Abstract
Polymorphisms in interleukin (IL)-4/IL-13 pathway genes have previously been reported to be associated with glioma susceptibility, although results are inconsistent. We therefore performed an updated meta-analysis to determine a more precise estimation of this relationship. Twelve eligible studies were identified by searching PubMed, EMBASE, Web of Science, and the Cochrane Library electronic databases. Nine polymorphisms in genes within the IL-4/IL-13 pathway (IL-4 rs2243250, rs2070874, rs2243248, IL-4R rs1805011, rs1805012, rs1805015, rs1801275, and IL-13 rs20541 and rs1800925) were assessed for their relationship with glioma risk by computing odds ratios (ORs) and corresponding 95 % confidence intervals (CIs). Akaike’s information criterion (AIC) was used to identify the best genetic model for each polymorphism. No association between IL-4/IL-13 pathway genetic polymorphisms and glioma risk was observed in the overall population, although a significant association was found between rs2234248 and glioblastoma when stratified by histological subtype (log-additive model, OR 1.57, 95 % CI 1.11–2.24). This meta-analysis therefore suggested that IL-4/IL-13 pathway genetic polymorphisms are not associated with glioma risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is an uncommon and rapidly fatal disease, for which the etiology remains largely unknown [1]. Exposure to therapeutic doses of ionizing radiation is the strongest known environmental risk factor for glioma but only accounts for a small proportion of cases [2]. Recent studies have consistently indicated an inverse association between self-reported allergies or atopic disease and glioma risk [3–6]. Moreover, evidence has also suggested that elevated immunoglobulin E (IgE) levels are associated with lower glioma risk [7, 8]. Given the consistent epidemiological data supporting the relationship between allergy and glioma, it has been hypothesized that inflammation-related cytokines, which are critical for allergy and IgE production, and their single nucleotide polymorphisms (SNPs) may alter the risk of glioma by affecting the body’s immune status.
Interleukin (IL)-4 and IL-13 are cytokines that share immunoregulatory functions and a common IL-4R chain on their receptors. They both play a central role in allergy by stimulating IgE synthesis in B lymphocytes and reducing production of pro-inflammatory cytokines by macrophages [9, 10]. Previous studies have also shown that they have strong antitumor activity in mice and inhibit proliferation of astrocytoma and low-grade glioma in human cell lines [11, 12]. Furthermore, polymorphisms of IL-4R and IL-13 have been reported to be associated with asthma risk [13, 14]. Considering the strong biological rational, several epidemiological studies have investigated the associations between IL-4/IL-13 pathway genetic polymorphisms and risk of glioma but have yielded contradictory results [15–17].
To date, two meta-analyses have been conducted to derive a precise estimation of this relationship [18, 19]. However, these findings have also been inconsistent and evaluated only a limited number of SNPs. Guo et al. reported that IL-4R rs1801275 was associated with a decreased risk of glioma [18], but this was not demonstrated in another meta-analysis [19]. Additionally, the published meta-analyses were based on the testing of multiple models (homozygote comparison, heterozygote comparison, recessive model, and dominant model), which are not independent so may increase the risk of false-positive results. Furthermore, some studies were not included in previous meta-analyses [20, 21], including those published later with larger sample sizes and rigorous designs [22, 23]. Considering that these factors could contribute to bias in the final results, we carried out an updated meta-analysis to provide a more reliable correlation between polymorphisms in IL-4/IL-13 pathway genes and glioma risk.
Materials and methods
Identification of eligible studies and SNPs
We performed a systematical search in PubMed, EMBASE, Web of Science, and the Cochrane Library databases for eligible articles with the following terms “glioma,” “polymorphism,” “IL-4” or “interleukin-4,” “IL-4R” or “interleukin-4R,” and “IL-13” or “interleukin-13” (last search updated on 10 April 2014). There were no language restrictions. We also reviewed reference lists and papers presented at international conferences for potentially relevant publications. If studies conducted in the same population existed, the most recent one or research with the most complete data was chosen. Inclusion criteria were as follows: (a) case–control or cohort design, (b) associations between relevant polymorphisms and glioma risk already assessed, and (c) sufficient data available for the computation of odds ratios (ORs) and 95 % confidence intervals (CIs). Studies not on human subjects, without available data, or with overlapping data were excluded. SNPs assessed in more than two studies were included for analysis.
Data extraction and assessment of methodological quality
Two reviewers (PQ Chen and K Chen) independently extracted the following data from selected articles: author, year of publication, country of origin, ethnicity, sample size, source of controls, matching criteria, and genotype distribution in cases and controls. We also contacted primary authors for genotype frequency data that were not reported in their articles.
After the concealment of authors, journals, supporting organizations, and funds to avoid subjective bias, two reviewers (K Chen and C Chen) assessed the methodological quality of the respective studies based on a set of criteria modified from previous research (Online Resource 1) [24, 25]. Quality scores ranged from 0 to 10, and studies scoring 6 or more points were considered eligible. Disagreements were resolved by discussion.
Statistical analysis
The strength of the association between IL-4/IL-13 pathway genetic polymorphisms and glioma risk was estimated by ORs with corresponding 95 % CIs. The common genetic models used for evaluating gene–disease associations are dominant, co-dominant, recessive, over-dominant, and log-additive. To avoid the problem of multiple model comparisons, we used Akaike’s information criterion (AIC) to determine the best genetic model for each SNP, and then pooled ORs were only calculated for this model. Between-study heterogeneity was determined by the chi-squared (χ 2)-based Q test [26]. P < 0.10 was considered to be an indicator of a substantial level of heterogeneity, and data were pooled using a random effects model (the DerSimonian and Laird method) [27]; otherwise, the fixed effects model (the Mantel-Haenszel method) was applied [28]. Publication bias was assessed visually using Begger’s funnel plots and formally tested using Egger’s test [29]. Hardy–Weinberg equilibrium (HWE) of controls was examined by the χ 2 test. Analyses were stratified by glioma histological types to address the differences in genetics and biology of glioblastoma (WHO grade IV) versus medium-grade (WHO grade III) and low-grade (WHO grade II) tumors. All statistical analyses were performed with STATA 12.0 (Stata Corporation, College Station, TX), and all P values were two-sided.
Results
Study characteristics
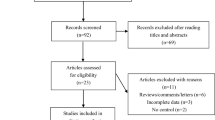
The selection procedure resulted in the inclusion of 12 published studies in the final meta-analysis (Fig. 1). Eleven studies used a case–control design, and only one used a cohort design [23]. The included studies were carried out in both Caucasian (n = 10) and Asian (n = 2) populations. Control subjects were mostly population-based rather than hospital-based and were all frequency-matched to the cases by age and gender. The quality score of included studies ranged from 6 to 9 with the majority scoring 7.5 or more points (Table 1). DNA used for genotyping was mostly extracted from blood samples, although some samples are derived from buccal cells [16]. The distribution of genotypes in the controls was consistent with HWE in all studies. Finally, nine polymorphisms in IL-4/IL-13 pathway genes were evaluated in this study: IL-4 (rs2243250, rs2070874, and rs2243248), IL-4R (rs1805011, rs1805012, rs1805015, and rs1801275), and IL-13 (rs20541 and rs1800925).
Meta-analysis results
Overall, no significant association was found between IL-4/IL-13 pathway genetic polymorphisms and glioma risk (Fig. 2). In the case of the IL-13 rs20541, eight studies were included in the present meta-analysis. The results suggested no association with glioma risk (recessive model, OR 0.88, 95 % CI 0.63–1.24), and a moderate heterogeneity across studies (I 2 = 46.3 %). Sensitivity analysis found that one study was the main source of heterogeneity [20]. After exclusion of this study, heterogeneity was reduced but there was no significant change to the results (recessive model, I 2 = 0; OR 0.79, 95 % CI 0.62–1.01).
The meta-analysis of three well-designed studies (including 2312 glioma cases and 2324 controls) found no significant association between IL-4 rs2243248 and glioma risk (log-additive model, OR 1.31, 95 % CI 0.90–1.91), but strong heterogeneity was evident (I 2 = 75 %). We considered that publication bias was a possible explanation for this high heterogeneity among studies. Analysis stratified by histological type showed that rs2234248 was associated with an increased risk of glioblastoma (log-additive model, OR 1.57, 95 % CI 1.11–2.24). This was the only IL-4/IL-13 genetic polymorphism to show a significant association with glioblastoma (Table 2 and Online Resource 2).
The meta-analysis of ten studies of IL-4R rs1801275 yielded no significant association with glioma risk (dominant model, OR 0.97, 95 % CI 0.88–1.08). When we restricted the study type to an analysis of Caucasians, high-quality score (quality score >8), or case sample >300, the results did not substantially alter.
Publication bias
Publication bias was detected by Egger’s test in IL-4 rs2243248 (P = 0.01) and IL-4R 1805012 (P = 0.03). We found no evidence of publication bias in the other SNPs using Begger’s or Egger’s test (Fig. 3).
Discussion
Since the IgE-glioma or allergy-glioma association was discovered, the role of polymorphisms in cytokine genes, which are critical for allergy and IgE synthesis, has been extensively studied on glioma risk but with mixed findings. The motivation of the present study is to define a precise estimation of this association. Our results revealed that IL-4/IL-13 pathway genetic polymorphisms were not associated with glioma risk in the overall population.
In recent years, genome-wide association studies (GWAS), which are not contingent on prior information concerning candidate genes or pathways, have made great progress in discovering the underlying architecture of genetic susceptibility to glioma. Previous GWAS have demonstrated that genetic variant mapping to 9p21.3 (CDKN2A/CDKN2B) [30], 7p11.2 (EGFR) [31], 5p15.33 (TERT) [32], 20q13.33 (RTEL1) [30], 8q24.21 (CCDC26) [33], and 11q23.3 (PHLDB1) [33] contribute to the heritable risk of glioma. However, they have thus far not identified a significant association between glioma and allergy-related genes, which further supports our present work.
In our study, rs2243248 was found to have a significant association only when analyses were restricted to the glioblastoma subgroup, which indicated that this variant might have a subtype-specific effect on the risk of glioma development. Although the specific functional relevance of rs2243248 is not completely understood, a previous study identified it as a risk factor for sporadic Alzheimer’s disease in the Chinese Han population [34]. Moreover, it has also been associated with a decreased risk of juvenile idiopathic arthritis [35]. Thus, an opposite association between rs2243248 and glioblastoma appears to be consistent with expectations from previous epidemiologic studies [36, 37]. Nevertheless, the significant publication bias (P = 0.01) observed for this polymorphism in the present study may have introduced serious bias to the pooled results. Thus, the association warrants replication in a larger population.
A previous meta-analysis including 1609 cases and 3016 controls identified IL-13 rs20541 as a protective factor for glioma [19], but this was not replicated in our study. Our meta-analysis had a larger sample size than the previous one, which might have altered the overall results. In addition, the previous meta-analysis tested multiple genetic models and did not correct for multiple comparisons, which may have increased the risk of false-positive results. By contrast, pooled ORs were only calculated for the recessive model in our study, which was determined from AIC to avoid the problems of multiple comparisons. Our findings are therefore likely to be more reliable than those of the previous study. Although rs20541 has been reported to play a role in IgE synthesis and asthma risk, our results suggest that it is unlikely to affect glioma development.
Four IL-4R polymorphisms (rs1805011, rs1805012, rs1805015, and rs1801275) were assessed in our study. No previous studies identified a significant association between rs1805011, rs1805012, or rs1805015 and glioma risk, and the results were still insignificant when we pooled data from these studies. An earlier meta-analysis revealed a significant association between rs1801275 and glioma risk [18], but this was not replicated in another meta-analysis [19] or our own. Even after restricting the study type to a Caucasian population, high-quality score (>8), or case sample >300, the OR for an association between rs1801275 and glioma risk was barely altered, strengthening the possibility that the lack of association was not caused by chance.
Consistent evidence supports a correlation between allergic conditions and glioma, so it is conceivable that genetic susceptibility to allergies is also related to the risk of glioma. However, this is not borne out by our current findings. One reason for this is that maybe the SNPs assessed in our study are not functional variants in the IL-4/IL-13 pathway, such that single base substitutions at these loci have no effect on the functions of target genes. Thus, other functional SNPs not in linkage disequilibrium could be investigated in future analyses.
Another potential reason is that although certain SNPs assessed are good candidates for affecting IgE levels and allergies such as rs20541 and rs1801275, they only account for some of the factors that affect IgE levels. For example, IL-13 haplotypes are estimated to be responsible for only 0.59 % of total IgE levels [38]. The point measurement of total IgE is also determined by other factors such as seasonality, diet, and circadian rhythms, so an individual SNP in the IL-4/IL-13 pathway might not substantially alter glioma risk through its small influence on the immune system.
To our knowledge, this is the most comprehensive study of polymorphisms in the IL-4/IL-13 pathway and glioma risk. Most of the nine polymorphisms assessed in our study have never been included in a meta-analysis before. Additional strengths of our study included the use of AIC to determine the best genetic model for each SNP and performing a methodology assessment to ensure the quality of included studies. However, there were some limitations that merit attention. First, because of the lack of original data, the effects of gene–gene and gene–environment interactions were not taken into consideration, yet these could modulate glioma risk. Second, although we undertook an extensive search to limit bias in the review process, publication bias was still detected for polymorphisms such as IL-4 rs2243248 and IL-4R rs1805012, so these results should be interpreted with caution. Third, only two studies conducted in Asian populations were included in our meta-analysis, so we were unable to perform a subgroup analysis by ethnicity. Finally, our meta-analysis was subject to the same multiple testing issues as a single-sample study. However, because correction for multiple testing is usually applied to eliminate false-positive results, our results are unlikely to differ after adjusting for multiple testing as we did not observe any significant associations.
In conclusion, our study revealed that IL-4/IL-13 pathway genetic polymorphisms are not associated with glioma risk in the overall population. However, studies with larger sample sizes, standardized unbiased homogenous patients, and well-matched controls are required to draw more comprehensive conclusions. Differences in genetic risk profiles between histological subtypes and tumor grades should also be considered in future studies.
References
Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi:10.1007/s00401-005-0991-y.
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503. doi:10.1038/ncpneuro0289. quiz 1 p following 16.
Schoemaker MJ, Swerdlow AJ, Hepworth SJ, McKinney PA, van Tongeren M, Muir KR. History of allergies and risk of glioma in adults. Int J Cancer. 2006;119(9):2165–72. doi:10.1002/ijc.22091.
Wigertz A, Lonn S, Schwartzbaum J, Hall P, Auvinen A, Christensen HC, et al. Allergic conditions and brain tumor risk. Am J Epidemiol. 2007;166(8):941–50. doi:10.1093/aje/kwm203.
Linos E, Raine T, Alonso A, Michaud D. Atopy and risk of brain tumors: a meta-analysis. J Natl Cancer Inst. 2007;99(20):1544–50. doi:10.1093/jnci/djm170.
Chen C, Xu T, Chen J, Zhou J, Yan Y, Lu Y, et al. Allergy and risk of glioma: a meta-analysis. Eur J Neurol. 2011;18(3):387–95. doi:10.1111/j.1468-1331.2010.03187.x.
Calboli FC, Cox DG, Buring JE, Gaziano JM, Ma J, Stampfer M, et al. Prediagnostic plasma IgE levels and risk of adult glioma in four prospective cohort studies. J Natl Cancer Inst. 2011;103(21):1588–95. doi:10.1093/jnci/djr361.
Schlehofer B, Siegmund B, Linseisen J, Schuz J, Rohrmann S, Becker S, et al. Primary brain tumours and specific serum immunoglobulin E: a case–control study nested in the European Prospective Investigation into Cancer and Nutrition cohort. Allergy. 2011;66(11):1434–41. doi:10.1111/j.1398-9995.2011.02670.x.
Akdis M. Healthy immune response to allergens: T regulatory cells and more. Curr Opin Immunol. 2006;18(6):738–44. doi:10.1016/j.coi.2006.06.003.
de Vries JE. The role of IL-13 and its receptor in allergy and inflammatory responses. J Allergy Clin Immunol. 1998;102(2):165–9. doi:10.1016/S0091-6749(98)70080-6.
Barna BP, Estes ML, Pettay J, Iwasaki K, Zhou P, Barnett GH. Human astrocyte growth regulation: interleukin-4 sensitivity and receptor expression. J Neuroimmunol. 1995;60(1–2):75–81. doi:10.1016/0165-5728(95)00055-7.
Liu H, Jacobs BS, Liu J, Prayson RA, Estes ML, Barnett GH, et al. Interleukin-13 sensitivity and receptor phenotypes of human glial cell lines: non-neoplastic glia and low-grade astrocytoma differ from malignant glioma. Cancer Immunol Immunother. 2000;49(6):319–24. doi:10.1007/s002620000110.
Loza MJ, Chang BL. Association between Q551R IL4R genetic variants and atopic asthma risk demonstrated by meta-analysis. J Allergy Clin Immunol. 2007;120(3):578–85. doi:10.1016/j.jaci.2007.05.019.
Cui L, Jia J, Ma CF, Li SY, Wang YP, Guo XM, et al. IL-13 polymorphisms contribute to the risk of asthma: a meta-analysis. Clin Biochem. 2012;45(4–5):285–8. doi:10.1016/j.clinbiochem.2011.12.012.
Schwartzbaum J, Ahlbom A, Malmer B, Lonn S, Brookes AJ, Doss H, et al. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005;65(14):6459–65. doi:10.1158/0008-5472.CAN-04-3728.
Wiemels JL, Wiencke JK, Kelsey KT, Moghadassi M, Rice T, Urayama KY, et al. Allergy-related polymorphisms influence glioma status and serum IgE levels. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1229–35. doi:10.1158/1055-9965.EPI-07-0041.
Schwartzbaum JA, Ahlbom A, Lonn S, Malmer B, Wigertz A, Auvinen A, et al. An international case–control study of interleukin-4Ralpha, interleukin-13, and cyclooxygenase-2 polymorphisms and glioblastoma risk. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2448–54. doi:10.1158/1055-9965.EPI-07-0480.
Guo J, Shi L, Li M, Xu J, Yan S, Zhang C, et al. Association of the interleukin-4Ralpha rs1801275 and rs1805015 polymorphisms with glioma risk. Tumour Biol. 2013. doi:10.1007/s13277-013-1080-9.
Sun G, Wang X, Shi L, Yue X, Fu L, Chen C, et al. Association between polymorphisms in interleukin-4Ralpha and interleukin-13 and glioma risk: a meta-analysis. Cancer Epidemiol. 2013;37(3):306–10. doi:10.1016/j.canep.2013.01.003.
Brenner AV, Butler MA, Wang SS, Ruder AM, Rothman N, Schulte PA, et al. Single-nucleotide polymorphisms in selected cytokine genes and risk of adult glioma. Carcinogenesis. 2007;28(12):2543–7. doi:10.1093/carcin/bgm210.
Amirian E, Liu Y, Scheurer ME, El-Zein R, Gilbert MR, Bondy ML. Genetic variants in inflammation pathway genes and asthma in glioma susceptibility. Neuro Oncol. 2010;12(5):444–52. doi:10.1093/neuonc/nop057.
Walsh KM, Anderson E, Hansen HM, Decker PA, Kosel ML, Kollmeyer T, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013;37(2):222–8. doi:10.1002/gepi.21707.
Backes DM, Siddiq A, Cox DG, Calboli FC, Gaziano JM, Ma J, et al. Single-nucleotide polymorphisms of allergy-related genes and risk of adult glioma. J Neurooncol. 2013;113(2):229–38. doi:10.1007/s11060-013-1122-6.
Al-Moundhri MS, Al-Nabhani M, Al-Bahrani B, Burney IA, Al-Madhani A, Ganguly SS, et al. Interleukin-1beta gene (IL-1B) and interleukin 1 receptor antagonist gene (IL-1RN) polymorphisms and gastric cancer risk in an Omani Arab population. Gastric Cancer. 2006;9(4):284–90. doi:10.1007/s10120-006-0392-5.
Gao LB, Pan XM, Li LJ, Liang WB, Zhu Y, Zhang LS, et al. RAD51 135G/C polymorphism and breast cancer risk: a meta-analysis from 21 studies. Breast Cancer Res Treat. 2011;125(3):827–35. doi:10.1007/s10549-010-0995-8.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi:10.1002/sim.1186.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi:10.1016/0197-2456(86)90046-2.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48. doi:10.1093/jnci/22.4.719.
Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–80. doi:10.1001/jama.295.6.676.
Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–8. doi:10.1038/ng.408.
Sanson M, Hosking FJ, Shete S, Zelenika D, Dobbins SE, Ma Y, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20(14):2897–904. doi:10.1093/hmg/ddr192.
Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–5. doi:10.1038/ng.3004.
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi:10.1038/ng.407.
Li W, Qian X, Teng H, Ding Y, Zhang L. Association of interleukin-4 genetic polymorphisms with sporadic Alzheimer’s disease in Chinese Han population. Neurosci Lett. 2014;563:17–21. doi:10.1016/j.neulet.2014.01.019.
Cinek O, Vavrincova P, Striz I, Drevinek P, Sedlakova P, Vavrinec J, et al. Association of single nucleotide polymorphisms within cytokine genes with juvenile idiopathic arthritis in the Czech population. J Rheumatol. 2004;31(6):1206–10.
Schwartzbaum J, Jonsson F, Ahlbom A, Preston-Martin S, Lonn S, Soderberg KC, et al. Cohort studies of association between self-reported allergic conditions, immune-related diagnoses and glioma and meningioma risk. Int J Cancer. 2003;106(3):423–8. doi:10.1002/ijc.11230.
Schlehofer B, Blettner M, Preston-Martin S, Niehoff D, Wahrendorf J, Arslan A, et al. Role of medical history in brain tumour development. Results from the international adult brain tumour study. Int J Cancer. 1999;82(2):155–60. doi:10.1002/(SICI)1097-0215(19990719)82:2<155::AID-IJC1>3.0.CO;2-P.
Maier LM, Howson JM, Walker N, Spickett GP, Jones RW, Ring SM, et al. Association of IL13 with total IgE: evidence against an inverse association of atopy and diabetes. J Allergy Clin Immunol. 2006;117(6):1306–13. doi:10.1016/j.jaci.2005.12.1354.
Acknowledgments
We thank Dr. Melissa L. Bondy from the Texas MD Anderson Cancer Center and Dr. Kyle M. Walsh from the Helen Diller Family Comprehensive Cancer Center for generously providing their data.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Peiqin Chen and Chao Chen contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Scale for methodological quality assessment (DOC 39 kb)
Online Resource 2
Meta-analysis for the association between IL-4/IL-13 pathway genetic polymorphisms and glioblastoma risk (GIF 122 kb)
Rights and permissions
About this article
Cite this article
Chen, P., Chen, C., Chen, K. et al. Polymorphisms in IL-4/IL-13 pathway genes and glioma risk: an updated meta-analysis. Tumor Biol. 36, 121–127 (2015). https://doi.org/10.1007/s13277-014-2895-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2895-8