Abstract
Long non-coding RNAs (lncRNAs) are emerging as crucial regulators of cancer. To identify novel targets for further study in esophageal squamous cell carcinoma (ESCC), we performed a genome-wide analysis of lncRNA expression in 12 ESCC tumor and normal tissues. Publicly available RNA-seq data were downloaded from the NCBI, GEO, and Co-LncRNA databases, and lncRNA and messenger RNA (mRNA) expression profiles were analyzed. In total, 127 lncRNAs were found to be differentially expressed, with a greater than fourfold change in ESCC tumor tissues compared with normal tissues. Among these lncRNAs, 98 were upregulated and 29 downregulated. Moreover, 1469 network nodes and 1720 connection edges between 119 lncRNAs and 1350 coding genes were integrated into the lncRNA and mRNA co-expression network. Bioinformatic analysis using GO terms revealed that these dysregulated lncRNAs are associated with developmental processes, proteinaceous extracellular matrix, and protein binding activity, with ECM-receptor interaction and the PI3K-Akt signaling pathway enrichment. Lastly, qRT-PCR results verified two significantly upregulated lncRNAs and three significantly downregulated lncRNAs in 50 pairs of ESCC tissues and adjacent normal tissues. These results reveal the landscape of ESCC-associated lncRNAs and co-expression networks, providing important insight regarding the lncRNAs involved in ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal carcinoma (EC) is the eighth most common and the sixth most lethal cancer worldwide [1]. Despite advances in multidisciplinary treatment of esophageal squamous cell carcinoma (ESCC), the disease generally has a very poor prognosis, with a 5-year survival rate ranging from 10 to 25 % [1, 2]. Esophageal adenocarcinoma (EAC) and ESCC [1] are the two main clinical subtypes of EC. Approximately 70 % of the worldwide cases of ESCC occur in China [1]. Although the application of targeted therapy is primarily limited to EAC, ESCC remains the dominant histological type of esophageal cancer both in China and worldwide [3]. Thus, there is an urgent need for novel strategies aimed at improving our understanding of ESCC biology and for providing targets for therapy or early detection of the disease.
Multiple important signaling pathways in tumorigenesis have been uncovered via expression profiling of coding genes. More recently, actively transcribed long non-coding RNAs (lncRNAs), endogenous cellular RNA transcripts longer than 200 nucleotides in length and without protein-coding capacity [4], identified by high-throughput platforms have been shown to be involved in even more complex genome regulatory networks in cancer. lncRNAs are emerging as crucial regulators of cancer biology; these molecules are generally expressed at lower levels than coding genes but display higher tissue specificity [4–6].
When located at or near the same genomic locus, lncRNAs are involved in the cis regulation of gene expression [7]. lncRNAs can also regulate distal gene expression through a trans-acting mechanism by associating with multiple protein partners, such as chromatin modifiers, transcription factors and splicing factors, or by serving as decoys, guides, or scaffolds [4, 8]. As lncRNAs appear to be involved in nearly all aspects of gene regulation, analysis of the co-expression of lncRNAs, and messenger RNA (mRNAs) can help predict their roles in the development of various diseases including cancer and lay a foundation for uncovering their mechanisms of activity.
Altered lncRNA profiles have been identified in breast cancer [9, 10], lung cancer [11, 12], colorectal cancer [13], renal cell carcinoma [14], and hepatocellular carcinoma [15–17], indicating that aberrant expression of certain lncRNAs contributes to carcinogenesis. Over the past 3 years, studies on lncRNAs have become common in esophageal cancer biology research. For example, Wu et al. found that the long non-coding RNA transcript AFAP1-AS1 is highly expressed in esophageal adenocarcinoma, and functional experiments showed that AFAP1-AS1 promotes invasion and metastasis in esophageal cancer cells [18]. More recently, HOTAIR [19, 20], ANRIL [21], UCA1 [22], PCAT1 [23], and MALAT1 [24] were reported to be upregulated in ESCC and were significantly associated with disease prognosis. Despite the considerable progress in understanding lncRNAs that has accompanied over a decade of research, only a few have been identified. Indeed, most lncRNAs remain largely unstudied, particularly with regard to ESCC.
Therefore, to investigate the potential role of lncRNAs in ESCC, we performed a comprehensive analysis of lncRNA and mRNA profiles in ESCC tissue. In particular, we evaluated the lncRNA and mRNA co-expression network during the genesis of ESCC.
Materials and methods
Data curation and processing
Transcriptomic sequencing data under the accession number GSE32424 [25] were downloaded from publicly available Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). Normalized reads were downloaded from the Co-LncRNA database (http://www.bio-bigdata.com/Co-LncRNA/) [26]. In brief, raw RNA-seq reads were aligned and mapped using TopHat v2.0.9, and transcriptome assemblies were generated using Cufflinks v2.1.1 with the default parameters. Only expressed genes were considered, and the threshold of the expression value for inclusion in the analysis was set to 0.001 [26]. In this study, human lncRNA and protein-coding gene annotations were directly download from GENECODE v22. All of the categories in the “long non-coding RNA gene annotation” GTF file were considered to be lncRNAs. To obtain genome-wide lncRNA and protein-coding gene expression profiles, normalized expression data were subsequently analyzed for differently expressed lncRNAs and protein-coding genes using the Bioconductor package (limma, version 3.26.1) [27] in R (version 3.2.2) with default parameters. Differentially expressed lncRNAs and mRNAs were identified through fold change filtering.
Construction of the lncRNA and mRNA co-expression network
Spearman’s correlation test was used to estimate co-expression relationships between lncRNAs and protein-coding genes. Moreover, the P value of the correlation coefficient was estimated. Finally, a set of co-expressed genes for each lncRNA was identified by applying a coefficient threshold of 0.95 and a significance threshold of 0.001. The filtered co-expressed genes were defined as potential targets of the lncRNAs in this study. Using Cytoscape (version 3.2.1), the resulting network was defined as an lncRNA-mRNA regulatory network. A direct connection between an lncRNA and an mRNA is represented as a solid line.
Bioinformatic analysis
Gene Ontology (GO) analysis is a functional analysis associating differentially expressed mRNAs with GO categories. The predicted target genes were uploaded into the Database for Annotation, Visualization and, Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/), which utilizes GO to identify the molecular function(s) represented in the gene profile. Furthermore, we also used the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (http://www.genome.ad.jp/kegg/) to analyze the potential functions of these target genes in pathways. The lower the P value, the more significant the correlation, and we used the recommended P value cutoff of 0.05.
Patient samples
We retrospectively collected paired tumor and adjacent normal tissues from 50 patients with ESCC and examined the expression of selected lncRNAs with validating RNA sequencing data. All patients had surgically proven primary ESCC and underwent surgery at National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College between June 2008 and June 2009. The clinical and pathological information for the patients is listed in Table 1. Samples were obtained with informed consent, and the study was approved by the medical ethics committee of the National Cancer Center/Cancer Hospital.
Quantitative RT-PCR
PCR analysis was performed on 50 pairs of ESCC and matched adjacent normal tissues. As an independent validation, the three top-ranked upregulated lncRNAs (RP11-334E6.12, DNM3OS, and RP11-150O12.6) and downregulated lncRNAs (AC103563.9, RP11-7 K24.3, and RP11-351 J23.1) with only one transcript were chosen as candidates. Total cellular RNA was isolated from ESCC tissues using the Oligotex mRNA mini kit (QIAgen) and then reversely transcribed using TransScript II One-Step gDNA removal and cDNA Synthesis SuperMix (Transgen) in accordance with the manufacturer’s instructions. The expression of selected lncRNAs was assayed by SYBR Green-based qRT-PCR using a 7900HT fast real time PCR system (Applied Biosystems/Life Technologies). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control; the primers used are listed in Table 2.
Statistical analyses
The expression levels of lncRNAs and mRNAs that were differentially expressed between ESCC and normal tissues were compared using the Bioconductor package limma (version 3.26.1) and R (version 3.2.2) software. Co-expression relationships between the lncRNAs and the protein-coding genes were estimated by Spearman’s correlation test. The false discovery rate (FDR) was also calculated to correct the P value for multiple testing, and unless otherwise stated, statistical significance was considered at P < 0.05.
Results
Differentially expressed lncRNAs and mRNAs in ESCC tissues
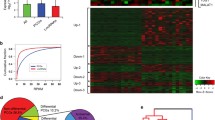
Fragments per kilobase of exon per million mapped fragments (FPKMs) were calculated for normalization of the expression level of each lncRNA and mRNA. We identified 127 lncRNAs that were differentially expressed (fold change ≥ 4, P < 0.01) between ESCC and normal tissues (Fig. 1a, b, Table S1). Among them, 98 lncRNAs were upregulated, and 29 lncRNAs were downregulated (Fig. 1a, b, Tables 3 and S1). RP11-334E6.12, VCAN-AS1, DNM3OS, AC093850.2, and RP11-150O12.6 were the five most significantly upregulated lncRNAs in ESCC; CYP4F35P, HCG22, LINC00675, C5orf66-AS1, and AC103563.9 were the five most significantly downregulated lncRNAs (Table 3).
Differentially expressed lncRNAs and mRNAs in ESCC. a Heatmap of expression profiles for the 127 lncRNAs that showed significant expression changes (29 downregulated and 98 upregulated). The red to green color gradient indicates a high to low level of expression. b Volcano plot of P values as a function of the weighted fold change for lncRNAs in five normal and seven tumor tissues. Black dots represent lncRNAs that are not significantly differentially expressed (fold change < 4, P > 0.01), and red dots represent lncRNAs that are significantly differentially expressed (fold change ≥ 4, P < 0.01). c Heatmap of expression profiles for the 3077 mRNAs that showed significant expression changes (219 downregulated and 2858 upregulated). The red to green color gradient indicates a high to low level of expression. d Volcano plot of P values as a function of the weighted fold change for mRNAs in five normal and seven tumor tissues. Black dots represent mRNAs that were not significantly differentially expressed (fold change < 4, P > 0.001), and red dots represent lncRNAs that were significantly differentially expressed (fold change ≥ 4, P < 0.001)
mRNA expression profiles in ESCC tissues were also compared with those in non-cancerous tissues. A total of 3077 mRNAs were found to be differentially expressed (fold change ≥ 4, P < 0.001) between ESCC and non-cancerous tissues: 219 were downregulated, and 2858 were upregulated (Fig. 1c, d, Table S2).
Next, we investigated whether the 127 non-coding and 3077 coding RNAs could distinguish ESCC from normal tissues. The heatmaps showed that the seven ESCC samples clustered together in one group, clearly separated from the normal tissue samples (Fig. 1a, c). The overall changes from normal to cancer state were also observed to be separated by a difference in the expression profile of either lncRNAs or mRNAs (Fig. 1). These observations suggest that a potential dynamic interaction between lncRNAs and coding RNAs may be reshaping the landscape of the entire transcriptome during ESCC development.
Significantly co-expressed mRNAs in ESCC tissues
Genome-wide gene expression profiling of both lncRNAs and coding genes from ESCC and normal tissues was conducted to detect possible associations of lncRNAs with ESCC. We predicted the potential target mRNAs of 127 differentially expressed lncRNAs using Spearman’s correlation test, revealing 1720 mRNAs (Coef > 0.95, P < 0.001) targeted by 119 lncRNAs (8 had no targets). Among them, 165 mRNAs were negatively correlated with lncRNAs, and 1555 mRNAs were positively correlated (Table 3, Table S3).
Construction of the co-expression network
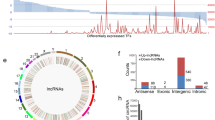
We constructed a co-expression network of the dysregulated lncRNAs and their target mRNAs; differently expressed lncRNAs and their significantly correlated mRNAs were used to draw the network with Cytoscape (version 3.2.1). The co-expression network was composed of 1469 network nodes and 1720 connection edges between 119 lncRNAs and 1350 coding genes (Fig. 2). Within this co-expression network, 1555 pairs were positively correlated and 165 pairs negatively correlated (Table S3). Interestingly, by sharing of the same mRNAs, approximately two thirds (76 of 119) of the lncRNAs and their correlated mRNAs were integrated into one complex network. This co-expression network indicates that one lncRNA could target up to 122 coding genes and that one coding gene could correlate with up to 5 lncRNAs (Fig. 2).
Predicted lncRNA and mRNA co-expression network in ESCC. The co-expression network was established between 119 significantly expressed lncRNAs and 1350 co-expressed mRNAs that had a Spearman correlation coefficient equal to or greater than 0.95. Within this co-expression network, 1555 pairs were positively correlated, and 165 pairs were negatively correlated. The diamonds represent lncRNAs, and the circles represent mRNAs. The red to green color gradient indicates a high to low level of expression
GO and KEGG pathway analyses
A GO enrichment analysis was conducted to explore the function of the co-expressed mRNAs identified in this study. Genes were organized into hierarchical categories to uncover gene regulatory networks on the basis of biological process, cellular component, and molecular function. Specifically, a two-sided Fisher’s exact test was used to determine the GO category and GO annotation list, which was greater than expected by chance (using the recommended P value cutoff of <0.05). Through GO analysis, we found that these dysregulated lncRNA transcripts are associated with developmental process and multicellular organismal development (ontology: biological process), proteinaceous extracellular matrix and extracellular matrix (ontology: cellular component), and protein binding and binding activity (ontology: molecular function). Among the genes corresponding to the identified mRNAs, 1040 are involved in biological processes, 1164 in cellular components, and 1098 in molecular functions (Fig. 3, Table S4).
To further specify and identify target mRNAs among the 1350 identified genes, significant pathways of co-expressed mRNAs were compared using the KEGG database. Without FDR correction, this analysis revealed 12 significantly enriched pathways among the transcripts (Table S5). Among these pathways, extracellular matrix (ECM)-receptor interaction (hsa04512) and chondroitin sulfate biosynthesis (hsa00532) were the only significantly enriched networks remaining after FDR correction. Some of the identified pathways, such as the classical gene category “PI3K-Akt” and “TGF-beta” signaling, have been reported to be involved in the induction of neoplasms in ESCC, but the enrichments were not significant after FDR correction (Table S5).
qRT-PCR validation of lncRNA expression
Based on the fold change, significance, and number of transcripts, three upregulated lncRNAs (RP11-334E6.12, DNM3OS, and RP11-150O12.6) and three downregulated lncRNAs (AC103563.9, RP11-7 K24.3, and RP11-351 J23.1) with only one transcript were chosen as candidates for further validation. We verified the expression of these lncRNAs by qRT-PCR using GAPDH as the reference gene with the 2-ΔΔCT method. Log2-transformed fold changes and dot plots of expression in tumor tissues vs. adjacent normal tissues are shown in Fig. 4. The results of qRT-PCR were consistent with the RNA sequencing data (Fig. 4). RP11-334E6.12 and RP11-150O12.6 were significantly upregulated in ESCC tissues (P < 0.05, Fig. 4a, c), though DNM3OS was not (P > 0.05, Fig. 4b), and AC103563.9, RP11-7 K24.3, and RP11-351 J23.1 were significantly downregulated in ESCC tissues compared to adjacent normal tissues (P < 0.05, Fig. 4d–f).
qRT-PCR validation of expression of selected lncRNAs. Both log2-transformed fold changes and dot plots of lncRNA expression in 50 pairs of tumor tissues vs. normal tissues are presented for each selected lncRNA. a, c RP11-334E6.12 and RP11-150O12.6 were significantly upregulated in ESCC tissues compared to normal tissues (P < 0.05). b Expression of DNM3OS showed no significant difference between ESCC tissues and normal tissues. d–f AC103563.9, RP11-7 K24.3, and RP11-351 J23.1 were significantly downregulated in ESCC tissues compared to normal tissues (P < 0.05)
Discussion
During the past two decades of molecular biological studies of human cancer, a number of coding genes have been determined to be genetically or epigenetically responsible for ESCC development. However, the pathogenesis of the disease remains poorly understood, and much of the alterations in gene expression and regulation involved in ESCC remain to be clarified. Moreover, the majority of lncRNAs described to date are thought to be functional, though few lncRNAs have been experimentally confirmed to be biologically relevant. For example, lncRNAs have been demonstrated to be involved in basal transcription machinery, RNA splicing and translation, and epigenetic regulation in cells [9, 14]. Overall, the cellular functions of lncRNAs remain largely unstudied. Thus, we conducted the current study to better understand the role of lncRNAs and co-expressed mRNAs in the development of ESCC. Recently, increasing evidence has confirmed that lncRNAs are important regulatory factors of gene expression either in a cis (neighboring genes) or trans (distant genes) manner, which is not easily predicted based on the lncRNA sequence [28, 29]. Therefore, predicting potential cancer-related lncRNAs by integrating various types of biological data represents an extremely important topic in such research and is attracting much attention.
Previous studies have shown that the long non-coding RNA CCAT1 promotes gall bladder cancer development via negative modulation of miRNA-218-5p [30], regulates long-range chromatin interactions at the MYC locus [31], and promotes hepatocellular carcinoma progression by functioning as a let-7 sponge [32]. CCAT1 has been reported to be a biomarker that is significantly associated with prognosis in colorectal cancer [31], hepatocellular carcinoma [33], and gastric cancer [34], as well as smoking in ESCC [35]. In the present study, significant CCAT1 upregulation was also observed in ESCC tissue relative to normal tissue and was correlated with U2AF2, HNRNPK, and SLC4A1AP, which according to GO functional annotation analysis, are associated with RNA binding and splicing. Additionally, lncRNA DNM3OS (dynamin 3 opposite strand), which is located within an intron of the Dnm3 gene, has been identified as possibly regulated by the transcription factor Twist-1 during mouse embryonic development [36]. In humans, DNM3OS encodes a miR-199a and miR-214 cluster, supporting the role of these miRNAs as novel intermediates in the pathways that control the development of hepatic stellate cells [37] and specific neural cell populations [38]. DNM3OS is also associated with Se’zary Syndrome [39]. In the present study, DNM3OS was upregulated in ESCC tissue compared with normal tissue and may play an important role in the development of ESCC. Moreover, upregulation of the lncRNA PCAT-1, which was also upregulated in ESCC compared with normal tissue, has been reported to be correlated with an advanced clinical stage and a poor prognosis in ESCC [23]. Thus, aberrant expression of the above lncRNAs has been linked to ESCC development.
To date, a few studies of differentially expressed lncRNAs in ESCC tissues have been reported. However, these studies were based on microarray technology, which tends to yield false positives and/or false negatives [40, 41]. Our study is the first to show by RNA sequencing a total of 127 differentially expressed lncRNAs, with a fold change of at least four, in ESCC tissues. A total of 119 differentially expressed lncRNAs and 1350 potential mRNA targets were then integrated into the lncRNA and mRNA co-expression network, and bioinformatic analysis revealed that these dysregulated lncRNAs are associated with cellular processes (ontology: biological process), cell (ontology: cellular component), and binding (ontology: molecular function). These lncRNAs are also associated with 12 gene pathways corresponding to transcripts involved in the cell cycle, ECM-receptor interaction, and focal adhesion, which were also enriched in another microRNA array study of ESCC [18] that was highly consistent with our study. ECM-receptor interaction leads to direct or indirect control of cellular activities, such as adhesion, migration, differentiation, proliferation, and apoptosis [42]. Our qRT-PCR results showed that the levels of RP11-334E6.12, RP11-150O12.6, AC103563.9, RP11-7 K24.3, and RP11-351 J23.1 expression were highly consistent with the RNA sequencing data. In contrast to the RNA sequencing data, DNM3OS was not significantly upregulated in ESCC tissues based on qRT-PCR, which may be due to either a false positive result or the relatively small number cohort in our validation. Overall, our results demonstrate that lncRNAs have a probable role in ESCC development and progression.
ESCC is a common malignant neoplasm worldwide, with an especially high incidence in China. The etiology, pathophysiology, and underlying molecular mechanisms of ESCC are largely unknown, and additional functional studies of candidate lncRNAs are needed to fully understand the roles of these molecules in ESCC and to effectively control this disease. This proof-of-principle with regard to the potential link between lncRNAs and ESCC presents a novel area for further investigation into the target genes of such lncRNAs, which may lead to the development of new therapeutic strategies for this disease.
References
Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–12. doi:10.1016/S0140-6736(12)60643-6.
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262.
Okines A, Cunningham D, Chau I. Targeting the human EGFR family in esophagogastric cancer. Nat Rev Clin Oncol. 2011;8:492–503. doi:10.1038/nrclinonc.2011.45.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi:10.1146/annurev-biochem-051410-092902.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-38.
Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, Foley JW, Witten DM, Giacomini CP, Flynn RA, Pollack JR, Tibshirani R, Chang HY, van de Rijn M, West RB. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi:10.1186/gb-2012-13-8-r75.
Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–75. doi:10.1038/nsmb.2428.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi:10.1038/nrg3606.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, RB W, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi:10.1038/nature08975.
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81. doi:10.1016/j.ccell.2015.02.004.
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi:10.1186/s13046-015-0123-z.
Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Chen J, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi:10.1371/journal.pone.0104044.
Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, Yan L, Wang K, Feng J. Downregulated long noncoding RNA BANCR promotes the proliferation of colorectal cancer cells via downregualtion of p21 expression. PLoS One. 2015;10:e0122679. doi:10.1371/journal.pone.0122679.
Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–31. doi:10.1158/0008-5472.CAN-14-2931.
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–96. doi:10.1016/j.molcel.2013.01.010.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi:10.1016/j.ccr.2014.03.010.
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Jin Y, Zheng X, Wu Y, Fu H. The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PLoS One. 2014;9:e101707. doi:10.1371/journal.pone.0101707.
Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, Suzuki M, Yu Y, Chen W, Eng C, Greally J, Verma A, Meltzer SJ. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–66 e4. doi:10.1053/j.gastro.2013.01.019.
Chen FJ, Sun M, Li SQ, QQ W, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, Wang CM, Lv J, De W, Wu M, Cao XF. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–15. doi:10.1002/mc.21944.
Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8:e63516. doi:10.1371/journal.pone.0063516.
Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin Y, Wu S, Mou X, Zhu Y. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289:91–6. doi:10.1016/j.cellimm.2014.03.015.
Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–44.
Shi WH, QQ W, Li SQ, Yang TX, Liu ZH, Tong YS, Tuo L, Wang S, Cao XF. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumor Biol. 2015;36:2501–7. doi:10.1007/s13277-014-2863-3.
Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Li Y, Zhao B. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2015:1–8. doi:10.1007/s13277-015-4223-3.
Tong M, Chan KW, Bao JYJ, Wong KY, Chen JN, Kwan PS, Tang KH, Fu L, Qin YR, Lok S, Guan XY, Ma S. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012;72:6024–35. doi:10.1158/0008-5472.CAN-12-1269.
Zhao Z, Bai J, Wu A, Wang Y, Zhang J, Wang Z, Li Y, Xu J, Li X. Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-seq data. Database (Oxford). 2015;2015:bav082. doi:10.1093/database/bav082.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi:10.1093/nar/gkv007.
Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One. 2010;5:e10316. doi:10.1371/journal.pone.0010316.
Hung T, Chang HY. Long noncoding RNA in genome regulation prospects and mechanisms. RNA Biol. 2010;7:582–5.
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi:10.1038/cddis.2014.541.
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–31. doi:10.1038/cr.2014.35.
Deng L, Yang SB, FF X, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi:10.1186/s13046-015-0136-7.
Zhu HQ, Zhou X, Chang H, Li HG, Liu FF, Ma CQ, Lu J. Aberrant expression of CCAT1 regulated by c-Myc predicts the prognosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:5181–5.
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–45. doi:10.1007/s00432-012-1324-x.
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q, Yin R, Xu L. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36:5523–8. doi:10.1007/s13277-015-3220-x.
Loebel DAF, Tsoi B, Wong N, Tam PPL. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–9. doi:10.1016/j.ygeno.2005.02.001.
Chen L, Chen R, Kemper S, Charrier A, Brigstock DR. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: role of exosomes in horizontal transfer of Twist1. Am J Physiol Gastrointest Liver Physiol. 2015;309:G491–9. doi:10.1152/ajpgi.00140.2015.
Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–8. doi:10.1093/nar/gkn920.
Qin Y, Buermans HP, van Kester MS, van der Fits L, Out-Luiting JJ, Osanto S, Willemze R, Vermeer MH, Tensen CP. Deep-sequencing analysis reveals that the miR-199a2/214 cluster within DNM3os represents the vast majority of aberrantly expressed microRNAs in Sezary syndrome. J Investig Dermatol. 2012;132:1520–2. doi:10.1038/jid.2011.481.
Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–42.
Wu J, Lenchik NI, Gerling IC. Approaches to reduce false positives and false negatives in the analysis of microarray data: applications in type 1 diabetes research. BMC Genomics. 2008;9(Suppl 2):S12. doi:10.1186/1471-2164-9-S2-S12.
Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209:139–51. doi:10.1530/JOE-10-0377.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Samples were obtained with informed consent, and the study was approved by the medical ethics committee of the National Cancer Center/Cancer Hospital.
Funding
The work was supported by National Natural Science Foundation of China (81301851, U1302223), The National Key Basic Research Development (973) Plan (2014CB542006) and PUMC Youth Fund (33320140169).
Rights and permissions
About this article
Cite this article
Li, Y., Shi, X., Yang, W. et al. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumor Biol. 37, 13091–13100 (2016). https://doi.org/10.1007/s13277-016-5227-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5227-3