Abstract
Metastasis is one of the main causes of breast cancer (BCa)-related deaths in female. It has been reported that cancer stem cell played an important role in metastasis. Here we first revealed a specific role of pyruvate kinase isozymes M2 (PKM2) in the stemness of breast cancer cells. Breast cancer tissue analysis confirmed the upregulation of PKM2 in breast cancer, and high PKM2 levels were associated with poor prognosis of breast cancer patients. Holoclone assay and colony formation assay significantly elucidated the role of PKM2 in the self-renewal of breast cancer cells. Moreover, PKM2 elevated the proportion of stem cell and the ability of sphere formation in breast cancer cells. PKM2 played its functional role in stemness by regulating β-catenin. Collectively, we identified critical roles of PKM2 in the stemness of breast cancer cells which may elevate the therapeutic effect on breast cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BCa) is one of the most common malignancies around females in the world. As shown in reports, it is roughly 55 % in women with BCa metastasis [1, 2]. At the initial period of BCa metastasis, cells of solid tumors changed to cancer stem-like cells (CSCs) which could permit these cells to leave the tissue parenchyma and enter the systemic circulations [3]. Of note, the existence of CSCs was first proved in the studies on acute myeloid leukemia [4], and subsequent studies demonstrated that CSCs were also present in solid tumors [5–8]. CSCs are intrinsically resistant to therapy [9, 10], so tumor recurrence may happen because of the unaffected CSCs [11, 12].
In recent years, some specific surface and intracellular biomarkers have been elucidated to distinguish CSCs from other tumor cells and normal stem cells. Fluorescence-activated cell sorting (FACS) is the most common method used to identify CSCs. Breast cancer stem cells (BCSC) have been identified as CD44+, CD24−/low, and ESA+ (epithelial specific antigen) as well as lacking expression of specific lineage markers (ESA+CD44+CD24−/low Lin−) [13]. Upon the specific biomarkers, we could identify BCSC and further study the regulatory network of self-renewal, pluripotency, highly tumorigenic potential, and resistance to therapy of BCa cells.
In few years, several mechanisms that control the survival and proliferation of CSC have been elucidated. As shown in reports, the Wnt/β-catenin [14, 15], Hedgehog [15], and Notch [16] pathways have all been identified in normal stem cell and cancer stem cell biology. In the Wnt/β-catenin signaling pathway, β-catenin functions as a key signaling mediator. When the canonical Wnt signaling is activated, levels of β-catenin both in the cytoplasm and the nucleus can increase and ultimately activate target genes such as cell cycle regulators Cyclin D1 [17] and c-MYC [18], stem cell gene Bmi-1 [19], and the Wnt pathway component Axin2 [20]. In other words, β-catenin plays a critical role in the proliferation and stemness of cancer cells.
Here we focused on pyruvate kinase isozymes M2 (PKM2) which has been proven to be overexpressed in some tumor cells [21, 22]. PKM2 was reported to be associated with cellular proliferation and apoptosis, tumor metastasis, and immunological responses [23]. In this report, overexpression of PKM2 was discovered in BCa and acted as a transcriptional factor to activate β-catenin. Collectively, PKM2 is possibly essential for the stemness of the BCa cell.
Results
Overexpression of PKM2 in BCa is positively correlated with metastasis and poor prognosis
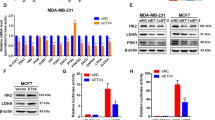
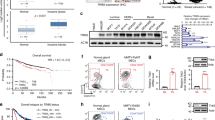
To confirm whether PKM2 has been overexpressed in BCa, we examined PKM2 levels in 96 BCa samples. We found that significantly elevated PKM2 levels were discovered in 81 BCa samples (84.375 %) compared with those in adjacent tissue samples (Fig. 1a, b). Moreover, we identified that PKM2 was expressed significantly higher in metastatic BCa samples than in non-metastatic BCa samples (Fig. 1c). These results revealed that higher levels of PKM2 were correlated with metastasis of BCa. In order to confirm whether PKM2 promotes metastasis of BCa, we examined PKM2 levels in multiple BCa cell lines. As shown in Fig. 1d, PKM2 was expressed higher in invasive BCa cell lines than in non-invasive BCa cell lines. Quantification analyses of protein levels (Fig. 1e) and RNA levels (Fig. 1f) of PKM2 further confirmed the PKM2-induced invasion of BCa cells. Moreover, we also confirm the ectopic PKM2 levels in a BCa tissue chip by immunohistochemistry. As shown in Fig. 2a, in normal breast tissue, PKM2 expression was hardly detectable, but PKM2 was overexpressed in BCa tissue and was most highly expressed in metastatic BCa tissue. Quantification analyses of PKM2 expression levels showed significant differences among different statuses of BCa (Fig. 2b).
Elevated PKM2 levels were examined in BCa and related with metastasis. a Quantification of PKM2 RNA in human breast tumors (Tumor) and adjacent breast tumor tissues (Adjacent). b Quantificational analysis of PKM2 levels in human breast tumors (Tumor) and adjacent breast tumor tissues (Adjacent). c Quantificational analysis of PKM2 levels in non-metastatic breast tumors (No-met tumor) and metastatic breast tumor tissues (Met tumor). d Western blot for PKM2 in breast cancer cell lines. e Quantificational analysis of protein levels of PKM2 in breast cell lines. f Quantificational analysis of RNA levels of PKM2 in breast cell lines. **p < 0.01 based on the Student’s t test. Error bars, SD
Elevated PKM2 levels were related with prognosis of BCa. a Immunohistochemistry of PKM2 in BCa tissue array. Scale bars: 100 μm in top, 20 μm in bottom. b Quantification of PKM2 in non-metastatic breast tumors (No-met tumor), metastatic breast tumor tissues (Met tumor), and normal breast tissues (Normal). c Correlation analysis of expression data and patient survival data from BCa tissue array showing that PKM2 levels are a risk indicator for survival. **p < 0.01 based on the Student’s t test. Error bars, SD
To evaluate whether PKM2 was associated with prognosis of BCa patients, we carried out the bioinformatics analysis of the data from the BCa tissue chip. We found that fewer patients with high PKM2 expression survived than did those with low PKM2 levels (Fig. 2c). These data suggested that PKM2 levels were significantly associated with prognosis of BCa patients.
PKM2 is essential for stemness of BCa cells
CSCs play an important role in BCa metastasis. To confirm whether PKM2 was related with CSCs, we examined the ability of BCa cells to form holoclones. We first retrovirally established BCa cell lines overexpressing or silencing PKM2. The expression levels of PKM2 in subsequent cells were examined by Western blot (Fig. 3a, b). Then, we examined the abilities of holoclone formation in indicated cells. As shown in Fig. 3c, PKM2 overexpression significantly increased the numbers of holoclones in three consecutive experiments while silencing PKM2 decreased the numbers of holoclones (Fig. 3d). In order to further confirm the ability of holoclones to self-renew, we carried out colony formation. As shown in Fig. 3e, overexpression of PKM2 significantly increased the numbers of colonies in two consecutive experiments while silencing PKM2 decreased the numbers of colonies (Fig. 3f). These results suggested that PKM2 promoted self-renewal of BCa cells.
PKM2 promoted self-renewal of BCa cells. a Western blot for PKM2 in BT20 overexpressing PKM2. pBabe: BT20 cell transfected with vector pBabe used as a control. b Western blot for PKM2 in MDA-MB-231 silencing PKM2. pSuper: MDA-MB-231 cell transfected with vector pSuper used as a control. c Holoclone assay of BT20-PKM2 and control cells. d Holoclone assay of MDA-MB-231-shPKM2 and control cells. e Colony formation assay of BT20-PKM2 and control cells. f Colony formation assay of MDA-MB-231-shPKM2 and control cells. In c and d, Exp. I: test in 96-well, Exp. II: test in a 60-mm dish, Exp. III: test in a 100-mm dish. In e and f, Exp. I: test in a 60-mm dish, Exp. II: test in a 100-mm dish. **p < 0.01 based on the Student’s t test. Error bars, SD
To further confirm the role of PKM2 in CSCs, we examined the proportion of stem cells by FACS. As shown in Fig. 4a, overexpression of PKM2 significantly increased the proportion of stem cell while silencing PKM2 decreased this proportion (Fig. 4b). Moreover, in the sphere formation assay, PKM2 significantly increased the numbers of spheres both in primary and secondary sphere formation assays (Fig. 4c), while silencing PKM2 inhibited the ability of sphere formation (Fig. 4d). Results above revealed that PKM2 played an important role in the self-renewal of CSCs in BCa.
PKM2 increased proportion of stem cell in BCa. a Analysis of stem cell with CD44high/CD24low in BT20-PKM2 cell. b Analysis of stem cell with CD44high/CD24low in MDA-MB-231-shPKM2 cell. c Sphere formation assay of BT20-PKM2 cell. d Sphere formation assay of MDA-MB-231-shPKM2 cell. **p < 0.01 based on the Student’s t test. Error bars, SD
PKM2 activates expression of β-catenin in BCa
To better understand the mechanisms by which PKM2 engaged in CSCs, we performed gene expression profiling on BT20-PKM2 and its control cells. Microarray analysis identified numbers of genes significantly differentially expressed after PKM2 overexpression (Fig. 5a). Moreover, gene set enrichment analysis indicated that cell movement, cell invasion, cancer metastasis, and β-catenin targets were significantly enriched in the cell with an overexpression of PKM2 (Fig. 5b). Moreover, expression levels of β-catenin were positively correlated with PKM2 expressions in a linear correlation analysis (Fig. 5c). To further confirm whether β-catenin was regulated by PKM2, we examined β-catenin levels in BT20-PKM2 and MDA-A-231-shPKM2 cells. As shown in the results, both the protein level (Fig. 5d) and the RNA level (Fig. 5e) of β-catenin were elevated in the cell with an overexpression of PKM2, while β-catenin expression was inhibited in the PKM2-silencing cell (Fig. 5f, g). Furthermore, we examined the downstream targets of β-catenin including c-MYC and Cyclin D1. As shown in Fig. 6, both the protein levels (Fig. 6a) and RNA levels (Fig. 6b) of C-MYC and Cyclin D1 were significantly elevated in BT20-PKM2 cell while downregulated c-MYC and Cyclin D1 were examined in MDA-MB-231-shPKM2 cell (Fig. 6c, d), suggesting that PKM2 transcriptionally regulated β-catenin.
PKM2 was correlated with β-catenin. a Supervised hierarchical clustering of the genes differentially expressed after PKM2 overexpression. b Gene set enrichment analysis was carried out using ConceptGen. Edge indicates significant overlap between two gene sets. The p values for enrichment between PKM2 signature and others determined with ConceptGen are shown. c β-catenin expression was positively correlated with PKM2 expression in BCa samples assay. d Western blot for β-catenin in BT20-PKM2 cell. e Quantification of β-catenin in BT20-PKM2 cell. f Western blot for β-catenin in MDA-MB-231-shPKM2 cell. g Quantification of β-catenin in MDA-MB-231-shPKM2 cell. **p < 0.01 based on the Student’s t test. Error bars, SD
PKM2 activated β-catenin signaling pathway. a Western blot for Cyclin D1 and c-MYC in BT20-PKM2 cell. b Quantification of Cyclin D1 and c-MYC in BT20-PKM2 cell. c Western blot for Cyclin D1 and c-MYC in MDA-MB-231-shPKM2 cell. d Quantification of Cyclin D1 and c-MYC in MDA-MB-231-shPKM2 cell. **p < 0.01 based on the Student’s t test. Error bars, SD
We then elucidated how PKM2 regulated β-catenin. We tested whether PKM2 occupied the promoter of β-catenin in the BCa cell. A quantitative chromatin immunoprecipitation (qChIP) assay was performed in BT20-PKM2 and MDA-MB-231-shPKM2 cells. We found that overexpression of PKM2 was associated with increased β-catenin levels at regions −410–180 and −2250–2000 in BT20-PKM2 cell (Fig. 7a–c). Less occupancy of those β-catenin gene promoter regions by PKM2 was detected in MDA-MB-231-shPKM2 cell (Fig. 7d, e). These results clearly indicated that PKM2 induces transcriptional activation of β-catenin.
PKM2 directly combined to the promoter of β-catenin. a Schematic presentation of three regions relative to the β-catenin transcriptional start site used as primers to test PKM2 occupied abundance. b, c qChIP was performed to assess PKM2 occupancy in BT20-PKM2 cells. IgG was used as negative control. d, e qChIP was performed to assess PKM2 occupancy in MDA-MB-231-shPKM2 cells. IgG was used as negative control. **p < 0.01 based on the Student’s t test. Error bars, SD
Silencing β-catenin in BT20-PKM2 cell restrains the proportion of CSCs
In order to confirm whether β-catenin is essential for PKM2-induced CSCs, we silenced β-catenin in BT20-PKM2 cell. As shown in Fig. 8a, downregulated c-MYC and Cyclin D1 were examined in BT20-PKM2 cell with a silent expression of β-catenin suggesting that β-catenin was successfully silenced. We then examined the proportion of stem cell in the BCa cell by FACS. We found that silencing β-catenin significantly reduced the proportion of stem cell in BT20-PKM2 cell (Fig. 8b, c). To further confirm whether β-catenin was essential for stemness, we tested the ability of spheres’ formation in BT20-PKM2 cell with silent expression of β-catenin. As shown in Fig. 8d, silencing β-catenin significantly decreased the number of spheres of BT20-PKM2 cell (Fig. 8d, e). Moreover, we found that XAV939 (inhibitor of β-catenin) also significantly inhibited the proportion of CSCs (Fig. 8f, g) and the number of spheres (Fig. 8h, i) in BT20-PKM2 cell. These data revealed that β-catenin was essential for the stemness of BCa cells induced by PKM2.
Silencing β-catenin decreased CSC in BT20-PKM2 cell. a Western blot for indicated proteins in BT20-PKM2 cell with silencing β-catenin. b Analysis of stem cell with CD44high/CD24low in BT20-PKM2 cell with silencing β-catenin by FASC. c Quantificational proportion of stem cell. d Sphere formation assay of BT20-PKM2 cell with silencing β-catenin. e Quantificational analysis of spheres. f Analysis of stem cell with CD44high/CD24low in BT20-PKM2 cell treated with XAV939 by FASC. g Quantificational proportion of stem cell. h Sphere formation assay of BT20-PKM2 cell treated with XAV939. i Quantificational analysis of spheres. **p < 0.01 based on the Student’s t test. Error bars, SD
Discussion
PKM2 is one isoenzyme of the pyruvate kinase family. In this study, we first indicated that PKM2 played a critical role in the stemness of BCa cells. We found that PKM2 was elevated in BCa, especially in metastatic BCa, and high PKM2 levels were associated with poor prognosis of BCa patients. Overexpression of PKM2 significantly promoted the abilities of holoclone formation and sphere formation. Of note, we found that PKM2 transcriptionally activated β-catenin. Silencing β-catenin in the BCa cell with an overexpression of PKM2 could suppress the functional ability of PKM2.
The Wnt-signaling pathway is an evolutionarily conserved and complex signaling cascade with important functions in both development and disease [24]. β-Catenin, as the core mediator of the canonical Wnt pathway, could translocate to the nucleus where it forms a transcriptionally active complex and drives the expression of Wnt target genes including CSC genes [25, 26]. Of note, self-renewal is the classical characteristic of CSCs [27, 28] which actively express telomerase and activate antiapoptotic and multidrug resistance pathways [29]. CSCs are considered to play an essential role in tumor initiation, progression, and relapse as well as metastasis and drug resistance [10], suggesting that CSCs were related with poor outcome [30]. In this study, we found a novel regulator of β-catenin. PKM2 could directly activate expression of β-catenin and promote self-renewal of CSCs.
In recent years, biotech companies paid attention to anticancer drugs targeting the Wnt/β-catenin signaling pathway. A series of preclinical experiments demonstrated that restraint of the Wnt/β-catenin signaling pathway can affect growth and survival of cancer cells [31]. Of note, most of the antidrugs are small molecule inhibitors, and monoclonal antibodies are designed to target cells with rapid proliferation but not CSCs. Due to the limitation above, some anticancer biological agents are failing to treat cancer. It has been reported that sorafenib fails to eradicate acute myeloid leukemia (AML) stem cells and primitive progenitors [32]. Moreover, trastuzumab has been shown to be ineffective in CSCs [33]. In this report, we revealed that PKM2 was a direct regulator of β-catenin which was important in CSCs, and anti-PKM2 biological agents may be effective in CSCs.
In summary, higher levels of PKM2 were examined in BCa and related with poorer prognosis of BCa patients. Ectopic PKM2 expression significantly elevated the proportion of CSCs in BCa. We also revealed that PKM2 directly regulated β-catenin and activated the Wnt/β-catenin signaling pathway. Together with all results, PKM2 may be a novel therapeutic target in BCa.
Materials and methods
Clinical specimens and cell culture
A total of 92 BCa specimens and their adjacent tissue samples frozen in liquid nitrogen were obtained from the First Affiliated Hospital of Medical College, Xi’an Jiaotong University. No patients had received any antitumor treatments before biopsy. All cell lines are cultured in an adaptive culture medium according to ATCC and are cultured at 37 °C in 5 % CO2.
Establishment of cell lines
A human gene PKM2 was cloned into a pBabe-puro vector, and shRNAs of PKM2 and β-catenin were cloned into a pSuper-puro vector. Retrovirus supernatants containing pBabe, pSuper, pBabe-PKM2, pSuper-sh. PKM2 and pSuper-sh. β-catenin were produced in Phoenix packaging cells. We respectively transfected indicated cell lines with these different viral supernatants containing 4 μg/ml of polybrene (Sigma). Then, cells were selected by puromycin (2 μg/ml).
RNA extraction, reverse transcription, and real-time RT-PCR
Total RNA was extracted from freshly frozen samples or cells with TRIzol reagent (Invitrogen). Total RNA was reverse-transcribed with the First Strand cDNA Synthesis Kit (Invitrogen). Real-time PCR reactions were conducted using Platinum SYBR Green qPCR SuperMix-UDG reagents (Invitrogen) on the PRISM 7900HT system (Applied Biosystems). All reactions were done in triplicate, and reactions without reverse transcriptase were used as negative controls. The GAPDH were used as the endogenous controls and the 2-ΔΔCT equation was used to calculate the relative expression levels.
Western blot analysis
A Western blot analysis was conducted using anti-PKM2, anti-β-catenin, anti-C-MYC, anti-clyclin D1, and anti-β-actin. All antibodies were purchased from Cell Signaling Technology.
Holoclone assay
A single human BCa cell was inoculated onto 96-well plates in a culture medium. During 10–12 days, primary colonies were counted. Each colony was then removed by trypsinization, and cells were transferred to 60-mm dishes with culture medium. After 12–14 days, secondary colonies were counted. Each colony was thirdly removed by trypsinization, and cells were transferred to 100-mm dishes with culture medium. After 12–14 days, secondary colonies were counted. Each plate was photographed, and the identity of the cell that founded each primary colony was determined as holoclone, meroclone, or paraclone, based on previously described methods [34].
Colony formation assay
Holoclones were plated at low density (1 × 103 cells seeded in a 60-mm dish) to achieve an initial density of 50 to 200 cells per cm2. After 6–21 days, colonies were counted under a light microscope. Each colony was secondly removed by trypsinization, and cells were transferred to 100-mm dishes (8 × 103 cells per dish) with cultured medium. After 6–21 days, colonies were counted under a light microscope.
Sphere formation assay
Cells were seeded in a 96-well low adhesion plate (Corning) at a density of 1000 cells/well and an initial volume of 200 μl. Spheres were grown in a DMEM F12 medium (Gibco) supplemented with 20 ng/ml of FGF and 4 μg/ml of heparin. Spheres were counted and photographed after 2 weeks. Spheres were monitored under the microscope daily to ensure that they were derived from single cells and that they did not become confluent during the experiment.
Isolation of stem cells
Cells were isolated using the Aldefluor kit (StemCell Technologies, Durham, NC, USA) according to the manufacturer’s instructions. Cells isolated with the Aldefluor kit were used for genetic and epigenetic analysis. Flow cytometry sorting was performed by FACS, and acquired data were analyzed using the Diva software (BD Bioscience).
Immunohistochemical analysis
Following deparaffinization, samples were immunohistochemically analyzed using antibodies for PKM2, respectively, and subsequently were pathologically confirmed for the tumor phenotype and specific immunostaining. The positive cells were counted and analyzed. The BCa tissue chip is purchased from Alenabio Company (BR2085c).
ChIP-qPCR
The ChIP kit was purchased from Millipore, and ChIP experiments were carried out essentially as described [35]. Immnuoprecipitated DNA was analyzed on the ABI PRISM 7900HT sequence detection system. The primers used for detection of promoters after ChIP are available upon request.
Statistical analysis
Statistical analysis data were described as the mean ± SD. Survival percent was estimated using the Kaplan–Meier method. The relationship between survival period and each of the variables was analyzed using the log-rank test for categorical variables. Comparisons between different groups were undertaken using the Student two-tailed t test. The criterion of statistical significance was p < 0.05. Statistical analysis was done with SPSS/Win11.0 software (SPSS Inc.).
References
Nemoto T, Vana J, Bedwani RN, Baker HW, McGregor FH, Murphy GP. Management and survival of female breast cancer: results of a national survey by the american college of surgeons. Cancer. 1980;45:2917–24.
Dings PJ, Elferink MA, Strobbe LJ, de Wilt JH. The prognostic value of lymph node ratio in node-positive breast cancer: a Dutch nationwide population-based study. Ann Surg Oncol. 2013;20:2607–14.
Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs. 2005;179:11–23.
Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7.
Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7.
Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–708.
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8.
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5.
Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–2.
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–5.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8.
Miki T, Yasuda SY, Kahn M. Wnt/beta-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev. 2011;7:836–46.
Takahashi-Yanaga F, Kahn M. Targeting wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–62.
Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409.
Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin d1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262–6.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12.
Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–56.
Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406.
Tolle SW, Dyson RD, Newburgh RW, Cardenas JM. Pyruvate kinase isozymes in neurons, glia, neuroblastoma, and glioblastoma. J Neurochem. 1976;27:1355–60.
Brinck U, Eigenbrodt E, Oehmke M, Mazurek S, Fischer G. L- and m2-pyruvate kinase expression in renal cell carcinomas and their metastases. Virchows Arch. 1994;424:177–85.
Gupta V, Bamezai RN. Human pyruvate kinase m2: a multifunctional protein. Protein Sci. 2010;19:2031–44.
Ring A, Kim YM, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 2014;10:512–25.
Moon RT. Wnt/beta-catenin pathway. Sci STKE. 2005;2005.
Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: a tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149–55.
Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–82.
Jamieson CH, Weissman IL, Passegue E. Chronic versus acute myelogenous leukemia: a question of self-renewal. Cancer Cell. 2004;6:531–3.
Armanios M, Greider CW. Telomerase and cancer stem cells. Cold Spring Harb Symp Quant Biol. 2005;70:205–8.
Hussenet T, Dembele D, Martinet N, Vignaud JM, du Manoir S. An adult tissue-specific stem cell molecular phenotype is activated in epithelial cancer stem cells and correlated to patient outcome. Cell Cycle. 2010;9:321–7.
Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4:a008052.
Parmar A, Marz S, Rushton S, Holzwarth C, Lind K, Kayser S, et al. Stromal niche cells protect early leukemic FLT3-ITD+progenitor cells against first-generation FLT3 tyrosine kinase inhibitors. Cancer Res. 2011;71:4696–706.
Lang J, Lan X, Liu Y, Jin X, Wu T, Sun X, et al. Targeting cancer stem cells with an 131i-labeled anti-AC133 monoclonal antibody in human colorectal cancer xenografts. Nucl Med Biol. 2015;42:505–12.
Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–6.
Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97.
Conflicts of interest
None
Authors’ contributions
Conception and design: Chengxue Dang
Development of methodology: Zheng Zhao, Chengxue Dang
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Zheng Zhao, Zhangjun Song, Zijun Liao, Haifeng Sun
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Zheng Zhao , Zijun Liao, Haifeng Sun, Baoxia Lei, Wenjuan Chen
Writing, review, and/or revision of the manuscript: Zheng Zhao, Chengxue Dang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Chengxue Dang
Study supervision: Chengxue Dang
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, Z., Song, Z., Liao, Z. et al. PKM2 promotes stemness of breast cancer cell by through Wnt/β-catenin pathway. Tumor Biol. 37, 4223–4234 (2016). https://doi.org/10.1007/s13277-015-4121-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4121-8