Abstract
CHEK2 gen encodes cell cycle checkpoint kinase 2 that participates in the DNA repair pathway, cell cycle regulation and apoptosis. Mutations in CHEK2 gene may result in kinase inactivation or reduce both catalytic activity and capability of binding other proteins. Some studies indicate that alterations in CHEK2 gene confers increase the risk of breast cancer and some other malignancies, while the results of other studies are inconclusive. Thus the significance of CHEK2 mutations in aetiology of breast cancer is still debatable. The aim of our study was to evaluate the relationship between the breast/ovarian cancer and CHEK2 variants by: i) the analysis of the frequency of selected CHEK2 variants in breast and ovarian cancer patients compared to the controls; ii) evaluation of relationships between the certain CHEK2 variants and clinico-histopathological and pedigree data. The study was performed on 284 breast cancer patients, 113 ovarian cancer patients and 287 healthy women. We revealed the presence of 430T > C, del5395 and IVS2 + 1G > A variants but not 1100delC in individuals from both study and control groups. We did not observe significant differences between cancer patients and controls neither in regard to the frequency nor to the type of CHEK2 variants. We discussed the potential application of CHEK2 variants in the evaluation of breast and ovarian cancer predisposition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
CHEK2 gene encodes cell cycle checkpoint kinase 2 that is regarded as a candidate tumor suppressor (Nevanlinna and Bartek 2006). CHEK2 participates in the DNA repair pathway, cell cycle regulation and apoptosis (Shieh et al. 2000). In response to DNA damage, CHEK2 is phosphorylated by ATM and ATR kinases. Active CHEK2 in turn phosphorylates p53 and BRCA1 (Brody 2002). Other substrates of kinase CHEK2 are the following: CDC25A, CDC25C phosphatases, PLK3 kinase and E2F1 transcription factor (Nevanlinna and Bartek 2006).
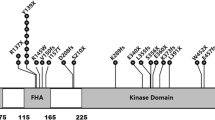
Mutations in CHEK2 gene were detected for the first time among TP53-negative LFS (Li-Fraumenni syndrom) and LFS-variant families (Bell et al. 1999). A number of rare alterations in CHEK2 which are defined as intermediate risk mutations, have been reported in breast cancer patients (Willems 2007; Turnbull and Rahman 2008). The more common mutations are the following: one missense variant (430T > C) and three truncating mutations (1100delC, IVS2 + 1G > A, del5395).
1100delC (c.1100delC/p.Thr367MetfsX15) disrupt the catalytic domain of CHEK2 resulting in an inactive kinase (Wu et al. 2001; Bell et al. 2007). In combined analysis of the ten case-control studies 1100delC was observed in 1.9% (201/10860) breast cancer cases and in 0.7% (64/9065) controls (CHEK2 Breast Cancer Case-Control Consortium 2004). It was estimated that CHEK2 1100delC confers about two-fold increased breast cancer risk in women (CHEK2 Breast Cancer Case-Control Consortium 2004). Nevertheless, the meta–analyses published by Weischer et al. (2008) showed that CHEK2 1100delC is an important breast cancer predisposition factor, increasing the risk by three- to five-fold. Studies carried out in Poland and neighboring countries shows that this mutation occurred in 0.0–5.2% breast cancer patients versus 0.0–0.92% in controls (Dufault et al. 2004; Kleibl et al. 2005; Rashid et al. 2005; Kwiatkowska et al. 2006; Cybulski et al. 2007a; Fedorova et al. 2007; Sokolenko et al. 2007; Suspitsin et al. 2009).
CHEK2 del5395 (originally described as del5567) has been observed by Walsh et al. (2006) in families with a high aggregation of breast and ovarian cancer. This mutation confers the deletion of two exons (9 and 10) (g.27417113_27422508del) and was detected in 0.9–1.3% of breast cancer cases while in 0.0–0.4% of controls (Walsh et al. 2006; Cybulski et al. 2006; Cybulski et al. 2007a).
Another CHEK2 protein truncating mutation found in breast cancer patients is a splice-site mutation IVS2 + 1G > A (c.444 + 1G > A) (Cybulski et al. 2004b; Dufault et al. 2004). The estimated incidence of this alteration was 0.0–1.1% for breast cancer patients and 0.0–0.4% for controls (Dufault et al. 2004; Bogdanova et al. 2005; Cybulski et al. 2007a; Kleibl et al. 2008; Sokolenko et al. 2007).
Another recurrent CHEK2 alteration found in breast cancer patients was missense variant 470T > C (c.470T > C/p.Ile157Thr) (Bell et al. 1999; Allinen et al. 2001; Schutte et al. 2003; Cybulski et al. 2004b; Friedrichsen et al. 2004; Kilpivaara et al. 2004; Bogdanova et al. 2005). In vitro studies showed that the CHEK2 470T > C variant encodes a protein with both reduced CDC25A catalytic activity and reduced capability of TP53 and BRCA1 binding (Falck et al. 2001a, b; Wu et al. 2001; Li et al. 2002; Bell et al. 2007). Kilpivaara et al. (2004) suggest that 470T > C variant may have negative effect on the pool of normal CHEK2 protein by formation of heterodimers with wild-type CHEK2. The occurrence of 470T > C variant was observed in 1.9–7.4% of cancer cases and in 0.6–4.8% of controls (Dufault et al. 2004; Kilpivaara et al. 2004; Bogdanova et al. 2005; Szymanska-Pasternak et al. 2006; Cybulski et al. 2007a; Kleibl et al. 2008). This variant confers lower risk of hereditary breast cancer than protein truncating mutation (Kilpivaara et al. 2004).
Another missense variant (1283C > T) associated with increased breast cancer risk was found in the Ashkenazi Jewish population (Shaag et al. 2005).
Summarizing, in some studies a strong correlation between specific CHEK2 variants and breast cancer risk has been observed (CHEK2 Breast Cancer Case-Control Consortium 2004; Dufault et al. 2004; Kilpivaara et al. 2004; Bogdanova et al. 2005; Górski et al. 2005; Cybulski et al. 2007a; Weischer et al. 2008), contrary to others, where weak or no correlation was found (Allinen et al. 2001; Schutte et al. 2003; Friedrichsen et al. 2004; Kleibl et al. 2005, 2008; Rashid et al. 2005). It has been shown that mutations in CHEK2 gene confers also a predisposition to some other malignancies like: prostate (Dong et al. 2003; Seppälä et al. 2003; Cybulski et al. 2004a, 2006), thyroid (Cybulski et al. 2004b), bladder (Złowocka et al. 2008) and colorectal cancer (Cybulski et al. 2004b, 2007b), as well as to sarcoma and brain tumors (Bell et al. 1999).
Since the results of studies on CHEK2 mutations in aetiology of breast cancer conducted by many authors are inconclusive and the significance of CHEK2 mutations in breast cancer risk is still debatable, the aim of our study was to search for the relationship between the breast/ovarian cancer and CHEK2 variants by: i) the analysis of the frequency of selected CHEK2 variants in breast and/or ovarian cancer patients negative for most common mutations in BRCA1 and BRCA2 genes in Polish population, comparing to the controls, ii) evaluation of relationships between the certain CHEK2 variants and clinico-histopathological and pedigree data.
Materials and methods
The study was performed on DNA isolated from peripheral blood lymphocytes obtained from 284 breast and 113 ovarian cancer patients (altogether 397 women). Patients were unrelated and came from the south-west region of Poland (Lower Silesia). The group was characterized in respect to family history of cancer, tumor histology (see Table 1) age of onset of the diseases as well as the presence of most common mutations (in Poland) in BRCA1 such as: 5382insC (c.5266dupC), 300T > G (c.181T > G), 4153delA (c.4034delA), 185delAG (c.68_69delAG) and BRCA2 gene: 6174delT (c.5946delT). Individuals with mutations in either BRCA1 or BRCA2 genes were excluded from the analysis. The average age was 49 ± 9 and 53 ± 13 years for breast and ovarian study groups, respectively. The diagnosis of hereditary breast cancer (HBC) and hereditary breast/ovarian cancer (HBOC) was evaluated based on pedigree analysis (according to Berliner and Fay 2007; Lynch et al. 2003). HBCs/HBOCs were diagnosed in 136 breast cancer cases. The matched control group comprised of 287 healthy women coming from the same population, with no family history of cancer. The average age of the controls was 46 ± 18 years. DNA was isolated using commercial kit (Qiagen). Molecular analysis of three truncating mutations: 1100delC, del5395, IVS2 + 1G > A and one missense variant 470T > C were carried out by using PCR-RFLP and ASA-PCR, employing primers as previously described by Cybulski et al. (2004a, 2007a). The study design was accepted by Ethical Committee of Wroclaw Medical University. Analysis of the frequency of selected variants in the CHEK2 gene in patients with breast and/or ovarian cancer in comparison to control group was assessed using the Fisher’s exact test. Logistic regression was used to estimate odds ratios. Confidence interval was 0.95 and p-value of less than 0.05 was considered as significant. Statistical analysis of data was performed using StatSoft, Inc. (2005) STATISTICA, version 7.0.
Results
Molecular analysis revealed the presence of 430T > C, del5395 and IVS2 + 1G > A variants but not 1100delC in individuals from both study and control groups. In the group of ovarian cancer patients the occurrence of all CHEK2 alterations was lower than in the control group, due to the relatively low incidence of missense variant (see Table 2). Therefore, ovarian cancer patients were excluded from further analysis. The frequency of all analyzed CHEK2 variants was higher in breast cancer patients (9.9%) than in the controls (7.3%), however the difference was not statistically significant (p = 0.28). We did not observe significant differences between cancer patients and controls neither in regard to the frequency nor to the type of CHEK2 variants (see Table 2). The missense variant 430T > C was the most often observed alteration in both breast cancer patients (7.0%) and in control group (6.3%) (the result was not statistically significant p = 0.71). The splice mutation IVS2 + 1G > A occured in 1.1 % of study in comparison to 0.7% of control groups (p = 0.65), while del5395 was observed in 2.1% of cancer cases versus 0.34% of the controls (p = 0.055). There were also no differences in the frequencies of CHEK2 mutations between the hereditary and sporadic breast cancer patients.
The analysis of association between the clinico-histopathological characteristics of disease and CHEK2 alterations revealed only an association between del5395 and breast cancer patients below/at 40-year-of-age (p = 0.043). Thus, further analysis was carried out for the whole breast cancer patients group versus controls. In the case of an odds ratio, the results were not statistically significant (the 95% confidence interval overlap 1.0), see Table 2.
Discussion
Despite the years of study, the importance of CHEK2 variants in prediction of individual breast/ovarian cancer risk is still controversial. Our study revealed that in ovarian cancer patients the frequency of CHEK2 mutations is lower than in control group. This observation supports the thesis of Nevanlinna and Bartek (2006) that the mutations in the CHEK2 gene are not associated with increased risk of ovarian cancer.
Our observation pointed out the missense variant 430T > C as the most often observed alteration in both breast cancer patients (7.0%) and in control groups (6.3%), as well as the correlation between the incidence of the following CHEK2 variants: del5395, and IVS2 + 1G > A are in agreement with the observations of other authors (Dufault et al. 2004; Bogdanova et al. 2005; Cybulski et al. 2007a; Kleibl et al. 2008). Nevertheless, all the correlations observed in our study have not been statistically significant.
Thus, when analyzing the results of present study in regard to still published data it can be noticed that our results are consistent with those, performed on groups similar in size to ours (see Table 3). The opposite results were obtained by authors studying a very large groups. They observed the strong correlation between certain CHEK2 gene variants and breast cancer (CHEK2 Breast Cancer Case-Control Consortium 2004; Dufault et al. 2004; Kilpivaara et al. 2004; Górski et al. 2005; Cybulski et al. 2007a; Weischer et al. 2008).
However, when discussing the potential inclusion of CHEK2 variants into the cancer predisposition tests, two facts have to be taken into account: i) “any small difference, no matter how clinically unimportant, will be statistically significant (p < 0.05) if the sample size is large enough” (Grunkemeier et al. 2009), ii) the presence of discussed above CHEK2 variants in a substantial part of control group (what was observed in all still published studies) limits the possibility of their application in testing of breast cancer predisposition. Therefore, an employment of CHEK2 variants in evaluation of cancer predisposition is doubtful.
References
Allinen M, Huusko P, Mäntyniemi S, Launonen V, Winqvist R (2001) Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br J Cancer 85:209–212
Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE et al (1999) Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528–2531
Bell DW, Kim SH, Godwin AK, Schiripo TA, Harris PL, Haserlat SM et al (2007) Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer 121:2661–2667
Berliner JL, Fay AM (2007) Risk assessment and genetic counseling for hereditary breast and ovarian cancer: recommendations of the National Society of Genetic Counselors. J Genet Couns 16:241–260
Bogdanova N, Enssen-Dubrowinskaja N, Feshchenko S, Lazjuk GI, Rogov YI, Dammann O et al (2005) Association of two mutations in the CHEK2 gene with breast cancer. Int J Cancer 116:263–266
Brody LC (2002) CHEKs and balances: accounting for breast cancer. Nat Genet 31:3–4
Chekmariova EV, Sokolenko AP, Buslov KG, Iyevleva AG, Ulibina YM, Rozanov ME et al (2006) CHEK2 1100delC mutation is frequent among Russian breast cancer patients. Breast Cancer Res Treat 100:99–102
CHEK2 Breast Cancer Case-Control Consortium (2004) CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10, 860 breast cancer cases and 9, 065 controls from 10 studies. Am J Hum Genet 74:1175–1182
Cybulski C, Huzarski T, Górski B, Masojć B, Mierzejewski M, Debniak T et al (2004a) A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res 64:2677–2679
Cybulski C, Górski B, Huzarski T, Masojć B, Mierzejewski M, Debniak T et al (2004b) CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 75:1131–1135
Cybulski C, Wokołorczyk D, Huzarski T, Byrski T, Gronwald J, Górski B et al (2006) A large germline deletion in the Chek2 kinase gene is associated with an increased risk of prostate cancer. J Med Genet 43:863–866
Cybulski C, Wokołorczyk D, Huzarski T, Byrski T, Gronwald J, Górski B et al (2007a) A deletion in CHEK2 of 5, 395 bp predisposes to breast cancer in Poland. Breast Cancer Res Treat 102:119–122
Cybulski C, Wokołorczyk D, Kładny J, Kurzawski G, Suchy J, Grabowska E et al (2007b) Germline CHEK2 mutations and colorectal cancer risk: different effects of a missense and truncating mutations? Eur J Hum Genet 15:237–241
Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK et al (2003) Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet 72:270–280
Dufault MR, Betz B, Wappenschmidt B, Hofmann W, Bandick K, Golla A et al (2004) Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer 110:320–325
Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J (2001a) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842–847
Falck J, Lukas C, Protopopova M, Lukas J, Selivanova G, Bartek J (2001b) Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene 20:5503–5510
Fedorova OE, Liubchenko LN, Paiadini IG, Kazubskaia TP, Amosenko FA, Gar'kavtseva RF et al (2007) Analysis of BRCA1/2 and CHEK2 mutations in ovarian cancer and primary multiple tumors involving the ovaries. Patients of Russian population using biochips. Mol Biol (Mosk) 41:37–42
Friedrichsen DM, Malone KE, Doody DR, Daling JR, Ostrander EA (2004) Frequency of CHEK2 mutations in a population based, case-control study of breast cancer in young women. Breast Cancer Res 6:629–635
Górski B, Cybulski C, Huzarski T, Byrski T, Gronwald J, Jakubowska A et al (2005) Breast cancer predisposing alleles in Poland. Breast Cancer Res Treat 92:19–24
Grunkemeier GL, Wu Y, Furnary AP (2009) What is the value of a p value? Ann Thorac Surg 87(5):1337–1343
Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D et al (2004) CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer 111:543–547
Kleibl Z, Novotny J, Bezdickova D, Malik R, Kleiblova P, Foretova L et al (2005) The CHEK2 c.1100delC germline mutation rarely contributes to breast cancer development in the Czech Republic. Breast Cancer Res Treat 90(2):165–167
Kleibl Z, Havranek O, Novotny J, Kleiblova P, Soucek P, Pohlreich P (2008) Analysis of CHEK2 FHA domain in Czech patients with sporadic breast cancer revealed distinct rare genetic alterations. Breast Cancer Res Treat 112:159–164
Kwiatkowska E, Skasko E, Niwinska A, Wojciechowska-Lacka A, Rachtan J, Molong L et al (2006) Low frequency of the CHEK2*1100delC mutation among breast cancer probands from three regions of Poland. Neoplasma 53:305–308
Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D et al (2002) Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9:1045–1054
Lynch HT, Snyder CL, Lynch JF, Riley BD, Rubinstein WS (2003) Hereditary breast-ovarian cancer at the bedside: role of the medical oncologist. J Clin Oncol 21:740–753
Nevanlinna H, Bartek J (2006) The CHEK2 gene and inherited breast cancer susceptibility. Oncogene 25:5912–5919
Rashid MU, Jakubowska A, Justenhoven C, Harth V, Pesch B, Baisch C et al (2005) German populations with infrequent CHEK2*1100delC and minor associations with early-onset and familial breast cancer. Eur J Cancer 41:2896–2903
Schutte M, Seal S, Barfoot R, Meijers-Heijboer H, Wasielewski M, Evans DG et al (2003) Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet 72:1023–1028
Seppälä EH, Ikonen T, Mononen N, Autio V, Rökman A, Matikainen MP et al (2003) CHEK2 variants associate with hereditary prostate cancer. Br J Cancer 89:1966–1970
Shaag A, Walsh T, Renbaum P, Kirchhoff T, Nafa K, Shiovitz S et al (2005) Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet 14:555–563
Shieh SY, Ahn J, Tamai K, Taya Y, Prives C (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14:289–300
Sokolenko AP, Rozanov ME, Mitiushkina NV, Sherina NY, Iyevleva AG, Chekmariova EV et al (2007) Founder mutations in early-onset, familial and bilateral breast cancer patients from Russia. Fam Cancer 6:281–286
Suspitsin EN, Sherina NY, Ponomariova DN, Sokolenko AP, Iyevleva AG, Gorodnova TV et al (2009) High frequency of BRCA1, but not CHEK2 or NBS1 (NBN), founder mutations in Russian ovarian cancer patients. Hered Cancer Clin Pract 7:5
Szymanska-Pasternak J, Szymanska A, Medrek K, Imyanitov EN, Cybulski C, Gorski B et al (2006) CHEK2 variants predispose to benign, borderline and low-grade invasive ovarian tumors. Gynecol Oncol 102:429–431
Turnbull C, Rahman N (2008) Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet 9:321–345
Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J et al (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388
Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG (2008) CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol 26:542–548
Willems PJ (2007) HotSpots Susceptibility genes in breast cancer: more is less? Clin Genet 72:493–496
Wu X, Webster SR, Chen J (2001) Characterization of tumor-associated Chk2 mutations. J Biol Chem 276:2971–2974
Złowocka E, Cybulski C, Górski B, Debniak T, Słojewski M, Wokołorczyk D et al (2008) Germline mutations in the CHEK2 kinase gene are associated with an increased risk of bladder cancer. Int J Cancer 122:583–586
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Myszka, A., Karpinski, P., Slezak, R. et al. Irrelevance of CHEK2 variants to diagnosis of breast/ovarian cancer predisposition in Polish cohort. J Appl Genetics 52, 185–191 (2011). https://doi.org/10.1007/s13353-010-0013-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-010-0013-1