Abstract
miR-221/222 are two highly homologous microRNAs that are frequently upregulated in solid tumors. However, the effects of miR-221/222 in malignant gliomas have not been investigated thoroughly. In this study, we found that miR-221/222 were significantly upregulated in human glioma samples and glioma cell lines. Both gain- and loss-of-function studies showed that miR-221/222 regulate cell proliferation, the cell cycle and apoptosis, in addition to, invasion, metastasis, and angiogenesis in glioma cell lines. Subsequent investigations revealed that TIMP2 is a direct target of miR-221/222, and overexpression of TIMP2 reduced the miR-221/222-mediated invasion, metastasis, and angiogenesis of glioma cells. Taken together, our results suggest that the suppression of miR-221/222 may be a feasible approach for inhibiting the malignant behaviors of glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of short noncoding endogenous RNAs that regulate the expression of numerous genes by translational repression and mRNA cleavage or decay [1]. miRNAs play vital roles in physiological and pathological processes by repressing their target genes [2, 3]. Calin et al. [4] have demonstrated that approximately 50 % of miRNA genes are located at fragile sites and cancer susceptibility loci, indicating the potential roles of miRNAs in tumorigenesis. miRNAs are often aberrantly expressed in cancer, and their function is linked to the regulation of oncogenes and/or tumor suppressor genes in cancer cells [5–8].
Glioblastoma is the most frequent brain tumor found in adults and is the most lethal form of human cancer. Despite the improvements in treatments with combinations of surgery, radiotherapy, and chemotherapy, the survival rate of patients remains poor [9]. Many studies have demonstrated that miRNAs regulate various oncogenes and tumor suppressor genes and participate in the formation, growth, migration, and invasion of glioma [10–13]. miR-221/222 have been shown to be deregulated in gliomas and are involved in a variety of biological processes in glioma cells such as proliferation, cell cycle distribution [14], apoptosis [15], and cell migration [16]. However, until recently, little was known about the role of miR-221/222 in angiogenesis in gliomas.
Angiogenesis is essential for tumor growth, invasion, and metastasis [17], and the angiogenic switch depends on the balance of pro- and antiangiogenic factors [18]. Pro-angiogenic factors, such as vascular endothelial growth factor (VEGF), activate downstream signaling pathways and promote angiogenesis, whereas the antiangiogenic factors, such as tissue inhibitors of metalloproteinases (TIMPs), can block the formation of tumor vessels by inhibiting the activity of metallopeptidases (MMPs) or by directly suppressing endothelial cell proliferation [36–38]. Previous studies have reported that VEGF and TIMPs are downregulated by several different miRNAs [41, 42].
In this study, we aimed to demonstrate the biological function of the miR-221/222 cluster in glioma cells. We found that miR-221/222 were more highly expressed in glioma samples and cell lines compared with healthy brain samples and cells. Furthermore, we confirmed the roles of these miRNAs in proliferation, cell cycle distribution, and apoptosis in glioma cell lines. More importantly, we found that miR-221/222 play vital roles in glioma cell invasion, migration, and angiogenesis by directly targeting TIMP2, which can influence tumorigenic processes and enhance the migration and invasion of glioma cells. Overall, our results indicate that suppression of miR-221/222 is a potential therapeutic strategy for treating glioma in the future.
Materials and methods
Human tissue samples
Fourteen human glioma tissue samples were collected from adult patients who were admitted and diagnosed at Xijing Hospital at Fourth Military Medical University. Two normal brain tissue samples were obtained from para-tumor areas. Tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C. This study was approved by the hospital Institutional Review Board, and written informed consent was obtained from all patients.
Cell culture and co-transfection
Glioma cell lines U87, U251, SHG-44, BT325, and A172 were acquired from the Department of Neurosurgery at Xijing Hospital. Human normal glia cell lines HEB were purchased from Biotechnology Research (Chuanglian, Beijing, China). The glioma cells, the normal glia cell line HEB2, and human umbilical vein endothelial cells (HUVECs) were all maintained in Dulbecco’s modified Eagle medium (Gibco, Los Angeles, CA, USA) supplemented with 10 % fetal calf serum (Gibco, Los Angeles, CA, USA) and were incubated at 37 °C in a 5 % CO2 atmosphere. The cell lines were used for in vitro experiments. Transfection of cells with oligonucleotides was performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) at a final concentration of 100 nM, according to the manufacturer’s instructions. Transfection efficiency was monitored by quantitative reverse transcription-PCR (qRT-PCR).
Plasmids and oligonucleotides
The TIMP2 expression vector was created by the ligation of TIMP2 cDNA into the EcoR I and Xho I sites of the pcDNA3.1 vector. The hsa-miR-221/222 mimics (miR-221/222), negative control (miR-control), hsa-miR-221/222 inhibitors (anti-miR-221/222), inhibitor negative control (anti-miR-control) were purchased from GenePharma (Shanghai, China). The primers utilized for PCR were as follows:
-
TIMP2-F: 5′-CGGAATTCATGGACTACAAGGATGACGATGACAAAGGCGCCGCGGCCCGCA-3′;
-
TIMP2-R: 5′-CCCTCGAGTTATGGGTCCTCGATGTCG-3′.
qRT-PCR analysis of mRNA and miRNA expression
Total RNA from glioma samples and cells was extracted with TRIzol reagent (Invitrogen) for both mRNA and miRNA analyses. Relative levels of mRNA and miRNA were examined by SYBR Green real-time qRT-PCR (TaKara, Dalian, China) according to the manufacturer’s protocols. Normalization was performed relative to U6 microRNA levels. A Bio-RAD cycler real-time PCR system was used to perform the quantitative PCR. A hot start protocol was used with the following parameters: annealing at 95 °C (10 s) and extension at 55 °C (30 s) for 40 cycles, followed by a melt curve analysis. Relative expression was calculated using the △△CT method. All qRT-PCRs were performed in triplicate, and the data are presented as the mean standard error of the mean (SEM). The following primers were used for qRT-PCR:
-
Hsa-miR-221-F: 5′-AGCTACATTGTCTGCTGGGTTTC-3′;
-
Hsa-miR-222-F: 5′-AGCTACATCTGGCTACTGGG-3′;
-
U6-F: 5′-CTCGCTTCGGCAGCACA-3′;
-
U6-R: 5′-AACGCTTCACGAATTTGCGT-3′;
-
TIMP2-F: 5′-GATATACAGGCACATTATG-3′;
-
TIMP2-R: 5′-TGAATAGAACAGGCTAAG-3′;
-
ACTIN-F: 5′-TGGCATCCACGAAACTACC-3′; and
-
ACTIN-R: 5′-GTGTTGGCGTACAGGTCTT-3′.
Cell proliferation assay
Cells were seeded into a 96-well plate at 2 × 103 cells per well the day before transfection and then were co-transfected with the hsa-miR-221/222 mimic/inhibitor or mimic/inhibitor negative control as described above. Cell proliferation was then measured at 0, 24, 48, 72, and 96 h after transfection. An MTT assay was used to detect viable proliferating cells. The absorbance at 570 nm (A570) was monitored by Bio-Rad Laboratories. Each experiment was performed in triplicate.
Cell cycle and apoptosis analysis
The effects of miR-221/222 on the cell cycle and apoptosis in gliomas cells were examined by flow cytometry. Briefly, pretreated glioma cells were harvested and washed twice with phosphate buffered saline (PBS) buffer, fixed with 70 % ethanol at −20 °C for 30 min and stored at 4 °C overnight. The cells were then washed with PBS again, treated with 100 mL of 100 mg/L RNase at 37 °C for 30 min, and stained with FITC-conjugated annexin V and PE-labeled PI at 4 °C for 30 min in the dark. Cell cycle and apoptosis were measured on an EPICS XL Flow Cytometer (Coulter, USA) at 488 nm, and the data were analyzed with CellQuest software. The percentages of cells in G0/G1 phase, S phase, or undergoing apoptosis were measured by calculating the ratio of the number of cells in each group and the number of total cells. For each sample, 1 × 104 cells were measured. The experiment was repeated at least three times.
Migration and invasion assays
Migration of the cells was assessed with 8-μm, 24-well Transwell chambers (Corning, NY, USA) according to the manufacturer’s instructions. For the invasion assay, the insert membranes were coated with diluted Matrigel (BD Biosciences, San Jose, CA, USA). Cells (2 × 105) in 200 μL serum-free medium were added to the upper chamber and cultured for 24 h. Migrated cells were stained with 0.1 % crystal violet for 10 min at room temperature and examined by light microscopy. Quantification of the migrated cells was performed according to published protocols [43].
Capillary tube formation assay
HUVECs were plated at a density of 1.5 × 104 cells per well and were grown in tumor cell-conditioned medium for 6 h at 37 °C in a 48-well plate coated with Matrigel. Images of the capillary-like structures that formed were captured with a light microscope. The branch points of the formed tubes, which represent the degree of in vitro angiogenesis, were imaged and quantified in five low-power fields (×100).
Bioinformatics
The miRNA targets predicted by computer-aided algorithms were obtained from TargetScan (http://www.targetscan.org) and Microcosm Targets (http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl).
Luciferase activity assay
Human TIMP2-3′UTR reporter constructs containing the putative binding site of miR-221/222 and its identical sequence with a mutation in the miR-221/222 seed sequence (mutant) were amplified by PCR, inserted between the EcoR I and Pst I restriction sites of the PGL3-MCS2 reporter vector and validated by sequencing. A number of 293 T cells were plated at 3 × 104 cells/well in a 48-well plate and transfected with PGL3-TIMP2-3′UTR or their mutant constructs (100 ng/well) and pRL-TK (10 ng/well), together with miR-221/222 negative control (NC)/mimics (100 nM). Forty-eight hours after transfection, the activities of firefly and Renilla luciferase were consecutively measured according to the Dual-Luciferase activity assay manual (Promega). The Renilla luciferase signal was normalized to the firefly luciferase signal for each individual analysis. Each experiment was repeated at least three times. The primers used for PCR were as follows:
-
TIMP2-F: 5′-CGGAATTCTAGACATGGTTGTGGGTC-3′;
-
TIMP2-R: 5′-GCTGCAGCATGTCCCTCTCAAGATG-3′;
-
TIMP2-F (mutant) : 5′-CGGAATTCTAGACATGGTTGTGGGTC-3′;
-
TIMP2-R (mutant) : 5′-GCTGCAGTAGGCCATGATGTCAGTCT -3′.
Western blot analysis
Isolated protein lysates were quantified with the BCA method. Proteins were resolved on a precast gradient gel (10 %) and then transferred to a NC membrane. Membranes were incubated with the primary antibody overnight at 4 °C, followed by incubation with the secondary antibody for 1 h at room temperature. The experiment was repeated at least three times. Antibodies against TIMP2 (1:250 dilution, AB21342b, Sangon, Shanghai, China) and β-actin (1:2000 dilution, A5441, Sigma) were used.
Statistical analysis
The data were analyzed using the Student’s t test. All data are presented as the mean ± standard deviation (mean ± SD). Statistical analysis was performed with software SPSS 18.0. P values less than 0.05 were considered statistically significant.
Results
MiR-221/222 are upregulated in glioma cancer tissues and glioma cell lines
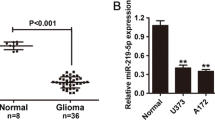
To investigate the expression levels of miR-221/222 in human gliomas, total mRNA from two normal brain tissue samples and 14 glioma tissue samples were extracted with TRIzol reagent and then qRT-PCR analyses were performed. We observed that miR-221/222 levels in glioma tissue samples were significantly higher than those in the normal brain tissue samples (Fig. 1a). Compared with HEB, a normal human brain cell line, the human glioma cell lines A172, BT325, SHG-44, U87, and U251 expressed higher levels of miR-221/222, which was in accordance with the data from tissue samples (P < 0.05, Fig. 1b). The high expression level of these miRNAs in both cancer tissues and cell lines suggests that miR-221/222 may be oncomiRs in human gliomas.
miR-221/222 are highly expressed in glioma tissue samples and glioma cell lines. a qRT-PCR analysis of miR-221/222 expression in normal brain and glioma tissue samples; N, normal brain tissue; G, glioma tissue. b qRT-PCR analysis of miR-221/222 expression in normal glia HEB cells and different glioma cell lines. The expression levels of miR-221/222 in A172, BT325, SHG-44, U87, and U251 cell lines are significantly higher than those in HEB cells, *P < 0.05
MiR-221/222 promote cellular proliferation and cell cycle and inhibit apoptosis in glioma cells
To assess the role of miR-221/222 in the biology of glioma cells, we transfected miR-221/222 mimics into A172 cells, which have a lower endogenous expression level of miR-221/222, and inhibited the expression of miR-221/222 by transfecting their inhibitors into U251 cells, which have a higher endogenous expression level (Fig. 1b). We achieved highly efficient transient transfections of the miR-221/222 mimics and inhibitors, as determined by qRT-PCR (P < 0.05, Fig. 2a). Functional assays were performed following these transfections. An MTT assay showed that the proliferation of A172 cells was increased when transfected with miR-221/222 mimics, which was consistent with the inhibitory effect observed when the miR-221/222 inhibitors were transfected into U251 cells (P < 0.05, Fig. 2b). Moreover, cell cycle distribution analysis showed that more A172 cells entered the S phase from G1 phase when treated with miR-221/222 mimics, and an inverse effect was found when miR-221/222 inhibitors were transfected into U251 cells, with more of them being arrested in S phase (P < 0.05, Fig. 2c). Furthermore, we found that miR-221/222 overexpression decreased the apoptosis rate of A172 cells (P < 0.01, Fig. 2d), while inhibitor miR-221/222 expression in U251 cells reversed this effect (P < 0.05, Fig. 2d). These results suggest that miR-221/222, as oncomiRs, participate in multiple malignant processes of glioma cells such as promoting proliferation and cell cycling and inhibiting tumor cell apoptosis.
miR-221/222 promote cellular proliferation and cell cycling and inhibit apoptosis of glioma cells. The controls, miR-221/222 mimics, and inhibitors were transfected at a final concentration of 100 nM into A172 and U251 cells; the following experiments and analyses were performed 48 h after transfection. a Transfection efficiencies of miR-221/222 mimics and inhibitors in A172 and U251 cells were quantified by qRT-PCR analysis. b, c, and d miR-221/222 mimics or control-treated A172 cells and miR-221/222 inhibitors or control-treated U251 cells were subjected to MTT assays. b *P < 0.05 and cell cycle distribution and apoptosis analyses by flow cytometry; c, d *P < 0.05, **P < 0.01
MiR-221/222 promote migration, invasion, and tube formation in glioma cells
To assess whether miR-221/222 regulate the invasiveness of glioma cells, Transwell assays were performed. The Transwell assay demonstrated that, in miR-221/222-overexpressing A172 cells, the number of invasive cells that were able to breakdown the collagen and migrate through the pores in the membrane was 40 % greater than those in the control group. An inverse effect was found when the miR-221/222 inhibitors were transfected into U251 cells (P < 0.001, Fig. 3a).
miR-221/222 promote cellular migration, invasion, and angiogenesis in glioma cells. a Transwell assays were performed 48 h after transfection of controls, miR-221/222 mimics and inhibitors at the indicated concentration in both A172 and U251 cells. b The capillary tube formation assay showed that angiogenesis of HUVECs is affected by secreted factors from miR-221/222 mimics or inhibit treated glioma cell-conditioned medium (*P < 0.05, ***P < 0.001)
To test whether miR-221/222 regulate angiogenesis of glioma, HUVECs were resuspended in glioma cell-conditioned medium and cultured in Matrigel to allow for tube formation. In the presence of cell-conditioned medium from miR-221/222-overexpressing A172 cells, HUVECs formed larger networks, with longer and more tube-like structures compared with the control group (P < 0.05, Fig. 3b). An inverse effect was observed when the HUVECs were resuspended in cell-conditioned medium from U251 cells that had miR-221/222 expression inhibited (P < 0.05, Fig. 3b). These results indicated that miR-221/222 play an important role in the angiogenesis of glioma.
TIMP2 is a target of miR-221/222 in glioma cells
To identify the effectors of miR-221/222, we first obtained the mature and seed sequences of miR-221/222. TIMP2 was identified as putative miR-221/222 target genes by prediction algorithms. TIMP2 contains seed sequences and regions of potential base-pairing with miR-221/222 in their 3′UTRs (Fig. 4a). The minimum free energy of miR-221/222 binding to the TIMP2 targeting site was calculated using an RNA hybrid for TIMP2-binding miR-221, it was −14.9 kcal/mol and for miR-222, −15.3 kcal/mol. These results further indicated that TIMP2 might be direct targets of miR-221/222.
TIMP2 is a direct target gene of miR-221/222. a TIMP2 mRNA 3′UTRs depicting target binding sites for miR-221/222. b Luciferase activity assays in 293 T cells, following co-transfection of WT or Mut 3′UTR constructs of TIMP2 (100 ng/well in 48-well plate) and miR-221/222 NC/mimics (100 nM), as indicated above. Data represent the fold change in activity (mean ± SD) of three replicates. WT, wild type; Mut, mutant. c qRT-PCR and d Western blot for TIMP2 expression 48 h post-transfection with miR-221/222 mimics and inhibitors (100 nM) in A172 and U251 cells, respectively. e qRT-PCR analysis of TIMP2 expression in normal brain and glioma tissue samples; N, normal brain tissue; G, glioma tissue. f qRT-PCR analysis of TIMP2 expression in the glioma cell lines A172, U87, and U251. (*P < 0.05, **P < 0.01, ***P < 0.001)
We next generated TIMP2 reporter constructs by inserting their 3′UTRs, containing the miR-221/222 binding sites, into the pGL3-mcs2 vector, downstream of the luciferase open reading frame (Fig. 4a). These reporter constructs were transfected into 293 T cells (100 ng/well in a 48-well plate) together with miR-221/222 NC/mimics (100 nM). Introduction of the miR-221/222 mimics into 293 T cells with the wild-type 3′-UTR constructs of TIMP2 significantly inhibited luciferase activity compared with the negative control (P < 0.001, Fig. 4b). In addition, mutation of the miR-221/222 binding sites in the TIMP2 3′-UTR reporter constructs abolished the ability of miR-221/222 to downregulate the luciferase activity (Fig. 4b).
Next, we enhanced or inhibited miR-221/222 expression in A172 and U251 cells, respectively. qRT-PCR and Western blot analyses showed that the level of TIMP2 was reduced in A172 cells upon miR-221/222 transfection, whereas, the TIMP2 level was upregulated in U251 cells upon inhibition of miR-221/222 (P < 0.05, Fig. 4c–d).
Finally, we examined the expression levels of TIMP2 in glioma specimens and cell lines by qRT-PCR. Glioma tissue samples expressed comparatively lower levels of TIMP2 than normal brain tissue samples (Fig. 4e), whereas the reverse was true of miR-221/222 levels in the same tissue samples (Fig. 1a). Consistent with this opposite expression pattern of TIMP2 and miR-221/222 in glioma tissue samples, the expression levels of TIMP2 in glioma cell lines were inversely correlated with miR-221/222 expression levels (Fig. 4f).
Taken together, several pieces of evidence, the direct binding assay, gain- and loss-of-function studies, and mRNA analyses, in both glioma tissue samples and cell lines indicate that miR-221/222 directly modulate TIMP2 level by binding to their 3′UTRs. Furthermore, this “silencing effect” of miR-221/222 on TIMP2 level may have a significant clinical impact in glioma.
Overexpression of TIMP2 partially rescues the miR-221/222-mediated increased invasion and angiogenesis in glioma cells
To confirm the function of TIMP2 in glioma cells, we first constructed vectors encoding TIMP2 on a pcDNA3.1 backbone. Upon transient transfection of these constructs into U251 cells, high levels of TIMP2 were detected by both qRT-PCR and Western blot (P < 0.05, Fig. 5a–b). Next, we examined whether the restoration of TIMP2 could rescue the carcinogenic effects of miR-221/222. TIMP2 overexpression in U251 cells was sufficient to inhibit cell invasion (P < 0.05, Fig. 5c) and angiogenesis (P < 0.01, Fig. 5d). If miR-221/222 mimics were co-transfected into TIMP2-overexpressing cells, the microRNA-mediated carcinogenic effects were partially rescued, as reflected by the lower number of invasive cells (P < 0.01, Fig. 5c) and the smaller complexes formed in the tube assay, with shorter and less tube-like structures compared with the control group (P < 0.05, Fig. 5d). These results further suggested that TIMP2 is a direct target of miR-221/222 and that miR-221/222 promote glioma cell invasion and angiogenesis, at least in part by inhibiting TIMP2 expression.
TIMP2 rescue miR-221/222-mediated changes in migration, invasion, and angiogenesis in glioma cells. a–b qRT-PCR and Western blot analyses of TIMP2 expression levels in U251 cells transfected with pcDNA3.1-TIMP2. c Transwell assays were performed 48 h after co-transfection of miR-221/222 NC/mimics (100 nM) and pcDNA3.1-TIMP2 (2 μg) at the indicated concentration in U251 cells. d Capillary tube formation assay showed that angiogenesis is regulated in HUVECs, by factors secreted by pcDNA3.1-TIMP2 and miR-221/222 NC/mimic-treated U251 cell-conditioned medium (*P < 0.05, **P < 0.01)
Discussion
This study focuses on the carcinogenetic effects of miR-221/222 as oncomiRs in glioma cells. We demonstrated the silencing effect of miR-221/222 on TIMP2 and demonstrated that the tumor-promoting effects of miR-221/222 on migration, invasion, and tube formation in glioma cells could be rescued by overexpressing TIMP2. For the first time, we discovered that miR-221/222 promote angiogenesis and metastasis of glioma cells by targeting TIMP2.
Many studies have reported on the carcinogenic role of miR-221/222 in the malignant processes of various cancers [19–26]. Among the tumor-suppressive target genes of miR-221/222, the cell cycle regulator p27Kip1 [19–22], cyclin-dependent kinase inhibitor 1C/p57 [23], estrogen receptor-α [24, 25], and the pro-apoptotic protein Bim [26] have all been reported to have an opposite expression pattern compared with miR-221/222. In glioma tissues and cells, researchers have found that miR-221/222 directly regulate apoptosis by targeting p53 upregulated modulator of apoptosis [15], and that overexpression of miR-221/222 can inhibit the expression of the tyrosine phosphatase PTPμ, which negatively regulates cell migration thus enhancing glioma tumorigenesis [16].
The tissue inhibitors of metalloproteinases, which are composed of four members [27], inhibit the activity of metallopeptidases by binding with a 1:1 stoichiometry to the active site [28]. Previous studies have shown that the third member of this family, TIMP3, is a clear target gene of miR-221/222. First discovered in non-small cell lung cancer and hepatocellular carcinoma, TIMP3, together with PTEN, is repressed by miR-221/222, resulting in cellular migration and invasion through AKT and ERK phosphorylation and the activation of MMP-3 and MMP-9 [29]. A similar effect was found in breast cancer [30], and more recently, Zhang and coworkers demonstrated that miR-221/222 directly regulate cell invasion by targeting TIMP3 and are a prognostic factor for glioma patients [31].
Similar to previous studies [32], we demonstrated that miR-221/222 target TIMP2, another member of the TIMP family that is widely distributed in the brain, to enhance cellular migration and invasion. In addition to the well-accepted fact that TIMPs can protect the extracellular matrix (ECM) from proteolytic degradation thus preventing the invasion and metastasis of tumor cells, we found a new function of the miR-221/222-meditated TIMP2 regulation, which is that rescues angiogenesis of glioma cells.
Angiogenesis, which is the formation of new blood vessels, requires endothelial cells to invade through an interstitial matrix and to proliferate [33, 34]. Accordingly, TIMPs have antiangiogenic activity either through inhibiting the activity of MMPs or by directly inhibiting endothelial cell proliferation. On one hand, because MMPs, such as MMP-2 and MMP-9 which are mostly reported to play a role in human glioma progression [35], can promote endothelial cell migration and trigger an angiogenic switch by releasing vascular VEGF from the extracellular matrix [36, 37], the inhibition of MMPs accounts for a major part of the antiangiogenic activities of TIMPs. On the other hand, researchers found that TIMP-2 can interact with integrin α3β1, which negatively regulates some tyrosine kinase receptor signal transduction pathways, leading to cell cycle arrest of endothelial cells and inhibition of angiogenesis [38, 39]. It has also been demonstrated that TIMP-3′ interacts with VEGF receptor 2, which competes with VEGF for binding to its receptor and thus results in a blockade of the VEGF signal transduction pathway and angiogenesis [40]. Therefore, the TIMP family directly inhibits endothelial cell proliferation.
Collectively, the above findings support our hypothesis that miR-221/222 promotes angiogenesis and metastasis of glioma cells by targeting TIMP2, which inhibit the activity of MMPs thereby protecting the ECM from proteolytic degradation and can directly downregulate the proliferation of endothelial cells. Thus, inhibition of miR-221/222 might be a potential therapeutic strategy for glioma treatment in the future.
Abbreviations
- VEGF:
-
Vascular endothelial growth factor
- TIMPs:
-
Tissue inhibitors of metalloproteinases
- MMPs:
-
Metallopeptidases
- ECM:
-
Extracellular matrix
- HUVECs:
-
Human umbilical vein endothelial cells
References
Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells. 2005;19(1):1–15.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105.
Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, et al. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS One. 2012;7(7):e40676.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci. 2004;101(9):2999–3004.
Yang TQ, Luo XJ, Wu TF, Ding DD, Zhao ZH, Chen GL, et al. miR-16 inhibits glioma cell growth and invasion through the suppression of BCL2 and NF-kappaB1/MMP-9 signaling pathway. Cancer Sci. 2014. doi:10.1111/cas.12351.
Lou YL, Guo F, Liu F, Gao FL, Zhang PQ, Niu X, et al. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol Cell Biochem. 2012;37(1–2):45–51.
Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls notch signalling in cancer cells. EMBO J. 2011;30(4):770–82.
Wang Y, Wang X, Zhang J, Sun G, Luo H, Kang C, et al. MicroRNAs involved in the EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 2012;106(2):217–24.
Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507.
Zhong Q, Wang T, Lu P, Zhang R, Zou J, Yuan S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J Neurosci Res. 2014. doi:10.1002/jnr.23339.
Guo M, Jiang Z, Zhang X, Lu D, Ha AD, Sun J, Du W, Wu Z, Hu L, Khadarian K, Shen J, Lin Z. miR-656 inhibits glioma tumorigenesis through repression of BMPR1A. Carcinogenesis. 2014
Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S, et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339(2):260–9.
Chen L, Wang X, Wang H, Li Y, Yan W, Han L, et al. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer. 2012;48(16):3104–11.
le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27 (Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26(15):3699–708.
Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, et al. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37(6):1621–6.
Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPμ. Oncogene. 2012;31(7):858–68.
Folkman J. Is angiogenesis an organizing principle in biology and medicine? J Pediatr Surg. 2007;42(1):1–11.
Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–10.
Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–24.
Fu X, Wang Q, Chen J, Huang X, Chen X, Cao L, et al. Clinical significance of miR-221 and its inverse correlation with p27Kip1 in hepatocellular carcinoma. Mol Biol Rep. 2011;38(5):3029–35.
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903.
Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–55.
Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–61.
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–86.
Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102(10):706–21.
Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276(12):3269–76.
Cruz-Munoz W, Khokha R. The role of tissue inhibitors of metalloproteinases in tumorigenesis and metastasis. Crit Rev Clin Lab Sci. 2008;45(3):291–338.
Bode W, Reinemer P, Huber R, Kleine T, Schnierer S, Tschesche H. The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J. 1994;13(6):1263–9.
Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509.
Lu Y, Roy S, Nuovo G, Ramaswamy B, Miller T, Shapiro C, et al. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J Biol Chem. 2011;286(49):42292–302.
Zhang C, Zhang J, Hao J, Shi Z, Wang Y, Han L, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med. 2012;10:119.
Blavier L, Henriet P, Imren S, Declerck YA. Tissue inhibitors of matrix metalloproteinases in cancer. Ann N Y Acad Sci. 1999;878:108–19.
Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–57.
Chun TH, Sabeh F, Ota I, Murphy H, McDonagh KT, Holmbeck K, et al. MT1-MMP-dependent neovessel formation within the confines of the three-dimensional extracellular matrix. J Cell Biol. 2004;167(4):757–67.
Levicar N, Nuttall RK, Lah TT. Proteases in brain tumour progression. Acta Neurochir (Wien). 2003;145(9):825–38.
Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2(10):737–44.
Fang J, Shing Y, Wiederschain D, Yan L, Butterfield C, Jackson G, et al. Matrix metalloproteinase-2 is required for the switch to the angiogenic phenotype in a tumor model. Proc Natl Acad Sci U S A. 2000;97(8):3884–9.
Seo DW, Li H, Guedez L, Wingfield PT, Diaz T, Salloum R, et al. TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell. 2003;114(2):171–80.
Seo DW, Li H, Qu CK, Oh J, Kim YS, Diaz T, et al. Shp-1 mediates the antiproliferative activity of tissue inhibitor of metalloproteinase-2 in human microvascular endothelial cells. J Biol Chem. 2006;281(6):3711–21.
Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003;9(4):407–15.
Chen L, Zhang K, Shi Z, Zhang A, Jia Z, Wang G, et al. A lentivirus-mediated miR-23b sponge diminishes the malignant phenotype of glioma cells in vitro and in vivo. Oncol Rep. 2014;31(4):1573–80.
Fan YC, Mei PJ, Chen C, Miao FA, Zhang H, Li ZL. MiR-29c inhibits glioma cell proliferation, migration, invasion and angiogenesis. J Neurooncol. 2013;115(2):179–88.
Valster A, Tran NL, Nakada M, Berens ME, Chan AY, et al. Cell migration and invasion assays. Methods. 2005;37:208–15.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (81172289 and 81472633).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
Fan Yang and Wei Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, F., Wang, W., Zhou, C. et al. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumor Biol. 36, 3763–3773 (2015). https://doi.org/10.1007/s13277-014-3017-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-3017-3