation at 488 nm, emission 525 nm) using an objective Clara 10 × as series of z-stacks of horizontal planar images with a z-step selected according to the Nyquist sampling (3.57 μm). The experiment was performed in three independent biological replicates.
Image and statistical analysis
All scans obtained by CLSM were analysed by the software Imaris 7.6.4 (Bitplane, UK). The biofilm structure was rendered using the Easy 3D view with auto-adjustment function to correct pixel intensities. Numerical data and 3D models of the biofilms were generated using the surface generator function. The quantity of the biofilm was evaluated as a volume of biomass stained by Syto-9. The statistical significance of the generated numerical data obtained from the Imaris software was determined by paired T-test. Results with a p-value lower than 0.01 were considered statistically significant.
Results
Preparation of Campylobacter jejuni 81–176 luxS mutants
To evaluate the effect of S-ribosylhomocysteine lyase (LuxS) and the mutagenic strategy on the production of AI-2 and biofilm formation of C. jejuni 81–176, several mutants were prepared. The luxS gene was inactivated by deletion (∆luxS), substitution (∙luxS), and insertion (\(\colon\colon\)luxS). All the mutants were complemented with a functional copy of the luxS gene carried on a complementation cassette and confirmed by PCR. Electropherogram confirmed that the sizes of all amplified cassettes in the luxS mutants are as predicted (Fig. 1). PCR specific for the inner sequence of the luxS, clearly distinguished the mutant with deleted ∆luxS gene and mutant with an enlarged PCR product with cat cassette in the middle of the \(\colon\colon\)luxS (insertion inactivation; Fig. 2). The results of sequencing showed that all cassettes were incorporated correctly, without unwanted mutations. Moreover, the sequencing directly confirmed the absence of the luxS gene in the deletion mutant, the presence of the cat in the middle of the luxS gene in the insertion mutant, and the point mutation ∙256A (A → G in the 256th nucleotide of luxS) in the substitution mutant (Fig. S2b, Online Resource 1). All sequences were deposited to the GenBank database (deletion cassette MN640707; substitution cassette MN738698; insertional inactivation cassette MN738697; complementation cassette MN711445).
Confirmation of C. jejuni 81–176 mutant/complementation strains by amplification of a characteristic 222 bp product formed within the inner region of the luxS by primer Set 12. (A) wild-type (wt); (B) substitution (∙luxS 256A); (C) insertional inactivation (::luxS); (D) deletion (ΔluxS); (E) complementation (∙luxSc,::luxSc, ΔluxSc)
Bioluminescence assay for AI-2
The presence of AI-2 in the tested cell-free supernatant (CFS) was determined by the bioluminescence of the Vibrio harveyi BB170 (reporter strain). The bioluminescence was evaluated as relative light units obtained from the tested sample of CFS in the moment of maximal stimulation of the reporter strain, with a subtraction of background bioluminescence obtained from the negative control (use of sterile cultivation medium instead of CFSs). CFSs prepared from the cultivation of the wild-type, mutant, and complementation strains of C. jejuni 81–176 in MH broth were tested for the presence of AI-2 and compared with a positive control (CFS from AI-2 producing strain Vibrio harveyi BB152). The results of the bioluminescence assay showed that the wild-type of C. jejuni produced significantly more AI-2 molecules (p < 0.05; Fig. 3) in comparison to the deletion (∆luxS), substitution (∙luxS), and insertion (\(\colon\colon\)luxS) mutants, which were not able to stimulate the reporter strain. However, after complementation of the luxS mutants with the functional copy of the luxS gene, the bioluminescence signal of the reporter strain was restored at least to the level of the wild-type (Fig. 3). These results suggest that the prepared luxS mutants of C. jejuni 81–176 are not able to produce the signal molecule AI-2.
Bioluminescence emitted by V. harveyi BB170 after stimulation with CFSs obtained from cultivation of the tested strains in MH broth. WT – C. jejuni 81–176 (wild-type); BB152 – V. harveyi BB152 (positive control); ΔluxS – deletion mutant;::luxS – insertional inactivation; ∙luxS 256A – substitution; ΔluxSc – deletion mutant complemented;::luxSc – insertional inactivation mutant complemented; ∙luxSc – substitution complemented. *statistically significant difference when compared to WT (p < 0.05)
As the quantity of produced QS molecules correlates with the cell density of the bacterial population, the C. jejuni strains were subjected to colony counting, in order to eliminate the influence of differential growth. The results clearly showed that all tested strains had comparable counts of colony-forming units per milliliter (CFU/mL) at the end of the cultivation time (from 9.71 ± 0.07 log10 CFU/mL for the wild-type to 9.87 ± 0.11 log10CFU/mL for the deletion luxS mutant, and 8.25 ± 0.06 log10 CFU/mL for the positive control).
Biofilm formation
The biofilms formed by all tested strains were stained with Syto-9 and the biofilm mass was quantified by CLSM. The results clearly showed that ∆luxS, ∙luxS, and \(\colon\colon\)luxS formed significantly lower amounts of biofilm mass when compared to the wild-type (p < 0.01; Fig. 4). Surprisingly, the ∆luxS mutant formed less biomass than both ∙luxS, and \(\colon\colon\)luxS mutants (p < 0.01; Fig. 4). Additionally, differences in the biofilm structure were observed between the strains. The wild-type C. jejuni 81–176 was able to form clusters of biofilm (Fig. 5). The ∆luxS strain formed only thin and sparse biofilms without the visible clusters observed in the ∙luxS and \(\colon\colon\)luxS mutants (Fig. 6). The complementation of all mutants, in particular the ∆luxSc, restored the production of the biofilm mass to the level of the wild-type. Moreover, after the complementation, increased biofilm mass containing small and medium clusters was observed in the ∆luxSc mutant (Fig. 6). These results suggest that, apart from the presence or absence of the luxS gene, the mutagenic strategy employed against the luxS has a critical impact on the formation of biofilm mass and the formation of microcolonies of C. jejuni 81–176.
Biofilm volume quantified by CLSM after the Syto-9 staining. WT – C. jejuni 81–176 (wild-type); ΔluxS – deletion mutant;::luxS – insertional inactivation; ∙luxS 256A – substitution; ΔluxSc – deletion mutant complemented;::luxSc – insertional inactivation mutant complemented; ∙luxSc – substitution complemented. *statistically significant difference when compared to WT (p < 0.01). **statistically significant difference when compared to ΔluxS (p < 0.01)
Early staged biofilm structure of the luxS mutant/complementation strains after 24 h of cultivation in Mueller Hinton medium (MH) statically in microaerobic conditions at 42 °C. Biofilm was analysed by CLSM after Syto-9 staining. Images of biofilm of ΔluxS C. jejuni 81–176 (deletion)/ ΔluxSc (complementation);::luxS C. jejuni 81–176 (insertional inactivation)/::luxSc (complementation); ∙luxS C. jejuni 81–176 (substitution)/ ∙luxSc (complementation) were processed in Imaris 7.6.4 software
Discussion
The common strategy of C. jejuni for survival and persistence in the external environment is the ability to form a biofilm. Within the biofilm, QS represents an effective way to coordinate the behaviour of the whole population of bacteria via gene expression regulation (Indikova et al. 2015). Since Elvers and Park (2002) described the existence of the luxS gene and AI-2 in C. jejuni, many studies were focused on the investigation of quorum-sensing system impact on different cell processes including biofilm formation, virulence, motility, and survival in the external environment (He et al. 2008; Ligowska et al. 2011; Plummer et al. 2012; Reeser et al. 2007). To explain discrepancies in the results provided by these studies, our work focused on two aspects that influence QS and therefore could affect biofilm formation — the presence/absence of the luxS gene, and the type of mutagenic strategy used to produce the luxS mutant strains. As it was previously suggested that the mutagenic strategy could have an impact on the results of the experiments (Adler et al. 2014), three different methods of genetic engineering were used to inactivate the luxS gene — deletion, substitution, and insertion. Indeed, even though none of the prepared luxS mutants was able to produce the signalling molecule AI-2, the deletion mutant formed significantly less biofilm than the substitution and insertion mutants. Also, the biofilm volume of all three mutants was significantly lower compared to the wild-type. A similar discrepancy was observed in the biofilm architecture, as biofilms of the deletion mutant lacked the visible clusters observed in other strains. These results suggest that apart from the presence/absence of the luxS gene itself, the mutagenic strategy used can also impact the biofilm formation and clustering ability of C. jejuni 81–176. In regards to biofilm structure, similar results were published by Balestrino et al. (2005), where the deletion luxS mutant of Klebsiella pneumoniae showed reduced ability to form microcolonies in the early stages of the biofilm formation. In addition, the luxS deletion mutant of Shewanella oneidensis formed an undifferentiated biofilm compared to the wild-type, which formed compact aggregates on the glass surface (Bodor et al. 2011). On the other hand, the deletion luxS mutant of Enterococcus faecalis formed distinct and complex clusters, opposite to the wild-type whose biofilm was more confluent (He et al. 2016). Insertion-inactivated luxS in Streptococcus oralis led to disruption of mutualistic interrelation with Actinomyces naeslundii resulting in a significant decrease of biofilm formation, which was restored after genetic or chemical (AI-2) complementation (Rickard et al. 2006). As described in the work of Golz et al. (2012), not all Campylobacter species encode the luxS gene, and the ability to form a biofilm differs between the strains of C. jejuni also due to different genetic backgrounds (Pascoe et al. 2015). Similarly, Shagieva et al. (2020) demonstrated that two water isolates of C. jejuni missing luxS gene formed significantly lower levels of biofilm biomass in comparison to other water isolates where the luxS gene was detected. In a different study, the deletion luxS mutant of C. jejuni M129 formed less biofilm mass than the wild-type, but its biofilm formation capacity was increased by the addition of prepared CFS containing the signal molecule AI-2. A similar effect was observed after treatment with CFS prepared from Pseudomonas aeruginosa and Arcanobacterium pyogenes BBR1, while the addition of CFS prepared from Clostridium perfringens and Chromobacterium violaceum did not affect biofilm formation of C. jejuni M129 (Reeser et al. 2007). The molecule of AI-2 is known as a universal signalling molecule (Bassler 2002), but at this time only a few forms of this molecule were identified and described (Chen et al. 2002; Miller et al. 2004). It seems that in the case of C. jejuni, the recognition of AI-2 can be dependent on a specific form of this molecule, as C. jejuni was not able to internalise commercially purchased AI-2 (Adler et al. 2015). According to a mathematical model provided by Emerenini et al. (2015), the translocation of QS molecules within the biofilm allows bacteria to control its size, structure, and dispersion of cells from the matured biofilm. The model was supported by a study performed by Auger et al. (2006), who treated Bacillus cereus with AI-2 extracted from Photorhabdus luminescens. Co-cultivation of Bacillus cells with the AI-2 inhibited its biofilm formation, while the addition of AI-2 to preformed biofilms led to the dispersion of cells from the structure of the biofilm (Auger et al. 2006).
It is important to mention that the proper functionality of quorum sensing is not solely based on the production of autoinducers but requires an appropriate sensor and transport mechanisms allowing the excretion of synthesised molecules to the extracellular space. In C. jejuni, a small non-coding RNA (CjNC110) located downstream of the luxS gene is described. Deletion of CjNC110 resulted in decreased levels of extracellular AI-2, while intracellular levels of AI-2 were significantly increased. Results suggest that CjNC110 may participate in the transport of AI-2 in C. jejuni (Kreuder et al. 2020). However, further research is needed to explain the details of quorum sensing machinery in C. jejuni.
Conclusion
This study clearly shows that the mutagenic strategy affects the results of experiments focused on the functionality of the luxS gene and should therefore be taken into consideration in any further research. Even though no mutant strain was able to produce a detectable amount of AI-2 and all formed lower levels of biofilm than wild-type, the volume and ultrastructure of biofilms differed between the particular mutants. As the deletion mutant formed meagre biofilm without distinct microcolonies, the process of biofilm formation is likely affected not only by the concentration of AI-2, but also by the presence of the gene in the genome. However, further research is needed to explain this phenomenon.
Data and materials availability
Datasets generated during the current study are available from the corresponding author on request.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
Datasets generated during the current study are available from the corresponding author on request.
Code Availability
This article does not contain any studies with human participants or animals performed by any of the authors.
References
Adler L, Alter T, Sharbati S, Golz G (2014) Phenotypes of Campylobacter jejuni luxS mutants are depending on strain background, kind of mutation and experimental conditions. PLoS ONE 9:e104399. https://doi.org/10.1371/journal.pone.0104399
Adler L, Alter T, Sharbati S, Golz G (2015) The signalling molecule autoinducer-2 is not internalised in Campylobacter jejuni. Berl Munch Tierarztl Wochenschr 128:111–116
Askoura M, Sarvan S, Couture JF, Stintzi A (2016) The Campylobacter jejuni ferric uptake regulator promotes acid survival and cross-protection against oxidative stress. Infect Immun 84:1287–1300
Auger S, Krin E, Aymerich S, Gohar M (2006) Autoinducer 2 affects biofilm formation by Bacillus cereus. Appl Environ Microbiol 72(1):937–941
Bae J, Oh E, Jeon B (2014) Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrobial Agents Chemotherapy 58:7573–7575. https://doi.org/10.1128/AAC.04066-14
Balestrino D, Haagensen JA, Rich C, Forestier C (2005) Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J Bacteriol 187:2870–2880. https://doi.org/10.1128/JB.187.8.2870-2880.2005
Bassler BL (2002) Small talk. Cell-to-Cell Communication in Bacteria Cell 109:421–424
Bodor AM, Jansch L, Wissing J, Wagner-Dobler I (2011) The luxS mutation causes loosely-bound biofilms in Shewanella oneidensis. BMC Res Notes 4:180. https://doi.org/10.1186/1756-0500-4-180
Bolton DJ (2015) Campylobacter virulence and survival factors. Food Microbiol 48:99–108
Bronowski C, James CE, Winstanley C (2014) Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol Lett 356:8–19. https://doi.org/10.1111/1574-6968.12488
Brown HL, Reuter M, Hanman K, Betts RP, van Vliet AH (2015) Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of Campylobacter jejuni. PLoS ONE 10:e0121680. https://doi.org/10.1371/journal.pone.0121680
EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) (2019) The European Union one health 2018 zoonoses report. EFSA J 2019;17(12):5926, 276 pp
Elvers KT, Park SF (2002) Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475–1481. https://doi.org/10.1099/00221287-148-5-1475
Emerenini BO, Hense BA, Kuttler C, Eberl HJ (2015) A mathematical model of quorum sensing induced biofilm detachment. PLoS ONE 10:e0132385. https://doi.org/10.1371/journal.pone.0132385
Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415(6871):545–549
Gambino M, Cappitelli F (2016) Mini-review: Biofilm responses to oxidative stress. Biofouling 32:167–178. https://doi.org/10.1080/08927014.2015.1134515
Golz G, Adler L, Huehn S, Alter T (2012) LuxS distribution and AI-2 activity of Campylobacter spp. J Appl Microbiol 112(3):571–578
He F (2011) E Coli Genomic DNA Extraction. Bio-Protocol 1:e97. https://doi.org/10.21769/BioProtoc.97
He Y, Frye JG, Strobaugh TP, Chen CY (2008) Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni strain 81–176. Foodborne Pathogens Disease 5:399–415. https://doi.org/10.1089/fpd.2008.0106
He Z, Liang J, Zhou W, Xie Q, Tang Z, Ma R, Huang Z (2016) Effect of the quorum-sensing luxS gene on biofilm formation by Enterococcus faecalis. Eur J Oral Sci 124(3):234–240
Indikova I, Humphrey TJ, Hilbert F (2015) Survival with a helping hand: Campylobacter and microbiota. Front Microbiol 6:1266. https://doi.org/10.3389/fmicb.2015.01266
Juers DH, Matthews BW, Huber RE (2012) LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21(12):1792–1807
Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI (2017) FastPCR: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319. https://doi.org/10.1016/j.ygeno.2017.05.005
Kreuder AJ, Ruddell B, Mou K, Hassall A, Zhang Q, Plummer PJ (2020) Small noncoding RNA CjNC110 influences motility, autoagglutination, AI-2 localization, hydrogen peroxide sensitivity, and chicken colonization in Campylobacter jejuni. Infect Immun 88(7):e00245-e220
Li Y-H, Tian X (2012) Quorum sensing and bacterial social interactions in biofilms. Sensors (basel, Switzerland) 12:2519–2538. https://doi.org/10.3390/s120302519
Ligowska M, Cohn MT, Stabler RA, Wren BW, Brondsted L (2011) Effect of chicken meat environment on gene expression of Campylobacter jejuni and its relevance to survival in food. Int J Food Microbiol 145(Suppl 1):S111-115. https://doi.org/10.1016/j.ijfoodmicro.2010.08.027
Miles AA, Misra SS, Irwin JO (1938) The estimation of the bactericidal power of the blood. Journal of Hygiene (lond) 38:732–749
Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM (2004) Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 15:677–687. https://doi.org/10.1016/j.molcel.2004.07.020
Pascoe B, Meric G, Murray S, Yahara K, Mageiros L, Bowen R, Jones NH, Jeeves RE, Lappin-Scott HM, Asakura H, Sheppard SK (2015) Enhanced biofilm formation and multi-host transmission evolve from divergent genetic backgrounds in Campylobacter jejuni. Environ Microbiol 17(11):4779–4789
Plummer P, Sahin O, Burrough E, Sippy R, Mou K, Rabenold J, Yaeger M, Zhang Q (2012) Critical role of LuxS in the virulence of Campylobacter jejuni in a guinea pig model of abortion. Infect Immun 80:585–593. https://doi.org/10.1128/IAI.05766-11
Plummer P, Zhu J, Akiba M, Pei D, Zhang Q (2011) Identification of a key amino acid of LuxS involved in AI-2 production in Campylobacter jejuni. PLoS ONE 6:e15876. https://doi.org/10.1371/journal.pone.0015876
Plummer PJ (2012) LuxS and quorum-sensing in Campylobacter. Front Cell Infect Microbiol 2:22. https://doi.org/10.3389/fcimb.2012.00022
Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA (2007) Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol 73:1908–1913. https://doi.org/10.1128/AEM.00740-06
Reuter M, Mallett A, Pearson BM, van Vliet AH (2010) Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 76:2122–2128. https://doi.org/10.1128/AEM.01878-09
Rickard AH, Colacino KR, Manton KM, Morton RI, Pulcini E, Pfeil J, Rhoads D, Wolcott RD, James G (2010) Production of cell-cell signaling molecules by bacteria isolated from human chronic wounds. J Appl Microbiol 108(5):1509–1522
Rickard AH, Palmer RJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE (2006) Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 60(6):1446–1456
Sambrook J, Russell DW (2006) Preparation and transformation of competent E. coli using calcium chloride. Cold Spring Harbor Protocols 2006:pdb.prot3932 doi:https://doi.org/10.1101/pdb.prot3932
Shagieva E, Teren M, Michova H, Strakova N, Karpiskova R, Demnerova K (2020) Adhesion, biofilm formation, and luxS sequencing of Campylobacter jejuni isolated from water in the Czech Republic. Frontiers in Cellular and Infection Microbiology 10(707)
Shang Y, Ren F, Song Z, Li Q, Zhou X, Wang X, Xu Z, Bao G, Wan T, Lei T, Wang N, Jiao X, Huang J (2016) Insights into Campylobacter jejuni colonization and enteritis using a novel infant rabbit model. Sci Rep 6:28737. https://doi.org/10.1038/srep28737
Turonova H, Briandet R, Rodrigues R, Hernould M, Hayek N, Stintzi A, Pazlarova J, Tresse O (2015) Biofilm spatial organization by the emerging pathogen Campylobacter jejuni: comparison between NCTC 11168 and 81–176 strains under microaerobic and oxygen-enriched conditions. Front Microbiol 6:709. https://doi.org/10.3389/fmicb.2015.00709
Turonova H, Neu TR, Ulbrich P, Pazlarova J, Tresse O (2016) The biofilm matrix of Campylobacter jejuni determined by fluorescence lectin-binding analysis. Biofouling 32:597–608. https://doi.org/10.1080/08927014.2016.1169402
Varsaki A, Murphy C, Barczynska A, Jordan K, Carroll C (2015) The acid adaptive tolerance response in Campylobacter jejuni induces a global response, as suggested by proteomics and microarrays. Microb Biotechnol 8:974–988. https://doi.org/10.1111/1751-7915.12302
Vondrakova L, Purkrtova S, Pazlarova J (2015) Demnerova K (2015) Species differentiation of thermotolerant Campylobacters based on distinctive banding patterns obtained by multiplex PCR. Czech Journal of Food Sciences 33:27–31
Wassenaar TM, Fry BN, van der Zeijst BA (1993) Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131–135
Xavier KB, Bassler BL (2003) LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol 6:191–197. https://doi.org/10.1016/s1369-5274(03)00028-6
Yao R, Alm RA, Trust TJ, Guerry P (1993) Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130
Acknowledgements
We would like to thank Ruth Ketley and Jana Terenova, both from the University of Oxford, for language corrections and critical comments on the manuscript. We also thank professor Elzbieta Katarzyna Jagusztyn-Krynicka from the University of Warsaw for providing us the plasmid pRY107 used in this study. The work was financially supported by The Czech Science Foundation through Project No. 18-16549S.
Funding
The work was financially supported by The Czech Science Foundation through Project No. 18-16549S.
Author information
Authors and Affiliations
Contributions
Conceptualization: [Martin Teren]; Methodology: [Martin Teren]; Formal analysis and investigation: [Martin Teren, Ekaterina Shagieva, Viviana Svarcova]; Writing – original draft preparation: [Martin Teren]; Writing – review and editing: [Hana T. Michova, Lucie Vondrakova, Jitka Viktorova, Katerina Demnerova]; Funding acquisition: [Katerina Demnerova]; Resources: [Hana T. Michova, Lucie Vondrakova, Jitka Viktorova, Katerina Demnerova]; Supervision: [Katerina Demnerova].
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Agnieszka Szalewska-Palasz
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Teren, M., Shagieva, E., Vondrakova, L. et al. Mutagenic strategies against luxS gene affect the early stage of biofilm formation of Campylobacter jejuni. J Appl Genetics 63, 145–157 (2022). https://doi.org/10.1007/s13353-021-00655-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-021-00655-y