Abstract
Lung carcinoma is a deadly malignant disease with poor prognosis and increasing incidence in recent years. However, the molecular mechanism underlying the initiation and progression of lung cancer is still not completely elucidated. Recently, myocyte enhancer factor 2D (MEF2D) has been reported to promote the growth of liver cancer, but its implication in lung cancer is still unknown. This study is aimed to determine the role of MEF2D in lung carcinoma. Quantitative PCR (qPCR) and immunoblot assays showed that MEF2D was overexpressed in lung cancer tissues and cell lines, compared with the matched normal tissues and cell lines. Small interfering RNA (siRNA) suppression of MEF2D was able to reduce the proliferation, survival, and invasion of lung carcinoma cells. The transfection of MEF2D-expressing constructs into normal lung fibroblast cells promoted their proliferation and motility. The role of MEF2D in the growth of lung cancer was also confirmed in mice. Further study revealed that miR-218, which was underexpressed in lung carcinoma, was predicted to bind the 3′-untranslated region (UTR) of MEF2D mRNA. miR-218 was shown to suppress the activity of luciferase with MEF2D 3′-UTR. The changes in miR-218 levels affected the expression of MEF2D in lung cancer cells and normal fibroblast cells. There is also an inverse association between miR-218 abundance and MEF2D levels in the lung carcinoma specimen. Furthermore, the transfection of a plasmid that expressed MEF2D resistance to miR-218 regulation abolished the inhibitory effect of miR-218 on lung cancer cells. Collectively, MEF2D overexpression participated in the growth of lung cancers and its aberrant expression may result from the reduction of tumor suppressor miR-218.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung carcinoma is a type of malignant tumor threatening human health and has increasing incidence in recent years. Meanwhile, the patients suffering from lung carcinoma always have a poor prognosis. One of major reason why patients’ survival benefits little from medical attentions is that the underlying molecular mechanisms of lung cancer progression have not been completely elucidated yet. Thus, the identification of novel genes or molecular signaling pathways involved with lung carcinoma is always of great interests for the oncologists.
Recently, myocyte enhancer factor 2 (MEF2) family of transcription factors was shown to be implicated in the initiation and progression of human cancers. MEF2C and MEF2D have been well established to play important roles in the progression of hepatocellular carcinoma (HCC) by promoting VEGF signaling pathway and increasing cell proliferation rates, respectively [1, 2]. The invasion and metastasis of HCC were also demonstrated to be enhanced by the MEF2 protein overexpression via promoting a TGF-β1 autoregulation circuitry [3]. In addition to solid tumors, leukemia was also verified to be associated with MEF2 family of transcription factors [4–6]. Interestingly, the possible link between lung carcinoma and MEF2 family has also been uncovered in a recently published research article. Zhao et al. reported that oleanolic acid, an antitumor natural compound with cytostatic activity, can restore the expression of miR-122 and suppress the expression of its downstream target, CCNG1 and MEF2D, in lung cancer cells [7]. This finding implies that MEF2 family may be associated with lung carcinoma. However, experimental evidence is still lacked so far.
In this study, we employed multidisciplinary approaches to investigate the function and involved regulatory mechanism of MEF2 family in lung cancers.
Materials and methods
Cell culture
A549, H450, and H1229 lung carcinoma cell lines were all purchased from the American Type Culture Collection (ATCC). MRC-5 and BJ normal lung fibroblasts were also from ATCC. The above cell lines were cultured in the recommended media supplemented with 10 % fetal bovine serum (FBS) in a 37 °C and 5 % CO2 condition.
Ethics statement
Lung cancer specimen was obtained from patients according to the procedures approved by the Ethical Review Board in the First Affiliated Hospital of Jilin University (Changchun, China). These patients have provided their written informed consent for this study. The resected lung carcinoma tissues were cut into small pieces, and collagenases were used to facilitate the preparation of single cell suspension by mechanical manipulation. The cells were initially cultured in DMEM containing 10 % FBS.
Immunofluorescent staining
A549 and MRC-5 cells (2 × 104) were seeded on slides. Overnight, the cells were fixed with 70 % alcohol for 1 h. MEF2D antibodies (BD, 1:200) and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (BD, 1:5000) were added to the media, followed by 4′,6-diamidino-2-phenylindole (DAPI) (1:10,000) staining for nuclei staining. The fluorescence was observed with an immunofluorescent microscope. The density of fluorescence signaling was quantified using ImageJ software.
Real-time quantitative PCR
RNAs were isolated with TRIzol solution (Invitrogen) following the protocols provided by the manufacturer. Then, the obtained RNA molecules were transcribed into complementary DNAs (cDNAs) with ReverTra Ace qPCR RT Kit (Toyobo, Japan) following the manufacturer’s procedures. Then, quantitative PCR (qPCR) assay was conducted with TaqMan® 2× Universal PCR Master Mix (Applied Biosystems) and CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, CA, USA). The involved primers were as follows: MEF2D-forward: 5′-AGGGAAATAACCAAAAAACTACCAAA-3′, MEF2D-reverse: 5′-GCTACATGAACACAAAAACAGAGACC-3′; GAPDH-forward: 5′-GCGAGATCGCACTCATCATCT-3′, GAPDH-reverse: 5′-TCAGTGGTGGACCTGACC-3′.
The experimental procedures for the detection of microRNA (miRNA) were described previously [8].
Lentiviral vectors, small interfering RNA, and plasmids
The used lentiviral vectors, Lv-shMEF2D and Lv-scrambled, were both generously gifted by Dr. Wang (Harvard Medical School, USA). MEF2D small interfering RNA (siRNA) as well as control siRNA were kindly provided by Dr. Zhao (General Hospital of Chengdu Military Area Command of Chinese PLA, China). A plasmid used for MEF2D overexpression (pcDNA-MEF2D) was constructed following the procedures. MEF2D open reading frame sequence was amplified from the extracted cDNAs from A549 cells using two primers (MEF2D-forward: 5′-CTAGCTAGCACCATGGGGAGGAAAAAGATT-3′; MEF2D-reverse: 5′-GCTCTAGATCACTTTAATGTCCAGGT-3′). The generated PCR product was then inserted into pcDNA3.1 at the sites of Nhe I and Xba I. A control plasmid, pcDNA-GFP, was purchased from Addgene.
Western blot analysis
Total proteins were isolated from the indicated cells using M-PER Mammalian Protein Extraction Reagent (Thermo). The obtained proteins were then separated by SDS-PAGE and transferred onto a 0.45-μm cellulose acetate membrane. After blocking with 5 % BSA-containing PBS, the membrane was incubated with primary antibodies recognizing MEF2D (BD, 1:1000) or GAPDH (CST, 1:2000), followed by the treatment of horseradish peroxidase (HRP)-conjugated secondary antibodies. Finally, the bands were visualized under ChemiDoc XRS imaging system (Bio-Rad).
The detection of proliferation rates
A549 and H460 lung carcinoma cells (3 × 103) and MRC-5 normal lung fibroblasts (3 × 103), which are pretreated with siMEF2D or control siRNA, and pcDNA-MEF2D or pcDNA-GFP, respectively, were seeded in each well of 96-well plates. At the indicated time points, the number of cells in each well was determined with Scepter Handheld Automated Cell Counter (Millipore).
Cell apoptosis detection
A549, H460, and MRC-5 cells (2 × 105) were seeded in 6-well plates, followed by the infection of the indicated lentiviral vectors. Forty-eight hours later, the cells were subjected to flow cytometrical analysis of apoptotic rates with FITC-labeled annexin V/PI staining apoptosis detection kit (BioVision, Palo Alto, CA, USA) on FACSAria II Cell Sorter (BD) according to the manufacturer’s instructions.
Transwell assay
Forty-eight hours after the transduction of MEF2D siRNA or pcDNA-MEF2D as well as their corresponding controls, 1 × 103 A549, H460, and MRC-5 cells were seeded on the surface of Matrigel-pretreated inserts and then 1600 and 600 μl media were added to the lower and upper chambers. Twenty-four hours later, the cells were stained with crystal violet (0.1 %) and, after washing and drying, observed under a CX22 microscope (Olympus). Infiltrating cells were counted in ten random fields (×200). The average values were expressed as means ± SD.
Animal studies
A549 cells (5 × 106) were subcutaneously injected into right flanks of 4–5-week-old BALB/c male nude mice (n = 14). When the tumors reached 6–8 mm, the mice were equally divided into two groups (n = 7) and Lv-shMEF2D (3 × 109 plaque-forming unit (pfu)) or Lv-scrambled (3 × 109 pfu) was then intratumorally injected at a single administration. The diameters of tumors were periodically measured with calipers, and the volumes were calculated according to the following formula: volume (mm3) = length × (width)2 / 2. Forty days later, all of the mice were euthanized and the tumors were collected for weighting and immunohistological analysis. The whole animal experiment was conducted according to Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry
Immunohistological staining was performed on the collected lung carcinoma sections from mice to detect the expression of MEF2D and Ki67 as well as apoptotic cells. Briefly, formalin and paraffin were used to fix and embed the tissues, respectively. The samples were heated to retrieve the antigen. Antibodies recognizing MEF2D (HPA004807; Sigma-Aldrich, MO, USA) and Ki67 (ab15580; Abcam, CA, USA) were employed to stain these two proteins, followed by counterstaining of nuclei with hematoxylin. To detect apoptosis in the same tumor sections, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL) staining was conducted using In Situ Cell Death Detection Kit (11684817910; Roche, Germany) following the manufacturer’s procedures.
Luciferase assay
A 259-bp DNA fragment MEF2D 3′-untranslated region (UTR) harboring miR-218 MRE (GACAATC) was synthesized and then inserted into psiCheck2 to generate psiCheck2-MEF2D3UTR-wt. Meanwhile, a mutant construct (GTGTTTC) was inserted to psiCheck2 to generate psiCheck2-MEF2D3UTR-mut as control. Twenty-four hours after the transfection of miR-218 mimics and inhibitors, psiCheck2, psiCheck2-MEF2D3UTR-wt, and psiCheck2-MEF2D3UTR-mut were transfected into A549 and MRC-5 cells. Forty-eight hours later, the cells were harvested and subjected to the evaluation of luciferase expression using Dual-Luciferase® Reporter Assay System/Dual-Luciferase® (Progema).
Statistics
All the experiments except animal experiments were conducted for at least three times. All the values in this study were shown as means ± SD. The difference between different groups was considered as significant and very significant when P < 0.05 (*) and P < 0.01 (**), respectively.
Results
MEF2D is overexpressed in lung carcinoma
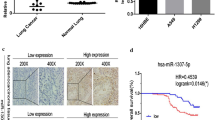
First of all, the expression profile of MEF2 family members, MEF2A, MEF2B, MEF2C, and MEF2D, was determined by qPCR assays in the lung cancer specimen (n = 30). The data revealed that the abundance of MEF2D mRNA was significantly elevated in lung cancer tissues and cells, compared with normal tissues and cells (Fig. 1a). There is no significant difference in MEF2A and MEF2C expression levels between cancerous and normal lung tissues (Fig. 1b, c). MEF2B expression was not detected in our study (data not shown). Furthermore, we confirmed the expression of MEF2D in lung carcinoma samples by immunoblot analysis. MEF2D proteins were found to be aberrantly expressed in lung cancer tissues and cells (Fig. 1d). Immunofluorescent staining further showed that MEF2D is mainly located in the nuclei of A549 lung carcinoma cells, while it is accumulated in the cytoplasm of normal lung fibroblast (Fig. 1e).
MEF2D is overexpressed in lung carcinoma. a The expression of MEF2D mRNA was detected by qPCR in the collected lung carcinoma specimen and the corresponding normal tissues (n = 30). Each value was shown as a dot, and the average values were presented as lines. P value was calculated with Student’s t test. b, c MEF2A and MEF2C levels were evaluated by qPCR in the same cohort of lung cancers. d Immunoblot analysis was used to examine the abundance of MEF2D proteins in three of the lung cancer samples and the matched normal tissues as well as in the lung cancer cells and normal lung fibroblasts. e The location of MEF2D was determined by immunofluorescent staining. DAPI was used to stain nuclei (×200)
MEF2D silencing affects the proliferation, survival, and invasion of lung carcinoma cells
To study the role of MEF2D in the progression of lung cancer cells, we employed a siRNA specifically targeting MEF2D (MEF2D siRNA) to suppress its expression (Fig. 2a). After the transfection of MEF2D siRNA, the proliferation rates of A549 and H460 lung cancer cells were shown to be greatly decreased, compared with that transfected with control siRNA (Fig. 2b). The apoptotic death was also found to be induced by the treatment of MEF2D siRNA in the two cell lines (Fig. 2c). In addition, transwell assays revealed that MEF2D siRNA impairs the invasiveness of A549 and H460 cells (Fig. 2d). The above findings indicated that MEF2D suppression leads to the reduction in the ability of lung cancer cells to proliferate, survive, and invade. BrdU assay and Ki67 expression profile showed that MEF2D suppression led to lower proliferation rates of lung cancer cells (Fig. 2a, e).
MEF2D silencing affects the proliferation, survival, and invasion of lung carcinoma cells. a MEF2D and Ki67 expressions were detected by immunoblotting 48 h after A549 and H460 cells were transfected with siMEF2D (50 nM) or control siRNA (50 nM). GAPDH was used as endogenous reference. b The number of A549 and H460 cells was determined at the indicated time points after siMEF2D (50 nM) or control siRNA (50 nM) was transfected. c The apoptotic rates of lung cancer cells were quantified by flow cytometrical analysis of sub-G0/G1 population 48 h after the above treatments. d Transwell assays were employed to examine the invasiveness of A549 and H460 cells under the same conditions (×100). e BrdU assay was conducted to detect the proliferation rates
MEF2D overexpression renders normal cells with increased proliferation rates and motility
In order to further confirm the role of MEF2D for the biological behaviors of cells, we transfected normal lung fibroblast with a plasmid expressing MEF2D (pcDNA-MEF2D, Fig. 3a). MEF2D overexpression was indicated to accelerate the proliferation of normal cells, while the delivery of a GFP-expressing vector has no influence on their proliferation (Fig. 3b). Also, transwell assays demonstrated that the elevation in MEF2D expression level promotes the invasiveness of lung fibroblasts (Fig. 3c). BrdU assay and Ki67 expression profile showed that MEF2D overexpression resulted in a higher proliferation rate in MRC-5 normal cells (Fig. 3a, d).
MEF2D overexpression renders normal cells with increased proliferation rates and motility. a MEF2D and Ki67 expressions were detected by immunoblotting 48 h after MRC-5 cells were transfected with pcDNA-MEF2D (1 ng/1 × 106 cells) or pcDNA-GFP (1 ng/1 × 106 cells). GAPDH was used as endogenous reference. b The number of MRC-5 cells was determined at the indicated time points after pcDNA-MEF2D (1 ng/1 × 106 cells) or pcDNA-GFP (1 ng/1 × 106 cells) was transfected. c Transwell assays were employed to examine the invasiveness of MRC-5 cells under the same conditions (×100). d BrdU assay was conducted to detect the proliferation rates
MEF2D suppression hampers the growth of lung cancer in mice
Given the fact that MEF2D is associated with the proliferation and survival of lung carcinoma cells in vitro, we aimed to investigate if MEF2D contributes to the growth of lung carcinoma in vivo. We established a mouse model harboring A549 lung carcinoma xenografts. Then, lentiviral vectors expressing a short hairpin for MEF2D (Lv-shMEF2D) or control (Lv-scrambled) vectors were directly injected into the established tumors. The sizes of tumors were periodically measured, and the volumes are presented in Fig. 4a. After sacrifice, the tumors were all harvested and weighted (Fig. 4b). The expression of MEF2D and Ki67 was then determined in these tumor sections by immunohistological staining. MEF2D expression was extensively detected in Lv-scrambled-injected tumors, while its expression was greatly suppressed in the group treated with Lv-shMEF2D (Fig. 4c). Ki67, a nuclear biomarker indicating a fast proliferation, was hardly detected in tumors injected with Lv-shMEF2D (Fig. 4c). Moreover, TUNEL assays were performed to examine apoptosis in the same tumors. Positive staining was easily found in the sections from Lv-shMEF2D-infected A549 human lung cancer xenografts (Fig. 4c). The above results verified the role of MEF2D in lung cancer progression in vivo.
MEF2D suppression hampers the growth of lung cancer in mice. a Lv-shMEF2D (3 × 109 pfu) or Lv-scrambled (3 × 109 pfu) was directly injected into the established A549 lung cancer xenografts in mice (groups = 2, n = 7). The tumor volumes at the indicated time points were shown as means ± SD. b The tumor weights were also measured and shown as dots. The average values were expressed as lines. c Immunohistological staining was conducted to detect the expression of MEF2D, Ki67, and TUNEL (×200)
Tumor suppressor miR-218 negatively regulates the transcription of MEF2D in lung cancer cells
Given that MEF2D overexpression contributes to the proliferation and survival of lung carcinoma cells, we are interested in the molecular mechanism by which MEF2D expression was elevated in lung carcinoma. Using online database, we found that there is a potential miRNA recognition element (MRE) of miR-218, a putative tumor suppressor for lung carcinoma [9, 10], within 3′-UTR of MEF2D mRNA (Fig. 5a). We inserted wild-type and mutant MEF2D 3′-UTR into a luciferase-expressing vector to generate psiCheck2-MEF2D3UTR-wt and psiCheck2-MEF2D3UTR-mut, respectively. Luciferase expression by psiCheck2-MEF2D3UTR-wt, but not psiCheck2-MEF2D3UTR-mut, was found to be greatly suppressed by synthetic miR-218 mimics in A549 cells (Fig. 5b). Similarly, miR-218 inhibitor increased psiCheck2-MEF2D3UTR-wt-regulated luciferase expression in normal lung fibroblasts, but not mutant constructs (Fig. 5c). Furthermore, we employed qPCR and western blot to detect MEF2D expression in A549 and normal lung fibroblast when miR-218 abundance was altered. The results revealed that miR-218 mimics were able to decrease the expression level of MEF2D in lung carcinoma cells (Fig. 5d, f), while miR-218 inhibitors elevate the expression of oncogenic MEF2D in normal cells (Fig. 5e, g). Moreover, we studied the association between the two molecules in the collected lung cancer specimen (n = 10) and confirmed that there is an inverse relationship between MEF2D and miR-218 expression levels (Fig. 5h). The above findings indicated that MEF2D is an authentic target of tumor suppressor miR-218.
Tumor suppressor miR-218 negatively regulates the transcription of MEF2D in lung cancer cells. a A potential miR-218 recognition site (MRE) is located within the 3′-UTR of MEF2D mRNA. b psiCheck2-MEF2D3UTR-wt, psiCheck2-MEF2D3UTR-mut, or psiCheck2 was transfected into A549 cells 24 h after the transduction of miR-218 mimics (50 nM). Another 48 h later, luciferase expression levels were determined in the cells. The bars represent the means ± SD of three independent experiments. c psiCheck2-MEF2D3UTR-wt, psiCheck2-MEF2D3UTR-mut, or psiCheck2 was transfected into MRC-5 cells 24 h after the transduction of miR-218 inhibitors (50 nM). Another 48 h later, luciferase expression levels were determined in the cells. The bars represent the means ± SD of three independent experiments. MEF2D mRNA was detected by qPCR in A549 cells treated with miR-218 mimics (d) and in MRC-5 cells treated with miR-218 inhibitors (e). The bars show the means ± SD of three independent experiments. f, g Immunoblot analysis was performed to examine the expression of MEF2D proteins under the above treatment. h The association between MEF2D and miR-218 levels was tested in lung cancer specimens
The inhibitory effect of miR-218 on lung cancer cells is dependent on MEF2D suppression
Regarding opposite roles of miR-218 and MEF2D in the progression of lung cancers and the link between miR-218 and MEF2D, we are interested if MEF2D suppression mediates the tumor suppressor activity of miR-218 on lung carcinoma. In order to answer the question, we transfected A549 cells with pcDNA-MEF2D (MEF2D mRNA expressed by this vector is resistant to the regulation of miR-218) and miR-218 mimics. After the treatment, we found that MEF2D overexpression abrogated the inhibitory effect of miR-218 on the proliferation rates of A549 cells (Fig. 6a). Meanwhile, flow cytometrical analysis revealed that apoptosis was also inhibited by MEF2D overexpression in A549 cells transfected with miR-218 (Fig. 6b). Transwell assays indicated that MEF2D introduction significantly enhanced the invasiveness of lung cancer cells under the treatment of synthetic miR-218 mimics (Fig. 6c).
The inhibitory effect of miR-218 on lung cancer cells is dependent on MEF2D suppression. a The number of A549 cells was determined at the indicated time points after the transfection of control (50 nM) or miR-218 mimics (50 nM), and pcDNA-GFP (1 ng/1 × 106 cells) or pcDNA-MEF2D (1 ng/1 × 106 cells). b The apoptotic rates of A549 cells were quantified by flow cytometrical analysis of sub-G0/G1 population 48 h after the above treatments. c Transwell assays were employed to examine the invasiveness of A549 cells under the same conditions (×100). d The illustration of this study is shown here. Briefly, MEF2D was found to be overexpressed in lung cancers and promotes the proliferation, survival, and invasion of lung cancer cells. MEF2D is a new target of tumor suppressor miR-218, and downregulation of miR-218 accounts for the aberrant expression of MEF2D in lung cancer
Discussion
Although MEF2D has been shown to contribute to cancer development in liver cancer [2], it is still unknown if this transcription factor is linked to the initiation and progression of lung carcinomas. In this study, we provided evidence that MEF2D is an oncogenic gene for human malignant lung tumors. This is the first time to establish the role of MEF2D in pulmonary malignancies. Thus, targeting MEF2 family members of transcription factors may be a reasonable therapeutic strategy for lung cancer treatment. However, the diagnostic and prognostic values of MEF2D for lung cancer are still determined yet. In future, some efforts should be made to address this issue.
We also explored how MEF2D affects the biological behaviors of lung cancer. Besides the enhancement in proliferation and invasion, the increase in survival of lung cancer cells was also attributed to the overexpression of MEF2D. Zhao et al. showed that MEF2D expression is suppressed by proapoptotic natural compounds, oleanolic acids [7]. Additionally, more evidence has been obtained from neurons to support the pro-survival role of MEF2D [11–13]. In fact, these findings have provided hints that MEF2D may be associated with the survival of cancer cells, regarding its overexpression in cancers. However, the experimental verification is lacked. To our knowledge, our work is the first study to confirm the anti-apoptotic effect of MEF2D on cancers.
After the establishment of the role of MEF2D in lung cancer, it is of interest what causes its overexpression in lung carcinoma cells. miR-122 downregulation has been demonstrated to be the major mechanism of MEF2D overexpression in liver cancer [2]. However, we did not detect a significant difference in miR-122 expression level between lung carcinoma tissues and the matched normal tissues (data not shown). This finding made us consider if there are other molecules regulating MEF2D expression in cancer cells. With the help of online TargetScan database, we found that there is a potential target site of miR-218 within the 3′-UTR of MEF2D. Subsequent experiments verified that MEF2D is an authentic target of miR-218, and the reduction in miR-218 abundance may be responsible for MEF2D overexpression in lung carcinoma. Therefore, our study originally added miR-218 to the list of MEF2D-regulating molecules.
miR-218 has been well demonstrated to serve as a tumor suppressor for lung cancers. The decline in miR-218 expression has been detected in squamous cell lung cancer [9]. Histone modification and ADAM9 overexpression may be responsible for the downregulation of miR-218 in lung cancer cells [14, 10]. Further studies showed that miR-218 can suppress the progression of lung cancers by targeting multiple oncogenes, such as HMGB1 [15] and N-cadherin [10]. Our study indicated that oncogenic MEF2D is also a target of miR-218, and at least partially, miR-218 exerts its antitumor effect through suppressing MEF2D expression.
Collectively, we experimentally demonstrated that MEF2D contributes to the proliferation, survival, and invasion of lung carcinoma cells and miR-218 downregulation is responsible to its overexpression (Fig. 6d). Our data suggests that MEF2D may be a promising therapeutic target for lung cancer therapy.
References
Bai XL, Zhang Q, Ye LY, Liang F, Sun X, Chen Y, et al. Myocyte enhancer factor 2C regulation of hepatocellular carcinoma via vascular endothelial growth factor and Wnt/beta-catenin signaling. Oncogene. 2014. doi:10.1038/onc.2014.337.
Ma L, Liu J, Liu L, Duan G, Wang Q, Xu Y, et al. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer Res. 2014;74(5):1452–62. doi:10.1158/0008-5472.CAN-13-2171.
Yu W, Huang C, Wang Q, Huang T, Ding Y, Ma C, et al. MEF2 transcription factors promotes EMT and invasiveness of hepatocellular carcinoma through TGF-beta1 autoregulation circuitry. Tumour Biol. 2014;35(11):10943–51. doi:10.1007/s13277-014-2403-1.
Cante-Barrett K, Pieters R, Meijerink JP. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33(4):403–10. doi:10.1038/onc.2013.56.
Lilljebjorn H, Agerstam H, Orsmark-Pietras C, Rissler M, Ehrencrona H, Nilsson L, et al. RNA-seq identifies clinically relevant fusion genes in leukemia including a novel MEF2D/CSF1R fusion responsive to imatinib. Leukemia. 2014;28(4):977–9. doi:10.1038/leu.2013.324.
Prima V, Hunger SP. Cooperative transformation by MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins generated by the variant t(1;19) in acute lymphoblastic leukemia. Leukemia. 2007;21(12):2470–5. doi:10.1038/sj.leu.2404962.
Zhao X, Liu M, Li D. Oleanolic acid suppresses the proliferation of lung carcinoma cells by miR-122/cyclin G1/MEF2D axis. Mol Cell Biochem. 2015;400(1-2):1–7. doi:10.1007/s11010-014-2228-7.
Ma L, Liu J, Shen J, Liu L, Wu J, Li W, et al. Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biol Ther. 2010;9(7):554–61.
Davidson MR, Larsen JE, Yang IA, Hayward NK, Clarke BE, Duhig EE, et al. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PLoS One. 2010;5(9):e12560. doi:10.1371/journal.pone.0012560.
Sher YP, Wang LJ, Chuang LL, Tsai MH, Kuo TT, Huang CC, et al. ADAM9 up-regulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells. PLoS One. 2014;9(4):e94065. doi:10.1371/journal.pone.0094065.
Kim MK, Kim SC, Kang JI, Hyun JH, Boo HJ, Eun SY, et al. 6-Hydroxydopamine-induced PC12 cell death is mediated by MEF2D down-regulation. Neurochem Res. 2011;36(2):223–31. doi:10.1007/s11064-010-0309-x.
Salma J, McDermott JC. Suppression of a MEF2-KLF6 survival pathway by PKA signaling promotes apoptosis in embryonic hippocampal neurons. J Neurosci. 2012;32(8):2790–803. doi:10.1523/JNEUROSCI.3609-11.2012.
Yao L, Li W, She H, Dou J, Jia L, He Y, et al. Activation of transcription factor MEF2D by bis(3)-cognitin protects dopaminergic neurons and ameliorates Parkinsonian motor defects. J Biol Chem. 2012;287(41):34246–55. doi:10.1074/jbc.M112.367540.
Wang B, Liu Y, Luo F, Xu Y, Qin Y, Lu X, et al. Epigenetic silencing of microRNA-218 via EZH2-mediated H3K27 trimethylation is involved in malignant transformation of HBE cells induced by cigarette smoke extract. Arch Toxicol. 2014. doi:10.1007/s00204-014-1435-z.
Zhang C, Ge S, Hu C, Yang N, Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Biophys Sin. 2013;45(12):1055–61. doi:10.1093/abbs/gmt109.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, L., Li, D., Zhao, Y. et al. miR-218 suppressed the growth of lung carcinoma by reducing MEF2D expression. Tumor Biol. 37, 2891–2900 (2016). https://doi.org/10.1007/s13277-015-4038-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4038-2