Abstract
Background and Aim
miR-21, a putative tumor oncomiR, is a frequently overexpressed miRNA in a variety of tumors. Because it targets tumor-suppressor genes it has been linked to tumor progression. In this study we investigated the role of miR-21 in esophageal squamous cell carcinoma (ESCC), and its possible mechanism.
Methods
Expression of miR-21 was detected by stem–loop RT-PCR in tissue from 76 invasive ESCC at stage I–IV and in their corresponding para-cancerous histological normal tissues (PCHNT). Thirty endoscopic esophageal mucosal biopsy specimens from non-tumor patients were used as controls. Expression of PTEN in 76 paired ESCC and PCHNT was investigated by real-time RT-PCR and an immunohistochemical method, respectively. Paired tumor and PCHNT specimens of 20 ESCC cases were randomly selected for western blot analysis. The effect of miR-21 on PTEN expression was assessed in the ESCC cell line with an miR-21 inhibitor to reduce miR-21 expression. Furthermore, the roles of miR-21 in cell biology were analyzed by use of miR-21 inhibitor-transfected cells.
Results
Stem–loop RT-PCR revealed miR-21 was significantly overexpressed in ESCC tissues and cell lines. Overexpression of miR-21 correlated with tumor status, lymph node metastasis, and clinical stage. We demonstrated that knockdown of miR-21 significantly increased expression of PTEN protein. Consequent PTEN expression reduced cell proliferation, invasion, and migration.
Conclusions
Our findings suggest that miR-21 could be a potential oncomiR, probably by regulation of PTEN, and a novel prognostic factor for ESCC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal squamous cell carcinoma (ESCC), the predominant histological subtype of esophageal cancer, is characterized by high mortality and with regional variation in incidence in China [1, 2]. ESCC is also characterized by striking geographic variation throughout the world. The so-called “Asian esophageal cancer belt” covers the Taihang Mountain region in northern China [3, 4]. Because ESCC is usually diagnosed at a relatively late stage, treatment options are limited and five-year survival is low [5].
Although multiple genetic and epigenetic alterations have been detected in ESCC [6–8], the precise molecular mechanisms of carcinogenesis and progression of ESCC are unknown. miRNA is a recently discovered microRNA with a length of 20–22 nucleotides. It participates in modulation of significant basic cellular physiological processes, for example cell growth and differentiation, energy metabolism, and apoptosis, by controlling translation or degradation by pairing with the 3′-UTR (3′-untranslated region) of the targeted mRNA [9, 10]. Upregulation of miR-21 has been found in a variety of malignant tumors, and could thus be functioning as an oncogene in tumor development [11]. It has been discovered that miR-21 may be involved in the development of hepatic carcinoma and ESCC by inhibiting PTEN, a tumor-suppressive gene [12, 13]. Although Ma et al. demonstrated that miR-21 can act as an “oncogene” in Kazakh’s ESCC [13], its role in Han Chinese’s ESCC, and how miR-21 acts in the development of ESCC remain to be elucidated. We postulated that miR-21 regulated PTEN as one of several miR-21 target genes in ESCC. This study was undertaken to identify the role of miR-21 in ESCC in Han Chinese, to clarify the regulation of PTEN by miR-21, and to determine possible mechanisms of this regulation. We used stem–loop real-time RT-PCR and immunohistochemistry to detect expression of miR-21 and PTEN protein, respectively, in 76 patients with ESCC, and analyzed their correlation and the relationship between miR-21 and clinical pathological features. We discuss the function and possible mechanism of miR-21 in ESCC progression.
Materials and Methods
Cell Culture and Tumor Specimens
The human esophageal epithelial cell line HEEC and the ESCC cell line (EC9706, EC-1, KYSE1170, KYSE410, KYSE180) were used in this study. They were maintained in a 1:1 mixture of RPMI 1640 (Invitrogen, Carlsbad, CA, USA) and Ham’s F12 (Nissui Pharmaceutical, Tokyo, Japan) containing 10 % fetal bovine serum (FBS; Gibco BRL Life technologies, Rockville, MD, USA) in humidified 5 % CO2–air at 37 °C. To avoid possible effects on gene expression, antibiotic and antimycotic drugs were not used in the cell culture. The Institutional Review Board on Medical Ethics, Zhejiang Province Cancer Hospital approved the method of tissue collection, including informed consent. Seventy-six ESCC specimens and their corresponding non-neoplastic tissues were acquired from surgery in Zhejiang Province Cancer Hospital between February and July in 2010. Corresponding non-neoplastic tissue was cut from areas 5 cm outside the edge region of tumors and confirmed by microscopy as normal tissue. After resection, a small portion was fixed in 10 % formalin for pathological diagnosis; the rest was immediately frozen in liquid nitrogen and stored at –80 °C in a refrigerator. Complete clinical and pathological information about each recruited case was collected. There were 61 male cases and 15 female cases. All were Han Chinese. Their ages ranged from 40 to 76 years (median age, 60 years). TNM staging was guided by the 7th edition of the AJCC Cancer Staging Manual published by the International Union Against Cancer (UICC) and the American Joint Committee on Cancer (AJCC), 2010 [14]. Of 76 cases with ESCC, five were stage I, 30 were stage II, 40 were stage III, and one was stage IV (Table 1). There were 46 cases with upper and middle esophagus lesions and 30 cases with lower lesions. Sixty cases were classified as well and/or moderately differentiated and 16 cases as poorly differentiated. Samples from fifteen cases of esophageal epithelium atypical hyperplasia, 10 cases of esophagitis, and five cases of esophageal varices were obtained by endoscopic biopsy; these were 24 male cases and six female cases, age from 43 to 65 years (median age, 53 years).
Transfection
EC9706 Cells (1 × 105) were plated to 50 % confluence and were transfected with 50 nmol/L GMR-miR microRNA-21 inhibitor (GenePharma, Shanghai, China) or inhibitor-negative control by use of Lipofectamine 2000 (Invitrogen) in Opti-Mem (Invitrogen), in accordance with the manufacturer’s procedure. The culture medium was changed and transfection efficiency was determined from fluorescence images after transfection for 24 h. Cells were harvested for analysis after transfection for 48 h. All experiments were performed in triplicate.
Luciferase Reporter Assays
The 3′-untranslated region (UTR) of human PTEN mRNA containing the miR-21-binding site was amplified by PCR from human genomic DNA and inserted into the Xba1-site of pGL3 vector (Promega, Madison, WI, USA), and named pGL3-PTEN-wt. Mutations in the predicted miR-21 binding sites were performed by use of a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), with pGL3-PTEN-wt as template, and named pGL3-PTEN-mut. EC9706 cells were co-transfected in 24-well plates with wild-type (wt) or mutant reporter plasmid vector by Lipofectamine 2000; 6 h after transfection the cells were transfected again with miR-21 inhibitor or negative control. Each transfection was conducted in four wells. Luciferase assays were performed 24 h after transfection by use of the Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Transwell Migration Assay
EC9706 cells were grown to ~60 % confluence in RPMI 1640 containing 10 % fetal bovine serum and 3.0 × 105 cells were transfected with miR-21 inhibitor or negative control. After 24 h, the cells were harvested by trypsinization and washed once with Hanks’ balanced salt solution (Invitrogen). To measure cell migration, 8-mm pore size culture inserts (Transwell; Costar, High Wycombe, UK) were placed in the wells of 24-well culture plates, separating the upper and the lower chambers. RPMI1640 (400 μl) containing recombinant human hepatocyte growth factor (HGF, 20 ng/mL) was placed in the lower chambers. HGF was purchased from R&D Systems (Minneapolis, MN, USA). Then, 1 μl containing 105 cells was added to the upper chamber. After incubation for 24 h at 37 °C in 5 % CO2, the number of cells that had migrated through the pores was quantified by counting 10 independent visual fields under a microscope (Zeiss) using a ×20 objective, and cell morphology was observed by staining with hematoxylin and eosin (H&E).
RNA Extraction
All experimental containers were treated with RNase inactivator, and all reagents were prepared in 0.11 % diethylpyrocarbonate (DEPC)–water. The tumor and non-tumor specimens of 76 cases were homogenized by use of a TissueLyser II (Qiagen, USA) for 2 min at 18 Hz, in accordance with the manufacturer’s instructions, at the Zhejiang Cancer Research Institute. Total RNA was isolated from the tumor and non-tumor specimens by use of MiRNeasy Mini Kits (Qiagen) and a modified acidic guanidinium phenol–chloroform method, in accordance with the manufacturer’s instructions. Total RNA was treated with DNase I (TaKaRa Bio, Otsu, Japan) to remove genomic DNA. The mRNA was purified by use of a poly(A) purification kit (Oligotex; Qiagen), in accordance with the manufacturer’s instructions. The quality of mRNA was assessed by measurement of A260/280 ratio. RNA integrity was checked by denaturing agarose gel electrophoresis, and contamination of genomic DNA was checked by use of PCR.
Real Time RT-PCR Analysis
According to sequence MI0000077 of miR-21, the RT primer of miR-21 with stem–loop was designed as 5′-GTC GTA TCC AGT GCG TGT CGT GGA GTC GGC AAT TGC ACT GGA TAC GAC TCA ACA TC-3′. 1 μg total RNA was reverse transcribed by use of the following reaction conditions: 16 °C for 30 min, 42 °C for 42 min, 85 °C for 5 min. The primer of U6 was: 5′-GCG GTA GCT TAT CAG ACT GA-3′ (forward); 5′-TGC GTG TCG TGG AGT C-3′ (reverse). PCR reactions were performed as follows: 95 °C for 5 min, 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s; the samples underwent 40 amplification cycles. Amplification and melt curve analysis were performed by use of SYBR Premix Ex Taq (Takara), in accordance with the manufacturer’s procedure, in the ABI7500 PCR system (Applied Biosystems). Each reaction was performed in triplicate parallel holes. C T value of the sample in each reaction tube was recorded. Data were analyzed by relative quantification of real-time PCR. \( 2^{{ - \Updelta \Updelta C_{\text{T}} }} \) stands for the rate of miR-21 expression in malignant tissues over the corresponding normal tissue, within which \( 2^{{ - \Updelta \Updelta C_{\text{T}} }} \) = (C TmiR-21 − C TU6) tumor − (C TmiR-2l − C TU6) normal. For analysis of PTEN mRNA expression, real-time RT-PCR was performed using SYBR Premix Ex Taq (Takara). GAPDH was used to normalize expression levels of PTEN mRNA. The primer sequences for PTEN mRNA were (F) 5′-GAG GGA TAA AAC ACC ATG-3′ and (R) 5′-AGG GGT AGG ATG TGA ACC AGT A-3′. The primers for GAPDH were (F) 5′-GAA GGT GAA GGT CGG AGT C-3′ and (R) 5′-GAA GAT GGT GAT GGG ATT TC-3′. All real-time RT-PCR was performed in triplicate.
Immunohistochemistry
The streptavidin-peroxidase (S-P) and diaminobenzidine (DAB) chromogenic methods were adopted for immunohistochemical analysis of PTEN. The tissue specimens were formalin-fixed and paraffin-embedded. Mouse anti-human monoclonal antibodies (Santa Cruz, CA, USA) of PTEN were used. The results were determined by the proportion of positive cells and the intensity of the color. Scoring of the proportion of positive cells was as follows:
-
1
scoring according to the proportion of positive cells: no positive cells was marked as 0 point, <10 % as 1 point, 10–50 % as 2 points, 51–80 % as 3 points, >80 % as 4 points
-
2
scoring according to staining intensity: PTEN was positive when the cytoplasm or nucleus was dyed as uniform brown or dark brown granules: not dyed was marked as 0 point; light yellow as 1 point; brown as 2 points; dark brown as 3 points.
A semi-quantitative measurement was obtained by summing the two scores, with 0–1 point as negative (−), 2–3 points as weak positive (±), ≥4 points as positive (+).
Western Blot Analysis
Paired tumor and PCHNT specimens of 20 cases were randomly selected from 76 esophageal cancer patients for western blot analysis. Total protein was extracted and then quantified using the Lowry method [15]. Western blot analysis was performed using anti-PTEN monoclonal antibodies (Santa Cruz) as reported elsewhere [16]. β-Actin served as internal control.
Statistical Analysis
miR-21 expression was shown as X ± S. Statistical analysis was performed by use of SPSS 14.0 software (SPSS, Chicago, IL, USA). p < 0.05 was considered statistically significant. The 2 test was used to compare groups of categorical data of PTEN. The correlation of miR-21 and PTEN was analyzed by Spearman Rank Correlation, with double-sided p < 0.05 as statistically significant.
Results
miR-21 Is Upregulated in ESCC Cell Lines and Clinical Specimens
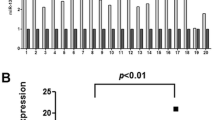
To determine if miR-21 was involved in the regulation of tumorigenesis of ESCC, we first examined the expression pattern of miR-21 in esophageal epithelium atypical hyperplasia, esophagitis, esophageal varices specimens, ESCC tissues and its corresponding normal esophagus tissues, as well as ESCC cell lines. Total RNA from all specimens were analyzed by the stem–loop RT-PCR. As a result, The relative expression of miR-21 in ESCC (2−ΔΔCT) was 5.583 ± 1.319, apparently higher than either the corresponding adjacent normal esophageal mucosa (1.089 ± 0.217) or the esophageal benign disease including esophageal epithelium atypical hyperplasia specimens, esophagitis and esophageal varices specimens (1.239 ± 0.224). The difference was statistically significant with both p values less than 0.0001 but the miR-21 expression between the latter two groups did not show significant difference (Fig. 1). Consistent with the results from specimens, miR-21 were dramatically upregulated in all 5 ESCC cell lines, as compared with HEEC cells (Fig. 1, all p < 0.0001). These results indicate that miR-21 expression is frequently upregulated in ESCC cells.
Overexpression of miR-21 in esophageal cancer tissues and cell lines compared to the corresponding normal controls. a Comparison of expression level of miR-21 between esophageal cancer tissue and normal tissue samples; b Comparison of expression level of miR-21 between esophageal cancer cell lines EC9706, EC-1, KYSE1170, KYSE410, KYSE180, and a control cell line (HEEC). Relative expression level of miR-21 was determined by stem–loop RT-PCR, and all data were normalized by U6 RNA. Mean ± SD are shown. **p < 0.01
miR-21 Overexpression Correlates with Advanced TNM Stage in ESCC
The clinical and pathological data of 76 ESCC patients are displayed in Table 1. The expression level of miR-21 was detected in 76 paired ESCC and adjacent non-tumor esophageal epithelium cells by the stem–loop RT-PCR. miR-21 was up-regulated in 61 tumor tissues compared with matched non-tumor tissues (Table 1). The difference of miR-21 expression between tumor and non-tumor tissues was statistically significant (Fig. 2a). Specifically, up-regulation of miR-21 in ESCC tissues was observed in 39 of 41 patients with stage III/IV. The difference of miR-21 expression in tumor specimens between patients with stage I/II and stage III/IV was statistically significant (Fig. 2b). Furthermore, the association of miR-21 with the clinicopathologic factors (Table 1) was examined in tumor tissues. No significant differences were found in miR-21 expression with respect to age at surgery, gender, localization, tumor diameter, histological differentiation, smoking, drinking and family history of cancer (all p > 0.05). It appeared that the miR-21 overexpression was associated with vascular invasion and advanced clinical TNM stage (all p < 0.05).
miR-21 Overexpression and Survival in ESCC Patients
Kaplan–Meier analysis showed that miR-21 expression in tumor tissues was an unfavorable predictor for the survival of ESCC patients after surgery, which was demonstrated by the significantly lower disease-free survival (DFS) of patients with miR-21 overexpression (≥5-folds) as compared with those with miR-21 expression (<5-folds) after surgery (Fig. 3). There was significant difference in the median survival (13 vs. 19 months) between patients with miR-21 overexpression (≥5-folds) and those patients with miR-21 expression (<5-folds) (p < 0.05). Retrospective analysis using Cox regression models indicated that miR-21 overexpression in tumor tissues is a promising independent predictor of survival in ESCC patients (p < 0.05). The risk ratio after adjustment for competing risk factors, sex, age, and stage of disease was found to be 3.925 (95 % confidence intervals, 3.674–4.176). The results showed that increases in miR-21 expression had a greater impact on the prognosis than tumor clinical stage.
Overexpression of miR-21 in ESCC tissues correlated with patients’ prognosis. Cumulative disease-free survival (Cum DFS) curves are plotted against miR-21 level in ESCC tissues. There was significant difference in the median survival (13 vs. 19 months) between patients with miR-21 overexpression (≥5-folds) and those patients with miR-21 expression (<5-folds) (p < 0.05)
The PTEN Protein Levels Have an Inverse Correlation with miR-21 Expression in ESCC Tissues
To identify gene targets of miR-21, we searched public algorithms, TargetScan (http://www.targetscan.org), for theoretical target genes whose downregulation could mediate the observed effects of miR-21. PTEN is a predicted target. Among the 76 pairs of matched ESCC specimens, 76 pairs were selected for analysis of PTEN mRNA (real-time RT-PCR) and PTEN protein (immunohistochemical staining), and 20 pairs were randomly selected for PTEN protein (western blotting). In comparison with the non-tumor counterparts, there was no significant difference in PTEN mRNA levels between tumor and non-tumor tissues (Fig. 4a, p > 0.05, χ 2 test). However, tumor tissues expressed significantly lower levels of PTEN protein (Fig. 4b–d); representative examples are shown in Fig. 4b, c. Next, we examined the association between PTEN protein and miR-21 in these 76 ESCC tumor samples. A statistically significant inverse correlation was observed between miR-21 and PTEN protein (Fig. 4e), with high expression of miR-21 correlating with low amounts of PTEN protein. No similar correlation was found between miR-21 and PTEN mRNA in these tumor samples (p = NS).
miR-21 and PTEN protein are inversely expressed in ESCC tissues. a PTEN-mRNA levels were evaluated by stem–loop RT-PCR. The results were normalized with GAPDH mRNA levels and are presented as relative PTEN-mRNA expression. There was no significant difference in PTEN mRNA levels between tumor and non-tumor tissues (p > 0.05, χ 2-test). b Relative expression of PTEN protein in ESCC tissues was detected by immunohistochemistry. (B-1) Absent of PTEN protein expression in ESCC tissue with miR-21 overexpression. (B-2) a representative positive, high expression of PTEN protein in ESCC tissue with miR-21 down-expression. Original magnification ×200. c 20 pairs of matched ESCC specimens were randomly selected to determine expression levels of PTEN protein by western blot and normalized to β-actin protein expression. Representative gels for PTEN protein levels in two pairs of tumor tissues and matched non-tumor tissues. d A summary of PTEN immunohistochemical staining results in 76 pairs of matched ESCC and non-tumor tissue specimens (**p < 0.01). e Correlation between miR-21 expression and PTEN protein levels in ESCC tissues (Pearson correlation, r = −0.967, p < 0.0001)
We performed immunohistochemical analysis for intratumoral heterogeneous expression of PTEN in individual ESCC specimens. Progressive reduction of PTEN protein expression was observed when tumor cells penetrated the deeper (invasive) part from the surface part. To delineate the clinical significance of PTEN downexpression, we analyzed the correlation between the downexpression of PTEN and clinicopathologic factors (Table 2). The downexpression of PTEN protein was significantly associated with gender (p = 0.004), vascular invasion (p < 0.001), T3/T4tumors (p < 0.001), lymph node metastasis (p < 0.001), stage III/IV disease (p < 0.001), and drinking (p = 0.005). However, PTEN downexpression was not associated with age at diagnosis (p = 0.484), tumor site (p = 0.815), tumor size (p = 0.113), histological differentiation (p = 0.260), smoking (p = 0.082), and family history of cancer (p = 0.575). Our data showed that down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to progression of esophageal cancer.
PTEN Is a Direct Target of miR-21
To validate whether PTEN is a direct target of miR-21, we constructed luciferase-reporter plasmids that contain the wt or mutant 3′-UTR segments of PTEN (Fig. 5a). We also mutated the miR-21 binding site in the 3′-UTR of PTEN (in the reporter plasmid). The wt or mutant reporter plasmid was cotransfected into EC9706 cells with miR-21 inhibitor or negative control. Compared with negative control, miR-21 inhibitor significantly increased the relative luciferase activity when cotransfected with the wt reporter plasmid. However, the mutant reporter plasmid abolished the miR-21 inhibitor-mediated increase in luciferase activity (Fig. 5b). These results all indicate PTEN is a direct target of miR-21 and that miR-21 suppresses PTEN by direct binding to the 3′-UTR of PTEN.
The 3′-UTR of PTEN mRNA is a target for miR-21. a Predicted miR-21 binding sites within the 3′-UTR of PTEN mRNA. The arrows indicate the mutated nucleotides. b The wt or mutant reporter plasmid was cotransfected into EC9706 cells with miR-21 inhibitor or negative control (NC). The normalized luciferase activity in the control group was set as relative luciferase activity 1. Luciferase activity of pGL3-PTEN-wt was significantly increased by miR-21 inhibitor (*p < 0.05). However, luciferase activity of pGL3-PTEN-mut was not affected by miR-21 inhibitor (p > 0.05). c miR-21 regulates PTEN expression at the post-transcriptional level and affects phosphorylation of AKT. Cells were transfected with miR-21 inhibitor, inhibitor-negative control (NC), or blank control culture medium (MOCK). Cell lysates were obtained after 48 h for analysis. miR-21 expression was detected by stem–loop RT-PCR. The results were normalized to U6 expression and expressed as fold change relative to the corresponding negative control (*p < 0.05). d Expression of PTEN and phosphorylated AKT was examined by western blot. The results were normalized to β-actin protein expression and expressed as fold change relative to the corresponding negative control (*p < 0.05)
We then assessed whether down-expression of miR-21 affected PTEN protein expression in esophageal cancer cells. EC9706 cells were transiently transfected with miR-21 inhibitor. Expression of PTEN protein was significantly increased and phosphorylated AKT expression was concomitantly reduced when endogenous miR-21 levels were reduced by miR-21 inhibitor (Fig. 5c, d), whereas PTEN mRNA was almost unaffected by alteration of miR-21 levels.
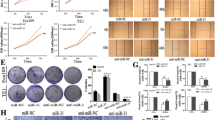
In an in vitro cell invasion assay (details are given in the section “Materials and methods”), we observed that cell invasion was significantly suppressed (p < 0.01; ~5-fold) by transfection of miR-21 inhibitor (miR-21 inhibitor group, 30 ± 5 cells/HPF; negative control group, 153 ± 11 cells/HPF) (Fig. 6).
miR-21 regulates the invasion ability of EC9706 cells. EC9706 cells that migrated through the pores: a after transfection with negative control miRNA and b after transfection with anti-miR-21 inhibitor. c Quantification of EC9706 cells that migrated through the pores after transfection with anti-miR-21 inhibitor or negative control
Taken together, these results demonstrated that the decrease of miR-21 with miR-21 inhibitor in EC9706 cells could target the PTEN gene and, in turn, suppress PTEN protein expression, leading to inhibition of cell invasion and migration.
Discussion
The onset and development of ESCC is affected by multiple genes [15, 17]. Abnormal or imbalanced expression of a variety of oncogenes and tumor-suppressor genes will result in uncontrolled cell proliferation, thus playing a key role in the onset, development, and consequence of ESCC [6–8]. Recent studies have revealed that almost 50 % of miRNAs target the tumor-associated sensitive sites in genomes, and function as oncogenes or tumor-suppressor genes by recognizing and combining with targeted molecules, leading either to degradation of mRNA or to regulation of translation of the mRNA after transcription. They play a significant role of gene regulation [9, 10].
Studies of miR-21 revealed it was up-regulated in a variety of malignant tumors [11], including breast cancer [18], gastric cancer [19], hepatocellular cancer [20], ovary cancer [21], and cervical cancer [22]. Nam et al. [23] screened the microRNAs in ovarian cancer tissues by the microarray method, and found 12 miRNAs were up-regulated, among which miR-21 was most outstanding. Huang et al. investigated the role of miR-21 in ESCC cells’ radioresistance and identified the possible mechanism. In this study, it was demonstrated that inhibition of miR-21 increased the radiosensitivity of esophageal cancer TE-1 cells, possibly as a result of activation of PTEN. Inhibition of miR-21 may be a novel therapeutic strategy to increase the radiosensitivity of esophageal cancer [24]. Ma et al. investigated the role of microRNA-21 (miR-21) and its regulation on phosphatase and tensin homolog deleted from chromosome-10 (PTEN) in esophageal cancer cell line Eca109, and 18 pairs of Kazakh’s ESCC and adjacent normal tissues by real-time quantitative PCR (qRT-PCR). The author concluded that MiR-21 was overexpressed in vitro and ESCC, and promoted cell proliferation, might target PTEN at post-transcriptional level, and regulated the cancer invasion in Kazakh’s ESCC [13]. Although it has been demonstrated that miR-21 can act as an “oncogene” in Kazakh’s ESCC, its role in Han Chinese’s ESCC, the correlation between its abnormal expression and the clinical pathological features of ESCC, remains to be elucidated. Our study detected the expression of miR-21 in 76 cases with ESCC tissues by stem–loop real time RT-PCR, and analyzed its correlation with clinical pathological features. We found that expression of miR-21 in ESCC was significantly higher than in both the corresponding normal tissues or in esophageal benign disease specimens (all p < 0.01). With the increase of clinical stage, miR-21 was apparently up-regulated and was not associated with patient’s age at surgery, gender, tumor position or length, histological differentiation, smoking or drinking behavior, and family history of cancer (all p > 0.05). The results indicated that high expression of miR-21 may be closely related with the development of esophageal cancer [25]. miR-21 may function by regulating a tumor-suppressive gene, thus affecting the biological features of the tumor cell, improving its invasiveness and metastasis ability, so it finally proceeds to lymph nodes and results in metastasis [23–27]. Because clinical stage, pathological classification, and lymph node metastasis are all important factors related to the prognosis of carcinoma patients [28–31], high expression of miR-21 is probably associated with poor prognosis for esophageal carcinoma [32–34].
PTEN is a tumor-suppressive gene with diphosphohydrolase activity [35]. It is important in cell growth, proliferation, migration, and apoptosis [36–39]. Research has shown that lack of PTEN protein expression is involved in the mechanism of ESCC. Many scholars have studied the mechanism of PTEN inactivation in esophageal carcinoma but could not identify gene mutation, methylation, or loss of heterozygosity (LOH) as the main cause [40]. Studies revealed that the rate of mutation of the PTEN gene was quite low and mainly occurred in patients at clinical stage I, which could not explain why expression of PTEN protein decreased as clinical stage and pathological classification increased [40, 41]. In recent years, studies have revealed that different miRNAs participated in the tumorigenesis [42, 43]. The new discovery that miRNA silenced the targeted genes after transcription is regarded as an important epigenetic regulatory mechanism [44, 45].
In this study it was found that loss of PTEN protein occurred in esophageal squamous cell carcinoma. Its expression level apparently decreased as clinical and pathological stage increased or when lymph node metastasis occurred. Loss of PTEN protein is significantly negatively correlated with miR-21 expression. This indicates that highly expressed miR-21 may combine with the PTEN gene, silence it after transcription, inhibit expression of the targeted gene at the translation level, regulate the malignancy of esophageal squamous cancer cells, and participate in the molecular mechanism of esophageal carcinoma, thus playing a significant role in the development of esophageal cancer [46, 47]. It has been reported that the site combining with miR-21 is in the 3′-UTR region, and that expression of PTEN protein would be improved, and cell proliferation, migration, and metastasis would be inhibited, when miR-21 was inhibited in hepatic carcinoma cells [48]. Studies have shown that miR-21 regulated PTEN as one of several miR-21 target genes, which result in alteration of the biological characteristics of some tumors, for example hepatic carcinoma, breast cancer, colorectal cancer, and gastric cancer [48–51]. Xiong et al. [49] reported that miR-21 targets PTEN at the post-transcriptional level and regulates cell proliferation and invasion in human colorectal cancer cells. Han et al. [50] demonstrated that antagonism of miR-21 reverses EMT and CSC phenotype by targeting PTEN, via inactivation of AKT and ERK1/2 pathways, and showed a novel mechanism which might relieve the malignant biological behavior of breast cancer. Yang et al. suggested that miR-21 may modulate the PTEN/PI3K/Akt pathway, thus providing a novel mechanism for cisplatin resistance in gastric cancer. This suggests that regulation of miR-21 on PTEN could be one of the important molecular mechanisms in the development of malignant tumors [48–51]. This postulation was verified by further experimental studies on EC9706 cells. As shown in Fig. 5b, luciferase activity of the wt, but not mutant, PTEN-3′-UTR reporter was significantly increased in EC9706 cells transfected with miR-21 inhibitor compared with negative control. Furthermore, transfection of EC9706 cells with miR-21 inhibitor resulted in a significant increase in PTEN protein without any change in PTEN mRNA and decreased phosphorylated AKT expression (Fig. 5c, d). These results show that miR-21 post- transcriptionally down-regulates PTEN via binding to the 3′-UTR of PTEN mRNA. PTEN has been shown to inhibit tumor cell growth and invasion by blocking the PI3 K/AKT pathway [52]. Our results demonstrated that miR-21 inhibitor restrains cell growth and invasion in EC9706 cells. Thus, restraint on EC9706 cell growth and invasion by miR-21 inhibitor may be partially mediated via up-regulation of PTEN. Last, our study suggests the relationship among miR-21, PTEN, and the prognosis of esophagus squamous cell carcinoma requires further investigation with a larger sample size and follow up of the survival years of the patients. In the future this will provide a basis for further study in the search for indicators of prognosis and treatment targets for esophagus squamous cell carcinoma.
References
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150.
Eslick G. Epidemiology of esophageal cancer. Gastroenterol Clin N Am. 2009;38:17–25.
Bader F, Anwar N, Mahmood S. Geographical variation in the epidemiology of esophageal cancer in Pakistan. Asian Pac J Cancer Prev. 2005;6:139–142.
Wu KS, Huo X, Zhu GH. Relationships between esophageal cancer and spatial environment factors by using geographic information systems. Sci Total Environ. 2008;393:219–225.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249.
Kuwano H, Kato H, Miyazaki T. Genetic alterations in esophageal cancer. Surg Today. 2005;35:7–18.
Sugimoto T, Seki N, Shimizu S, et al. The galanin signaling cascade is a candidate pathway regulating oncogenesis in human squamous cell carcinoma. Genes Chromosomes Cancer. 2009;48:132–142.
Qin YR, Wang LD, Fan ZM, Kwong D, Guan XY. Comparative genomic hybridization analysis of genetic aberrations associated with development of esophageal squamous cell carcinoma in Henan, China. World J Gastroenterol. 2008;14:1828–1835.
Davis BN, Hata A. microRNA in cancer—the involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer. 2010;1:1100–1104.
Nicolas FE, Lopez-Martinez AF. MicroRNAs in human diseases. Recent Pat DNA Gene Seq. 2010;4:142–154.
Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor-suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658.
Ma WJ, Lv GD, Tuersun A, et al. Role of microRNA-21 and effect on PTEN in Kazakh’s esophageal squamous cell carcinoma. Mol Biol Rep. 2011;38:3253–3260.
International Union Against Cancer (UICC). In: Sobin LH, Gospodarowicz MK, Wittekind Ch, eds. TNM Classification of Malignant Tumours, 7th ed. New York: Wiley-Liss; 2010.
Noble JE, Bailey MJ. Quantitation of protein. Methods Enzymol. 2009;463:73–95.
Ge MH, Chen C, Xu JJ, Ling ZQ. Critical regions and spreading of runt-related transcription factor-3 C-phosphate-G (CpG) island methylation in human salivary gland adenoid cystic carcinoma. Hum Pathol. 2011;42:1862–1872.
Mao WM, Zheng WH, Ling ZQ. Epidemiologic risk factors for esophageal cancer development. Asian Pac J Cancer Prev. 2011;12:2461–2466.
Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast. J Breast Cancer. 2011;14:269–275.
Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor-suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658.
Lou Y, Yang X, Wang F, Cui Z, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 2010;26:819–827.
Deftereos G, Corrie SR, Feng Q, et al. Expression of mir-21 and mir-143 in cervical specimens ranging from histologically normal through to invasive cervical cancer. PLoS One. 2011;6:e28423.
Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695.
Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus. 2012; doi:10.1111/j.1442-2050.2012.01389.x.
Sheth S, Jajoo S, Kaur T, et al. Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PLoS One. 2012;7:e51655.
Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45.
Zaravinos A, Radojicic J, Lambrou GI, et al. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol. 2012;188:615–623.
Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–276.
Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol. 2013;111:71–81.
Yang M, Shen H, Qiu C, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49:604–615.
Kjaer-Frifeldt S, Hansen TF, Nielsen BS, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107:1169–1174.
Zhao Y, Schetter AJ, Yang GB, et al. microRNA and inflammatory gene expression as prognostic marker for overall survival in esophageal squamous cell carcinoma. Int J Cancer. 2013;132:2901–2909.
Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300.
Komatsu S, Ichikawa D, Takeshita H, et al. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther. 2012;12:S53–S59.
Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378.
Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510.
Stewart AL, Mhashilkar AM, Yang XH, et al. PI3 K blockade by Ad-PTEN inhibits invasion and induces apoptosis in radial growth phase and metastatic melanoma cells. Mol Med. 2002;8:451–461.
Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617.
Castellino RC, Durden DL. Mechanisms of disease: the PI3K-Akt-PTEN signaling node-an intercept point for the control of angiogenesis in brain tumors. Nat Clin Pract Neurol. 2007;3:682–693.
Hu YC, Lam KY, Tang JC, Srivastava G. Mutational analysis of the PTEN/MMAC1 gene in primary oesophageal squamous cell carcinomas. Mol Pathol. 1999;52:353–356.
Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28:1379–1386.
Reis AH, Vargas FR, Lemos B. More epigenetic hits than meets the eye: microRNAs and genes associated with the tumorigenesis of retinoblastoma. Front Genet. 2012;3:284.
Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20.
Goeppert B, Schmezer P, Dutruel C, et al. Down-regulation of tumor suppressor A kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology. 2010;52:2023–2033.
Fabbri M, Calore F, Paone A, Galli R, Calin GA. Epigenetic regulation of miRNAs in cancer. Adv Exp Med Biol. 2013;754:137–148.
Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167.
Alder H, Taccioli C, Chen H, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736–1744.
Liu C, Yu J, Yu S, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53:98–107.
Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219–228.
Han M, Liu M, Wang Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One. 2012;7:e39520.
Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306C:162–168.
Vogt PK, Gymnopoulos M, Hart JR. PI 3-kinase and cancer: changing accents. Curr Opin Genet Dev. 2009;19:12–17.
Acknowledgments
This research was supported by a grant from the Program for New Century Excellent Talents in University, Ministry of Education, China (NCET-11-0949), a grant from the Science and Technology General Project of Zhejiang Province (no. 2009C33143), Key Research Projects of Medicine, Ministry of Health, China (no. WKJ2010-2-004), partly by a grant from the Backbone Talent of Zhejiang Provincial Medicine and Hygiene Platform Programs (no. 2011RCA009), a grant from the Scientific and Technological Innovations Fund of Henan Province Higher Education (no. 2009HAST1T001) and a grant from the Science and Technology Key Project of the Ministry of Education, China (no. 210130).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, P., Mao, WM., Zheng, ZG. et al. Down-Regulation of PTEN Expression Modulated by Dysregulated miR-21 Contributes to the Progression of Esophageal Cancer. Dig Dis Sci 58, 3483–3493 (2013). https://doi.org/10.1007/s10620-013-2854-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-013-2854-z