Abstract
The aim of the present study was to analyze the expression of Cyfip1 in acute lymphoblastic leukemia (ALL) and its correlations with clinical pathologic features. A total of 86 ALL samples and 32 normal peripheral blood lymphocyte (PBL) samples were enrolled in our study. mRNA expression of the Cyfip1 was assessed by real-time fluorescent relatively quantitative PCR, and Cyfip1 protein expression was evaluated by Western blot analysis. As a result, both mRNA and protein expression levels of Cyfip1 were significantly lower in ALL patients than those in the control samples (P = 0.025 and 0.000, respectively). Moreover, both mRNA and protein expression of both had an inverse relation with lymph node metastasis (P = 0.015 and 0.007, respectively), In conclusion, detecting mRNA and protein expression of Cyfip1 could provide clinically significant information relevant to diagnosis, progression, and treatment modalities for ALL, and Cyfip1 may serve as a potential biomarker for diagnosis and prognosis in ALL.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy which mainly affects children between 2 and 5 years old. According the reports and statistics, it affects approximately 1–4.75 individuals per 100,000 worldwide [1, 2]. European countries and North America and the Caribbean countries such as Switzerland, Italy, Costa Rica and United States occupy the highest incidence of ALL [3]. The present high incidence and mortality rate of ALL make a harmful effect on people’s living standard and survival quality. Although its 5-year survival rate has improved to approximately 90 % and a majority of children can be cured in recent years, the prognosis of infants and children remains poor [4].
The etiology of ALL has been considered complicated and multifactorial, including exogenous or endogenous exposures, environment factors, and genetic susceptibility. The occurrence of ALL is intricate and multistep with various genetic alterations, including activation of oncogene and inactivation of tumor suppressor genes (TSG). In recent years, TSG have been considered playing an important role in the development of ALL [5]. TSG prevent the tumor growth and progression by stopping cell proliferation, inducing cell apoptosis, allowing adhesion-independent cell survival, and promoting the normal differentiated state of a tissue. Cancer growth and progression are partly caused by suppressing or deleting the expression of TSG. It is likely that the human genome has some undiscovered TSG that may be a major goal of cancer research, as the TSG may become new drug targets.
With the goal in mind, Silva et al. applied RNAi to target genes found in chromosomes regions that frequently deleted in tumors [6]. They studied the regions of deletion that contained unknown TSGs in hopes of finding new ones. Of 29 new genes tested, they found that Cyfip1 was implicated in normal epithelial morphology, and they concluded that Cyfip1 was a potential tumor suppressor gene that regulates invasive behavior [6]. So far, the literatures on the function of Cyfip1 as a TSG are rare. Furthermore, Cyfip1 as a TSG is only linked with a few cancers including breast cancer, lung cancer, colon cancer, and bladder cancer [6]. To determinate whether there is a similar phenomenon in ALL, in the present study, we explore the mRNA expression of Cyfip1 in ALL and its correlation with clinical pathologic features.
Materials and methods
Patients and samples
All bone marrow samples were extracted from ALL patients who presented to Hematology Department of the First Affiliated Hospital of Guangxi Medical University (Nanning City, Guangxi Zhuang Autonomous Region, and China) during the period of September 1st 2011 to March 31st 2014. A total of 86 ALL patients were regarded as experimental group. In addition, 32 non-ALL patients were enrolled as control group. The diagnosis for ALL was established by internationally recognized criterion including morphology, histopathology, expression of leukocyte differentiation antigens, and the French American British (FAB) classification. Written consent was derived from patients and protocol was also approved by ethics committee of the First Affiliated Hospital of Guangxi Medical University.
Cyfip1 mRNA transcript expression
Real-time fluorescent relatively quantitative PCR analysis was used to detect the Cyfip1 expression level in ALL patients. The total RNA was extracted from bone marrow samples, and cDNA was synthesized by each RNA. The primer sequence of both target gene (Cyfip1) and housekeeping gene (18S) was originally derived from the published literatures. The specific sequence of corresponding primes was shown in Table 1. The present PCR reactions containing SYBR green dye were conducted and amplified in Corbett real-time PCR machine (Roche Diagnostics, USA). The 20-ul reaction mix consisted of 10-μl SYBR Green real-time PCR Master Mix, 2-μl cDNA, 2-μl ROX Reference Dye, 0.6-μl forward primer, 0.6-μl reverse primer, and 4.8-μl deionizer water. We make the above 20-ul reaction mix system in centrifuge for 3500 turns of 30 s. And then PCR reaction was performed in ABI7500. The PCR procedure was conducted as following steps: 10 min at 95 °C as pre-temperature, and then followed by 45 cycles of 5 s at 95 °C, 15 s of 60 °C, and 20 s of 72 °C. 18S gene was applied as an internal control and calculated to obtain the adjusted cycle threshold (∆CT) value for expression level of Cyfip1 gene. The calculation method for calculating the adjusted CT value is the relative expression of Cyfip1 = 2−∆CT. ∆CT = CT value Cyfip1 − CT value of 18S. The specific method for calculating relative expression level of tumor suppressor gene could be referenced in previous published literature [5].
Western blot analysis of Cyfip1 protein
The corresponding protein was separated by 10–12 % SDS-PAGE and transferred by membrane of PVDF. The acquisition of protein was dependent on 2× SDS sample buffer, including 4 % SDS, 10 % glycine, 10 mM EDTA, and 100 mM Tris-HCl (pH 6.8). The sample or protein should be incubated with specific primary antibodies overnight at four temperature levels. And then, it should be incubated with secondary antibodies for two hours. The following products were used in the present study: anti-Cyfip1 (ProteinTech Group) and anti-GAPDH (Proteintech Group, Inc., Cat# 10494-1-AP, dilution 1:100,000). Quantity One imaging software was applied to detect the band density of the corresponding protein.
Statistical analysis
All relevant statistical analyses were computed by the software of SPSS 20.0. Data were expressed as mean ± SD. P < 0.05 was considered statistically significant.
Results
Cyfip1 mRNA transcript expression
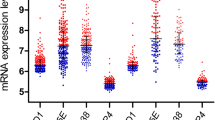
The Cyfip1 mRNA relative expression level of ALL and normal PBL samples were shown in Fig. 1. We found that the Cyfip1 mRNA expression level of ALL patients was significantly lower than that of non-ALL patients (P = 0.025). Furthermore, the Cyfip1 mRNA expression level of ALL patients was just 0.16 times of that the non-ALL patients. The detailed information about Cyfip1 mRNA expression level of ALL patients was summarized in Table 1. Based on the clinical data, we made classification for ALL patients. These clinical data consisted of age, gender, type of ALL patients, and lymph node metastasis. Consequently, significant differences were found in both type of ALL patients and lymph node metastasis. The detailed information about the relationship between Cyfip1 mRNA expression level and clinical data was summarized in Table 2.
Western blot analysis of Cyfip1 protein
The Cyfip1 protein expression of ALL patients and non-ALL patients was shown in Table 3. The expression of Cyfip1 protein was detected by Western blot in ALL tissues and normal PBL samples (Fig. 2). However, it was found only in 27 of the 86 ALL patients. In particular, a very faint Cyfip1 band could be detected in 18 of the 86 ALL patients only after a very long exposure of the blot. The 27 ALL samples showed robust expression of Cyfip1 protein that was alike in signal strength to the normal PBL samples. Almost the sample amount of Cyfip1 protein was loaded. The specific expression status of Cyfip1 protein in ALL patients and non-ALL patients were shown in Table 3. Furthermore, the detailed information about the relationship between Cyfip1 mRNA expression level and clinical data was summarized in Table 4. We found that reduced expression of Cyfip1 protein was significantly associated with lymph node metastasis of ALL.
Discussions
In recent years, more and more scientists focus their attention on genetic mechanism of ALL in order to control and prevent relatively high mobility and mortality of ALL. And it is widely accepted that tumor suppressor genes play an important role in tumor development. Cyfip1 is a new tumor suppressor gene which has been found reduced expression in several tumor types. Therefore, we think it is necessary to discuss whether there is a similar phenomenon in ALL. To the best of our knowledge, this is the first study that investigates the Cyfip1 expression in ALL.
In the present study, we found that the decreased expression of Cyfip1 mRNA and protein was significantly associated with the occurrence and lymph node metastasis of ALL. To some extent, the current study supports the viewpoint that Cyfip1 is a tumor suppressor gene and it may play an important role in the occurrence and development of ALL. It was reported that 59 % colon cancer, 63 % lung cancer, 31 % breast cancer, and 24 % bladder cancer showed negative Cyfip1 expression [6]. Our study shows 40.7 % (25/86) Cyfip1 negative expression, which does not have too much difference compared with previous studies.
Other tumor suppressor genes also show similar expression status in ALL such as WWOX, FHIT, and P73. These genes show altered or absent expression in 21–69 % cases of ALL, which is close to our results [4, 7].
Several limitations should be paid attrition to. Firstly, the present study methods consisting of real-time fluorescent relatively quantitative PCR and Western blot are not complete and sufficient. Immunohistochemistry is a good method, and we think it should be applied to investigate Cyfip1 protein expression in the next step, which contributes to make our result more reliable. Secondly, the present study only investigates the Cyfip1 expression in mRNA and protein. Although we have detected the reduced expression of Cyfip1 in ALL, its reduced mechanism remains unknown. Thus, several mechanisms such as DNA methylation, loss of heterozygosity, and point mutations should be explored in the future.
We conclude that decreased expression of Cyfip1 is associated with the occurrence and development of ALL, suggesting that Cyfip1 may serve as a potential biomarker for diagnosis and prognosis in ALL.
References
Iughetti L, Bruzzi P, Predieri B, Paolucci P. Obesity in patients with acute lymphoblastic leukemia in childhood. Ital J Pediatr. 2012;38:4.
Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48.
Dalmasso P, Pastore G, Zuccolo L, Maule MM, Pearce N, Merletti F, et al. Temporal trends in the incidence of childhood leukemia, lymphomas and solid tumors in north-west italy, 1967–2001. A report of the Childhood Cancer Registry of Piedmont. Haematologica. 2005;90:1197–204.
Chen X, Zhang H, Li P, Yang Z, Qin L, Mo W. Gene expression of WWOX, FHIT and P73 in acute lymphoblastic leukemia. Oncol Lett. 2013;6:963–9.
Chen X, Li P, Yang Z, Mo WN. Expression of fragile histidine triad (FHIT) and ww-domain oxidoreductase gene (WWOX) in nasopharyngeal carcinoma. Asian Pac J Cancer Prev : APJCP. 2013;14:165–71.
Silva JM, Ezhkova E, Silva J, Heart S, Castillo M, Campos Y, et al. Cyfip1 is a putative invasion suppressor in epithelial cancers. Cell. 2009;137:1047–61.
Ishii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, et al. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res : MCR. 2003;1:940–7.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Written consent was derived from patients, and protocol was also approved by ethics committee of the First Affiliated Hospital of Guangxi Medical University.
Rights and permissions
About this article
Cite this article
Chen, X., Qin, L., Li, P. et al. Cyfip1 is downregulated in acute lymphoblastic leukemia and may be a potential biomarker in acute lymphoblastic leukemia. Tumor Biol. 37, 9285–9288 (2016). https://doi.org/10.1007/s13277-016-4786-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4786-7