Abstract
In the present study, we made further investigation into the diversity of Trichoderma in China than previous ones utilizing comprehensive approaches of morphological microscopic observation and phylogenetic analysis by detecting molecular markers. One thousand nine hundred ten Trichoderma strains were isolated from soil or other materials in China: East (Anhui, Fujian, Jiangsu, Jiangxi, Shandong, Zhejiang province and Shanghai municipality), South-West (Guizhou, Qinghai, Shanxi, Sichuan and Yunnan province, Tibet Autonomous Region and Chongqing municipality), South-East (Guangdong, Guangxi, Hainan province), and Middle China (Henan, Hubei and Hunan province). Representative isolates were verified at the species level by morphological characters and the oligonucleotide barcode program TrichoOKey v.10 and the custom BLAST server TrichoBLAST, using sequence of the ITS 1 and 2 region of the rDNA cluster and partial sequences of translation elongation factor 1-alpha(tef1-α). A total of 23 Trichoderma species were identified : T.asperellum, T.atrioviride, T.aureovriride, T.brevicompactum, T.citrioviride, T.erinaceum, T.gamsii, T.hamatum, T.harzianum (H.1ixii), T.intricatum, T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.pleuroticola, T.reeseii (H.jecorina), T.sinensis, T.spirale, T.stromaticum, T.tomentosum, T.velutinum, T.vermipilum, T.virens (H.virens), T.viride. Among them, 3 species: T.intricatum, T.stromaticum, T.vermipilum were first reported in China; T.harzianum (H,1ixii) was the most widely distributed species in China. This study further shows that, the highest biodiversity of Trichoderma population appeared in South-West China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As cosmopolitan soil-borne fungus genus, Trichoderma (Ascomycetes, Hypocreales) are remarkable for their rapid growth, capability of utilizing diverse substrates and resistance to noxious chemicals (Klein and Eveleigh 1998). In terms of the ecological importance of their production of enzymes and antibiotics (Kubicek and Penttilä 1998; Sivasithamparam and Ghisalberti 1998) and its application as a biocontrol agent against plant diseases (Hjeljord and Tronsmo 1998; Harman et al. 2004; Bailey et al. 2006), it is vital to identify accurately Trichoderma species and understand its biodiversity and biogeography.
Until recently, most of the known species have been isolated from China, over 40 Trichoderma species have been identified in China (Wen et al. 1992; Wen et al. 1993; Zhang et al. 1996; Luo and Wen 2003; Zhang and Xu 2004; Zhao et al. 2004; Zhang and Xu 2005; Sun et al. 2006a, b; Yu et al. 2007; He et al. 2008; Shao et al. 2008; Jia et al. 2009; Gao et al. 2007; Wu et al. 2008; Yuan et al. 2008; Xia and Chen 2009; He et al. 2010a, b; Li et al. 2010; Pan et al. 2010; Yu et al. 2010). Among them, 21 species of Trichoderma which are isolated from soil samples in different ecological environments (Wen et al. 1992; Zhang et al. 1996; Luo and Wen 2003; Zhao et al. 2004; Sun et al. 2006a, b; Jia et al. 2009; Pan et al. 2010; Yu et al. 2010). Furthermore, nine Trichoderma species are obtained from different spawn and fruiting bodies of edible fungi based on morphological and colony characters (Gao et al. 2007; He et al. 2008; Shao et al. 2008; Wu et al. 2008). Besides, some species of Trichoderma are isolated from rhizosphere of plants (Ze et al. 2007; Yuan et al. 2008).
However previous studies in China lack systemic survey to Trichoderma population in East, South-East, South-West and Middle China, particularly in Sourth-East and South-West where the most regions are characterized with the most abundant biodiversity led by warm and moist climates, which thereby allow Trichoderma massive reproduction. The purpose of the present study was on a large scale to isolate Trichoderma from forest, garden and vegetable soil and other materials, and then to identify Trichoderma species and understand their geographic distribution state in China. The study would lead us to more reasonable exploration of Trichoderma sources, and also reveal partially the status of farming soil micro-ecological quality in China.
Materials and methods
Geography of sample sites
Trichoderma spp. were sampled in 20 regions of East, South-East, South-West and Middle China, which differ in their geographic location, altitude and climate: East (Anhui, Fujian, Jiangsu, Jiangxi, Shandong, Zhejiang province and Shanghai municipality), South-West (Guizhou, Qinghai, Shanxi, Sichuan and Yunnan province, Tibet Autonomous Region and Chongqing municipality), South-East (Guangdong, Guangxi, Hainan province), and Middle China (Henan, Hubei and Hunan province).
Substrates, storage and isolation of pure cultures
Samples of soil were kept in sterile polyethylene bags or kraft envelopes and stored at 4 °C until isolation. The soil dilution plating method was applied. PDAm (S Vargas Gil et al. 2009) was used as a selective medium for isolating Trichoderma. Putative Trichoderma colonies were purified by two rounds of subculturing on potato-dextrose agar (PDA) at 28 °C. Pure cultures were kept in 20 % (w/v) glycerol at -20 °C.
Morphological analysis
For morphological analysis, strains were grown on special nutrient agar (SNA), on 2 % (w/v) cornmeal dextrose agar (CMD), on 2 % (w/v) cornmeal sucrose agar (CMA), on 2 % (w/v) malt agar (MA), and on PDA at 25 °C under ambient daylight conditions for 7 days. Trichoderma species were identified according to Gams and Bissett (1998) and Samuels et al. (2002, 2009; http://nt.arsgrin.gov/taxadescriptions/keys/TrichodermaIndex.cfm).
PCR amplification and DNA sequencing
Mycelium for DNA extraction was obtained on PD through 3-day incubation at 28 °C on a rotary shaker (180 rpm). Mycelium was collected on filter paper in a Buchner funnel, washed with sterile water, frozen at -20 °C, and freeze-dried. Total DNA was extracted using the CTAB method (Doohan et al. 1998). Primary identification was based on the sequencing of internal transcribed spacer regions 1 and 2 (ITS1 and ITS2) of the rRNA gene cluster. In case of failure with ITS 1 and ITS 2 to take unambiguous species identification, we also sequenced a fragment of the translation-elongation factor 1-alpha (tef1-α) gene. The ITS region of the rDNA of 1910 isolates was amplified using primers ITS4, ITS5 (White et al. 1990). A fragment of tef1-α gene containing the 4th and 5th introns was amplified using the primers Ef728M (Carbone and Kohn. 1999) and tef1R (Kullnig-Gradinger et al. 2002). The PCR reaction was carried out in a 25-μl reaction mixture containing the following: 1 μl 50 ng/μl of DNA, 2.5 μl 10 × PCR buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl, pH 8.8, 0.1 % Triton X-100), 1.5 μl 10 mM dNTP Fermentas), 0.2 μl 100 mM of each primer, 19.35 μl MQ H2O, 0.25 μl (2 U/μl) DyNAzymeTM II DNA Polymerase (Fermentas). Amplifications were performed in a Eppendorf Mastercycler pro Gradient thermal cycler (Shanghai Jiaotong University, China) under the following conditions: initial denaturation 5 min at 94 °C, 35 cycles of 60 s at 94 °C, 45 s at 50 °C (for the ITS region), or 55 °C (for the tef1-α fragment), 50 s at 72 °C, with the final extension of 7 min at 72 °C. Amplification products were separated on 1.2 % agarose gel (Invitrogen) in 1 × TBE buffer (0.178 M Tris-borate, 0.178 M boric acid, 0.004 M EDTA) and stained with ethidium bromide. The 10-μl PCR products were combined with 2 μl of loading buffer (0.25 % bromophenol blue, 30 % glycerol). A 100-bp DNA Ladder Plus (Fermentas) was used as a size standard. PCR products were electrophoresed at 4 V·cm-1 for about 1.5 h, visualized under UV light, and photographed (Syngene UV visualizer). The 3-μl PCR products were purified with exonuclease I and shrimp alkaline phosphatase according to Chełkowski et al. (2003). The amplicons were sequenced with the aid of a MegaBACE 1000 DNA automatic sequencing system (Pharmacia), using cycle-sequencing with the DYEnamic ET Dye Terminator Cycle Sequencing Kit (Pharmacia). Sequences were edited and assembled using Chromas v.1.43 (Applied Pharmacia). CLUSTALW (Thompson et al. 1994) and MUSCLE (Edgar 2004) were used to align the sequences; the resulting alignments were inspected and refined manually.
Phylogenetic analysis
For species identification, ITS1 and ITS2 sequences were submitted to the BLAST interface in NCBI (http://blast.ncbi.nlm.nih.gov/) and TrichOKEY (http://www.isth.info; Druzhinina et al. 2005; Druzhinina and Kubicek 2005). In ambiguous cases, the result was rechecked using NCBI (http://blast.ncbi.nlm.nih.gov/) and the TrichoBLAST program based on tef1-α gene sequences (Druzhinina and Kopchinskiy 2004a, b). The NCBI GenBank accession numbers for some sequences obtained in this study are given in Table 1.
For phylogenetic analyses in MEGA4.0 (Tamura et al. 2004), 93 isolates included in 23 Trichoderma species, were selected randomly in different geographical locations. Both ITS1, ITS2 and tef1-α gene sequences were analyzed using the maximum parsimony (Eck and Dayhoff 1966) approach of close-neighbor-interchange algorithm with search level 3 (Nei and Kumar 2000), in which the initial trees were obtained with the random addition of sequences (1000 replicates). In total, there were 435 parsimony informative positions retained from an initial alignment of 996 for the ITS1, ITS2 sequences and 840 positions in the final dataset, of which 786 were parsimony informative for tef1-α gene sequences. In both cases, to infer the consensus, phylogenetic trees bootstrapping with 1000 data replicates was conducted (Felsenstein 1985).
A five-point sampling method was used. We collected soil core from each of the four corners and a center of the given field (10 m × 10 m) by using a hand probe, then combined soil cores, these soil core samples were combined and mixed evenly, and took the central part of each combined sample into bags. The calculation formula of Margalef Species richness index: dMa = (S-1)/lnN (S: the total number of species in the community, i.e., species richness) (Hill et al. 2003).
Results
Species identification
A total of 1910 strains of Trichoderma were isolated from 491 soil samples collected from 18 provinces and two municipalities in China and then identified by using morphological and molecular methods. All Trichoderma strains were identified at species level by the analysis of ITS rDNA and translation-elongation factor 1-alpha (tef1-α) gene sequences. Identifications of species were completed by searching the BLAST interface in TrichOKEY, TrichoBLAST (http://www.isth.info) and NCBI. Those strains belonging to 23 taxa were eventually classified into species as follows: Trichoderma asperellum (430 isolates), Trichoderma atrioviride (25), Trichoderma aureovriride (14), Trichoderma brevicompactum (101), Trichoderma citrioviride (15), Trichoderma erinaceum (10), Trichoderma gamsii (1), Trichoderma hamatum (346), Trichoderma harzianum (Hypocrea 1ixii) (443), Trichoderma intricatum (1), Trichoderma koningii (Hypocrea koningii) (53), Trichoderma koningiopsis (55), Trichoderma longibranchiatum (59), Trichoderma pleuroticola (22), Trichoderma reeseii (Hypocrea jecorina) (1), Trichoderma sinensis (1), Trichoderma spirale (54), Trichoderma stromaticum (3), Trichoderma tomentosum (1), Trichoderma velutinum (4), Trichoderma vermipilum (1), Trichoderma virens (Hypocrea virens) (115), Trichoderma viride (155). The most abundant species (23.2 %) isolated from all regions was T.harzianum (H.1ixii). Besides, T.intricatum, T.stromaticum, T.vermipilum were first reported in China.
Phylogenetic analysis
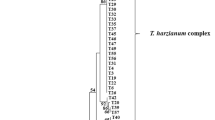
The phylogenetic relation of 93 regional representative Trichoderma isolates was constructed based on the analysis of ITS1 and ITS2 sequences, tef1-α sequences (Figs. 1, 2). According to the ITS tree, the Harzianum clade, with T.harzianum (H.1ixii), T.stromaticum, T.tomentosum, T.velutinum; the Longibrachiatum Clade, with T.citrinoviride, T.sinensis and T.longibrachiatum, T.reeseii (H.jecorina); the Viride Clade, with T.atrioviride, T.erinaceum, T.gamsii, T.koningii (H.koningii), T.koningiopsis, T.viride; the Pachybasium A Clade, with T.asperellum, T.hamatum, T.aureovriride, T.brevicompactum, T.intricatum, T.spirale, T.vermipilum and T.virens (H.virens) were distinguished in a single moderately supported branch with bootstrap support of 80 %. 569 Trichoderma strains were identified as T.harzianum (H.1ixii), but this species is known to include several ITS alleles (Hermosa et al. 2004; Migheli et al. 2009) and is considered to be a species complex (Chaverri et al. 2003). In the present research, 12 haplotypes of T.harzianum (H.1ixii) were found (Fig. 1). With bootstrap support of only 53 %, 12 haplotypes of T.harzianum (H.lixii) formed a moderately well-supported (80 %) clade with T.aureovriride, T.koningiopsis, T.pleuroticola, T.tomentosum and an unresolved polytomy with T.spirale, T.virens (H.virens). Two groups were distinguished within the Longibrachiatum clade with moderate to good bootstrap support. One group, with a bootstrap value of 70 %, contains four strains of T.longibrachiatum. The second group, with a bootstrap value of 73 % included one strain of T.longibrachiatum and one strain of T.reeseii (H.jecorina). Sixteen strains of Trichoderma belonging to the Viride clade, formed a polytomy.
Phylogenetic tree of the 93 regional representative Trichoderma isolates inferred by parsimony analysis of ITS1, ITS2 sequences. Sequences obtained during this study are listed by their GenBank numbers in Table 1. The numbers given over branches indicate bootstrap coefficient >50 %
Phylogenetic tree of the 93 regional representative Trichoderma isolates inferred by parsimony analysis of tef1-α sequences. Sequences obtained during this study are listed by their GenBank numbers in Table 1. The numbers given over branches indicate bootstrap coefficient >50 %
A phylogenetic analysis based on tef1-α sequences indicated four groups were distinguished within the Longibrachiatum clade with moderate to good bootstrap support (Fig. 2). One group, with a bootstrap value of 68 % includes one strain of T.longibrachiatum and one strain of T.reeseii (H.jecorina). The second group, with a bootstrap value of 100 %, contains two strains of T.citrinoviride. The third group, with a bootstrap value of 61 %, contains two strains of T.longibrachiatum. The fourth group, with a bootstrap value of 100 %, contains two strains of T.longibrachiatum. T.harzianum (H.1ixii) include several tef1-α alleles, so itis considered to be a species complex, which is in accordance with Chaverri's view (Chaverri et al. 2003). In the present research, 17 haplotypes of T.harzianum (H.1ixii) were found (according to Fig. 2). With bootstrap support of only 53 %, these haplotypes of T.harzianum (H.1ixii) formed a moderately well-supported clade with T.citrinoviride, T.erinaceum, T.hamatum, T.koningii (H.koningii), T.longibrachiatum, T.pleuroticola, T.reeseii (H.jecorina), T.tomentosum, and an unresolved polytomy with T.aureovriride, T.spirale, T.virens (H.virens). As a result, the eight species including T.aureovriride, T.citrinoviride, T.erinaceum, T.koningii, T.pleuroticola, T.reeseii (H.jecorina), T.spirale, T.tomentosum were resolved with high bootstrap support.
Species geographic distribution
Twenty three species of Trichoderma were identified among 1910 isolates collected from varied soil of 20 different regions in China by using both morphological and molecular analysis, those species were T.asperellum, T.atrioviride, T.aureovriride, T.brevicompactum, T.citrioviride, T.erinaceum, T.gamsii, T.hamatum, T.harzianum (H.1ixii), T.intricatum, T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.pleuroticola, T.reeseii (H.jecorina), T.sinensis, T.spirale, T.stromaticum, T.tomentosum, T.velutinum, T.vermipilum, T.virens (H.virens) and T.viride. Diversity distribution of the species according to their geographic origin revealed significant differences (Table 2, Table 3, Table 4 and Table 5). The species richness of Trichoderma according to their geographic origin, see Table 6 for details.
The 94 strains from Middle China (Henan, Hubei and Hunan province) comprised 11 species: T.asperellum, T.citrinoviride, T.erinaceum, T.hamatum, T.harzianum (H.1ixii), T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.spirale, T.virens (H.virens), T.viride, in which T.asperellum was a dominant species in this region. From East China 1281 strains (Anhui, Fujian, Jiangsu, Jiangxi, Shandong, Zhejiang province and Shanghai municipality) comprised 16 species: T.asperellum, T.atrioviride, T.aureovriride, T.brevicompactum, T.citrinoviride, T.erinaceum, T.hamatum, T.harzianum (H.1ixii), T.intricatum, T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.pleuroticola, T.sinensis, T.virens (H.virens), T.viride, in which T.asperellum was dominant species in this region. In South-East China (Guangdong, Guangxi, Hainan province), 218 strains comprised 15 species of Trichoderma: T.asperellum, T.atrioviride, T.aureovriride, T.brevicompactum, T.citrinoviride, T.hamatum, T.harzianum (H.1ixii), T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.reeseii (H.jecorina), T.spirale, T.velutinum, T.virens (H.virens), T.viride, in which T.harzianum (H.1ixii) was a dominant species. From South-West China 317 isolated (Guizhou, Qinghai, Shanxi, Sichuan and Yunnan province, Tibet Autonomous Region and Chongqing municipality) comprised 20 species: T.asperellum, T.atrioviride, T.aureovriride, T.brevicompactum, T.citrinoviride, T.erinaceum, T.gamsii, T.hamatum, T.harzianum (H.1ixii), T.koningii (H.koningii), T.koningiopsis, T.longibranchiatum, T.pleuroticola, T.spirale, T.stromaticum, T.tomentosum, T.velutinum, T.vermipilum, T.virens (H.virens) and T.viride, in which T.harzianum (H.1ixii) was a dominant species.
In terms of geographic distribution level, T.harzianum was the most abundant species (23.2 %) distributed widely in China, and followed successively by T.asperellum (22.5 %), T.hamatum (18.1 %) and T.viride (8.1 %). T.aureovriride, T.citrinoviride, T.erinaceum, T.gamsii, T.intricatum, T.reeseii (H.jecorina), T.sinensis, T.stromaticum, T.tomentosum, T.velutinum and T.vermipilum were barely isolated among identified species (≤1 %) of all isolates from the collection.
Discussion
The present study was a preliminary domestic assessment of the occurrence of Trichoderma diversity in China. Sampling regions were mainly distributed in East, South-East, South-West and middle China; Northern China was not included since a similar study has been completed by other authors in those areas. Our results showed that Trichoderma distribution were remarkably diversified, the dominant species and proportion of each species were significantly different in a given area, so much varied distribution might be associated with obviously different ecological environments of vast land in China. On the other hand, the variable character of Trichoderma made itself more adaptive to diversified environment. The similar results about Trichoderma biodiversity have been found in South-East Asia (Kubicek et al. 2003), Austria (Wuczkowski et al. 2003), South America (Druzhinina et al. 2005), on Sardinia in Italy (Migheli et al. 2009), and in Poland (Błaszczyk et al. 2011). The Trichoderma biodiversity extent depended upon the difference level among geographic areas (Hoyos-Carvajal et al. 2009).
Geographical distribution of T.harzianum (H.1ixii) in China
Based on previous study, T.harzianum (H.1ixii) has been commonly considered as predominant taxa (Kubicek et al. 2003; Wuczkowski et al. 2003; Druzhinina et al. 2005, 2010; Zhang et al. 2005; Migheli et al. 2009). Using gene sequence analysis, we identified 88 strains as T.harzianum (H.1ixii) from a total of 382 strains. T.harzianum (H.1ixii) was the most commonly reported species in the genus, occurring in diverse ecosystems and ecological niches. Since T.harzianum (H.1ixii) historically originated from China, thus it would be rational that the species distributes most widely in China. Besides, T.asperellum and T.harzianum (H.1ixii), the global distribution species (Danielson and Davey 1973; Chaverri and Samuels 2003), were also identified in our work.
Species difference between East and West China
Different Trichoderma species usually require different living environments, and different geographic regions and climate types determined features of Trichoderma population and its distribution. Apart from T.harzianum (H.1ixii), there was much significant difference in Trichoderma species distribution between East and West in China. For instance, T.gamsii, T.stromaticum, T.tomentosum, T.vermipilum and the majority of strains of T.aureovriride, T.erinaceum, T.koningiopsis were mainly distributed in South-West China. Moreover, T.reeseii (H.jecorina) and the majority of strains of T.citrinoviride, T.spirale were found in South-East China. In addtion, T.intricatum, T.sinensis and the majority of strains of T.asperellum, T.brevicompactum, T.hamatum, T.koningii (H.koningii), T.longibranchiatum, T.pleuroticola, T.virens (H.virens), T.viride were found in East China.
This was probably due to climatic preferences of these species. These species are representative of a temperate Trichoderma biota (Kubicek et al. 2008). For example, T.viride survived better at lower temperatures, however, T.koningii and T.hamatum seem to exhibit a somewhat higher stress-tolerance than the other species, and tend to be more adaptive to colder soils, winter conditions and acid soils (Widden and Scattolin 1988). In contrast, T.sinensis, a species ever isolated from tree bark in Taiwan, China (Bissett et al. 2003), was also found in samples collected from East China, which was demonstrated to be commonly distributed species in South-East Asia (Zhang et al. 2005).
High abundance of Trichoderma present in South-West China
We found that the abundance of Trichoderma was largest in South-West China among all investigated regions, the phenomenon is understandable because South-West China is a place where varied geographic environments offered plenty of ecological habitats to foster the abundant species resources (http://www.biodiversityhotspots.org/xp/Hotspots/China). Tai and Wen's earlier studies (Tai 1979; Wen et al. 1993) also stressed the abundant diversity of Trichoderma spp. in South-West China.
More Trichoderma species to be explored in China
Until now, 44 species of Trichoderma have been isolated from diversified habitats in China, however, most of the 23 species we identified in this study were mainly included in those known species. In our study, Trichoderma strains were mainly sourced from forest soil (22 species/698 isolates), garden soil (20 species/496 isolates) and vegetable soil (13 species/415 isolates), in which 20 species of Trichoderma again were isolated from 491 soil samples collected from 18 provinces and two municipalities in China. Furthermore, three species, T.intricatum, T.stromaticum, T.vermipilum were first reported in China.
Some habitats might nurture a very abundant Trichoderma population such as crop and forest roots, mushroom, grain crop soil, etc. On the other hand, Trichoderma population might dynamically change or vary with seasons, therefore for the next step we will collect samples in different seasons, which will allow us to find species of Trichoderma, and a global picture of Trichoderma population in China will be generated.
References
Bailey BA, Bae H, Strem MD, Roberts DP, Thomas SE, Crozier J, Samuels GJ, Choi I-Y, Holmes KA (2006) Fungal and plant gene expression during the colonization of cacao seedlings by endophytic isolates of four Trichoderma species. Planta 224:1449–1464
Bissett J, Szakacs G, Nolan CA, Druzhinina I, Kullnig-Gradinger C, Kubicek CP (2003) New species of Trichoderma from Asia. Can J Bot 81:570–586
Błaszczyk L, Popiel D, Chełkowski J, Koczyk G, Samuels GJ, Sobieralski K, Siwulski M (2011) Species diversity of Trichoderma in Poland. J Appl Genet 52:233–243
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Chaverri P, Samuels G (2003) Hypocrea/Trichoderma (ascomycota, hypocreales, hypocreaceae): species with green ascospores: Centraalbureau voor Schimmelcultures, pp 1–36
Chaverri P, Castlebury LA, Samuels GJ, Geiser DM (2003) Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol Phylogenet Evol 27:302–313
Chełkowski J, Golka L, Stępień Ł (2003) Application of STS markers for leaf rust resistance genes in near-isogenic lines of spring wheat cv. Thatcher. J App Genet 44:323–338
Danielson R, Davey C (1973) The abundance of Trichoderma propagules and the distribution of species in forest soils. Soil Biol Biochem 5:485–494
Doohan FM, Parry DW, Jenkinson P, Nicholson P (1998) The use of species-specific PCR-based assays to analyse Fusarium ear blight of wheat. Plant Pathol 47:197–205
Druzhinina I, Kopchinskiy A (2004a) TrichoBLAST version 1.0, Multiloci database of phylogenetic markers and similarity search. Published online by the International Subcommission on Trichoderma and Hypocrea Taxonomy (ISTH). Home page at: http://www.isth.info/tools/blast/index.php
Druzhinina IS, Kopchinskiy AG (2004b) TrichoBLAST version 1.0, Trichoderma oligonucleotide key. Published online by the International Subcommission on Trichoderma and Hypocrea Taxonomy (ISTH). Home page at: http://www.isth.info/tools/molkey/index.php
Druzhinina IS, Kubicek CP (2005) Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters. J Zhejiang Univ Sci B 6:100–112
Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42:813–828
Druzhinina IS, Kubicek CP, Komoń-Zelazowska M, Mulaw TB, Bissett J (2010) The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol Biol 10:94
Eck RV, Dayhoff MO (1966) Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Springs, MD
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gams W, Bissett J (1998) Morphology and identification of Trichoderma. In: Kubicek CP, Hannan GE (eds) Trichoderma and Gliocladium, vol 1: basic biology, taxonomy and genetics. Taylor and Francis, London, pp 3–34
Gao W, Baoju L, Sun J, Yanxia S (2007) Inderntificaotion of Trichoderma Speices from Contaminnated Substrate During Cultivation of Pleurotus ostreatus. Acta Edulis Fungi 14(3):81–85
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56
He Z, Sun H, Gao Y (2008) Identification of Trichoderma Species on Mushrooms. J Hebei Normal Univ Sci Technol 22(4):41–45
He Z, Gao Z, Gao Y, Zhao B, Zhang X (2010a) ITS sequence of Trichoderma species in soil planted vegetables in the greenhouse and UP-PCR analysis on genetic diversity. Acta Phytophylacica Sinica 37(5):459–465
He Z, Gao Z, Hou D (2010b) Study on species of Trichoderma on edible fungi and conidiospore production on the edible fungi residue with cotton seed shell as fermentation medium. Jiangsu Agric Sci 3:440–442
Hermosa MR, Keck E, Chamorro I, Rubio B, Sanz L, Vizcaíno JA, Grondona I, Monte E (2004) Genetic diversity shown in Trichoderma biocontrol isolates. Mycol Res 108:897–906
Hill TCJ, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43:1–11
Hjeljord L, Tronsmo A (1998) Trichoderma and Gliocladium in biological control: an overview. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium, vol 2: enzymes, biological control and commercial application. Taylor and Francis, London, pp 131–151
Hoyos-Carvajal L, Orduz S, Bissett J (2009) Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol 46:615–631
Jia DC, Yu ZF, Qiao M, Zhang KQ (2009) A new Chinese record of the genus Trichoderma. Mycosystema 28(6):860–862
Klein D, Eveleigh DE (1998) Ecology of Trichoderma. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium, vol 1: basic biology, taxonomy and genetics. Taylor and Francis, London, pp 57–74
Kubicek CP, Penttilä ME (1998) Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocladium, vol 1: basic biology, taxonomy and genetics. Taylor and Francis, London, pp 49–71
Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger C, Szakacs G (2003) Genetic and metabolic diversity of Trichoderma: a case study on South East Asian isolates. Fungal Genetics Biol 38:310–319
Kubicek CP, Komoń-Zelazowska M, Druzhinina IS (2008) Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. J Zhejiang Univ Sci B 9(10):753–763
Kullnig-Gradinger CM, Szakacs G, Kubicek CP (2002) Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res 106:757–767
Li G, Chen J, Liu T, Liu L (2010) Trichoderma koningiopsis: A New Chinese Record of the Genus Trichoderma. Microbiol China 37(11):1663–1665
Luo X, Wen C (2003) Studies on Eeology and Biological Control of Antagonisti Trichoderma spp. Against Phytophthora capsici in ya'an. Sichuan Agricultural University, ya'an Sichuan, China
Migheli Q, Balmas V, Komoñ-Zelazowska M, Scherm B, Fiori S, Kopchinskiy AG, Kubicek CP, Druzhinina IS (2009) Soils of a Mediterranean hot spot of biodiversity and endemism (Sardinia, Tyrrhenian Islands) are inhabited by pan-European, invasive species of Hypocrea/Trichoderma. Environ Microbiol 11(1):35–46
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Pan Z, Fu J, Ma L, Wang H (2010) Diversity of Trichoderma spp. isolated from medicinal plant rhizosphere soils in Liaoning Province. J Zhejiang Univ Agric and Life Sci 36(4):405–410
Samuels GJ, Chaverri P, Farr DF, McCray EB (2009) Trichoderma online systematic mycology and microbiology laboratory home page at: http://ntars-gringov/taxadescriptions/keys/TrichodermaIndexcfm
Samuels GJ, Dodd SL, Gams W, Castlebury LA (2002) Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94:146–170
Shao L, Shi Y, Guo L (2008) Trichoderma Species Isolated from Contaminated Mushrooms and Cultivation Substrste in the Beijing Region. Acta Edulis Fungi 15(1):86–90
Sivasithamparam K, Ghisalberti EL (1998) Secondary metabolism in Trichoderma and Gliocladium. In: Harman GE, Kubicek CP (eds) Trichoderma and Glicoladium, vol 1: basic biology, taxonomy and genetics. Taylor and Francis, London, pp 139–191
Sun J, Duan Y-X, Lǚ G-Z (2006a) Morphological Identification of Trichoderma Species in Liaoning Province. J Fungal Res 4(2):38–44
Sun J, Duan X-X, Lǚ G-Z (2006b) Primary Research On the Diversity Resource of Trichoderma in Liaoning Province. J Anhui Agri Sci 34(17):4193–4195, 4197
Tai FL (1979) Sylloge fungorum sinicorum. Science Press, Academia Sinica, Peking, China
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vargas Gil S, Pastor S, March GJ (2009) Quantitative isolation of biocontrol agents Trichoderma spp., Gliocladium spp. and Actinomycetes from soil with culture media. Microbiol Res 164(2):196–205
Wen C, Chen W, Tao J (1992) Distribution of Trichoderma spp. in the Cotton Soil and Their Biological Activities. Acta Phytopathologica Sinica 4:306
Wen C, Chen W, Tao J (1993) Studies on The Taxonomy of The Genus Trichoderma in Southwestern China. Acta Myeologlca Sinlea 12(2):118–130
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Widden P, Scattolin V (1988) Competitive interactions and ecological strategies of Trichoderma species colonizing spruce litter. Mycologia 80:795–803
Wu X, Wu X, Hu F, He H, Xie B (2008) Identification of Trichoderma Species Associated with Cultivated Edible Fungi. J Agric Biotochnol 16(6):1048–1055
Wuczkowski M, Druzhinina I, Gherbawy Y, Klug B, Prillinger HJ, Kubicek CP (2003) Species pattern and genetic diversity of Trichoderma in a mid-European, primeval floodplain-Forest. Microbiol Res 158:125–133
Xia C, Chen Y (2009) Biodiversity Research of the Genus Trichoderma from Yunnan Province: Data from Morphology and ITS Sequence. Southwest Forestry University, China
Yu H-l, Wei J-g, Su X-l (2010) Investigation on Trichoderma resources in forest soil of Northern Guangxi. Guangxi Agric Sci 41(7):703–706
Yu Z-F, Qiao M, Zhang Y, Zhang K-Q (2007) Two new species of Trichoderma from Yunnan, China. Antonie van Leeuwenhoek 92:101–108
Yuan Z-L, Chen Y-C, Zhang C-L, Lin F-C, Chen L-Q (2008) Trichoderma chlorosporum, a new record of endophytic fungl from Dendrobium in China. Mycosystema 27(4):608–610
Ze F, Qiao Y-M, Zhang Y, Zhang K-Q (2007) Two new species of Trichoderma from Yunnan, China. Antonie van Leeuwenhoek 92:101–108
Zhang C-L, Xu T (2004) Trichoderma species from China. J Zhejiang Univ Agric and Life Sci 30(4):464
Zhang C-L, Xu T (2005) Records of Trichoderma species from Hebei, Zhejiang, Yunnan and Tibet of China. Mycosystema 24(2):184–192
Zhang E, Zhou B, Yang L (1996) Ecological distribution of Trichoderma propagules in main vegetation soils of Yunnan. Chin J Appl Ecol 7:78–82
Zhang C-L, Druzhinina IS, Kubicek CP, Xu T (2005) Trichoderma biodiversity in China: Evidence for a North to South distribution of species in East Asia. FEMS Microbiol Lett 251:251–257
Zhao Z, Sun X, Yang R, Yang H, Lǚ G (2004) Diversity of Trichoderma in greenhouse soil. J Zhejiang Univ Agric and Life Sci 30(4):467
Acknowledgements
This work was supported by key grant from Shanghai Committee of Science and Technology (09391910900), 948 project of Ministry of Agriculture (2011 - G4), the National Department Public Benefit Research Foundation of China (200903052), and China Agriculture Research System (CARS - 2).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, Ry., Liu, Zc., Fu, K. et al. Trichoderma biodiversity in China. J Appl Genetics 53, 343–354 (2012). https://doi.org/10.1007/s13353-012-0093-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-012-0093-1