Abstract
It has been suggested that hOGG1 Ser326Cys polymorphism may be a risk factor for colorectal cancer. Published data on its association with colorectal cancer generated contradictory results; thus, we performed an updated meta-analysis of eligible published studies to estimate the effect of hOGG1 Ser326Cys polymorphism on colorectal cancer susceptibility. We reviewed many abstracts and finally included 18 eligible case–control studies comprising 5235 cases and 8438 controls. We pooled data with a fixed or random-effect model. Subgroup analysis by ethnicity was also performed. The overall data indicated a significant association of hOGG1 Ser326Cys polymorphism on colorectal cancer risk (allele model OR = 1.14, 95 %CI 1.02–1.27; homozygote model OR = 1.32, 95 %CI 0.92–1.92; recessive model OR = 1.12, 95 %CI 1.00–1.26; dominant model OR = 1.15, 95 %CI 1.00–1.32). Furthermore, in the subgroup analysis by ethnicity, increased cancer risk was observed among Caucasians under the allele, heterogeneity, recessive, and dominant models (allele model OR = 1.23, 95 %CI = 1.05–1.44; homozygote model OR = 1.49, 95%CI 1.05–2.12; recessive model OR = 1.40, 95 %CI 1.16–1.69; dominant model OR = 1.21, 95 %CI = 1.12–1.45). In summary, the present meta-analysis suggested that hOGG1 Ser326Cys polymorphism might modify the susceptibility to colorectal cancer among the total population, especially among Caucasians.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is one of the most common malicious tumors [1]. It is one of the major causes of mortality and morbidity, whose 5-year survival rate is low [2]. It is known to all that early diagnosis is very important which improves the chances of patient’s survival significantly [3]. The causes of colorectal cancer have not been established [4]. The pathogenesis of colorectal cancer has involved oxidative DNA damage [5]. We know that the base excision repair pathway is the major DNA repair pathway for oxidative DNA damage and genetic variation which is associated with impaired base excision repair and may increase the risk for colorectal cancer [6]. The possible relationship between oxygen-free radicals and cancer development has been reported chiefly in organs that are under a high burden of oxygen-free radicals. 7,8-Dihydro-8-oxoguanine (8oxoG) is one of the most important lesions, which has been produced in DNA by oxygen radical-forming causes. The mispairing of 8oxoG with deoxyadenosine causes mutagenic transversion of G:C to T:A in vitro and in vivo [7, 8]. The hOGG1 gene encodes a DNA glycosylase/AP-lyase that can catalyze the removal of 8oxoG adducts as part of the base excision repair pathway. The hOGG1 gene is divided as multiple alternatively spliced isoforms with only one а-form which have a nuclear localization signal [9, 10]. Many studies have found the presence of several polymorphisms at the hOGG1 locus. A C/G polymorphism at position 1245 in the 1a-specific exon 7 of the hOGG1 gene resulted to an amino acid change from serine to cysteine in codon 326 [11]. Studies on the association of hOGG1 Ser326Cys polymorphism with colorectal cancer generated controversial results, so we performed an updated meta-analysis of eligible studies to estimate the effect of hOGG1 Ser326Cys polymorphism on colorectal cancer risk [12–17].
Materials and methods
Literature search strategy
We systematically searched PubMed, Embase, Web of Science, and Chinese National Knowledge Infrastructure (CNKI) with no language restrictions, for studies on the association between hOGG1 Ser326Cys polymorphism and colorectal cancer risk. We covered all studies published up to June 2013. We used the following terms: hOGG1, OGG1, Ser326Cys, polymorphism, polymorphisms, colorectal cancer (CRC), colon cancer, rectal cancer, susceptibility, and risk act. All of the searched studies were retrieved, and the bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were searched by hand in order to find additional eligible studies.
Inclusion criteria
The following criteria were used for the literature selection: first, studies should concern the association of hOGG1 Ser326Cys polymorphism with colorectal cancer risk; second, studies should be observational studies (case–control or cohort); third, papers must report the size of the sample, odds ratios (ORs), and their 95 % confidence intervals (95 %CIs), genetic distribution, and ethnicity or must report sufficient data to estimate these correlated data. After rigorous searching, we reviewed all papers according with the criteria defined above for further analysis.
Data extraction
The data were extracted carefully from all eligible publications independently by two of the authors in accordance to the criteria mentioned above. This study used the following information: study design, publication year, matching factors, source of controls, frequencies of hOGG1 Ser326Cys polymorphism genotypes, ethnicity, country, patient characteristics, ORs, and 95 %CIs. The extracted information was entered into a database.
Statistical analysis
The pooled OR and 95 %CI were used to assess the association between hOGG1 Ser326Cys polymorphism and colorectal cancer risk for each case–control study. The pooled ORs were performed for a homozygote model (Cys/Cys vs. Ser/Ser), a dominant model (Cys/Cys + Ser/Cys vs. Ser/Ser), a recessive model (Cys/Cys vs. Ser/Cys + Ser/Ser), and an allele model (Cys vs. Ser). The I 2 value was used as an index for the heterogeneity test, with values less than 25 % indicating low, 25 to 50 % indicating moderate, and greater than 50 % indicating high heterogeneity. The I 2 statistic was used to estimate heterogeneity in the pooled studies [18]. When I 2 > 50 %, potential sources of heterogeneity were explored through a meta-regression analysis [19]. The data were pooled according to the fixed-effect model (Mantel–Haenszel) if I 2 < 50 %; otherwise, the random-effect model (DerSimonian and Laird) was used [20, 21]. The significance of the pooled ORs was determined by Z test. The Hardy–Weinberg equilibrium (HWE) was assessed by Fisher’s exact test. Publication bias was assessed by visual inspection of funnel plots [22], in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot indicates a possible publication bias. The symmetry of the funnel plot was further evaluated by Begg’s linear regression test [23]. Statistical analysis was performed using the program Stata 11.2 software (Stata Corporation, College Station, TX, USA). P values <0.05 were considered statistically significant. There was no funding source for this study. The corresponding author had full access to all the data in the study and had the final responsibility for the decision to submit for publication.
Results
Study characteristics
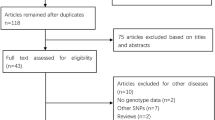
We retrieved and screened the publications relevant to the keywords originally. Finally, as shown in Table 1, a total of 18 case–control studies were included for further analysis (Table 1). All the eligible studies were written in English. The characteristics of the studies included in the present meta-analysis were shown in the figure principally. According to the figure, we could know the first author, the number, and characteristics of cases and controls for each study as well as other necessary information. As can be seen in Table 1, there were 12 groups of Caucasians [12–16, 24–30] and 6 groups of Asians [17, 31–35]. The distributions of the control groups of all the studies were in line with HWE.
Meta-analysis
The overall data contained a total of 5235 cases and 8438 controls. We analyzed the heterogeneities for the dominant model, recessive model, etc., respectively. As shown in Table 2 and Figs. 1 and 2, the overall data indicate a significant association of hOGG1 Ser326Cys polymorphism with colorectal cancer risk (allele model OR = 1.14, 95%CI 1.02–1.27; homozygote model OR = 1.28, 95 %CI 1.02–1.62).
Furthermore, as what Table 2 and Fig. 3 are showing, in the subgroup analysis by ethnicity, increased cancer risk was observed among the Caucasians under the allele, homozygote, recessive, and dominant models (allele model OR = 1.23, 95 %CI 1.05–1.44; homozygote model OR = 1.49, 95 %CI = 1.05–2.12; recessive model OR = 1.40, 95 %CI = 1.16–1.69; dominant model OR = 1.21, 95 %CI = 1.12–1.45) (Fig. 3). That means that an increased colorectal cancer risk was shown among Caucasians with 326Cys mutation of hOGG1 polymorphism.
Funnel plots were created to evaluate possible publication bias. Then, Egger’s linear regression tests were used to assess the symmetries of the plots. The data suggest that the funnel plots were symmetrical for the overall data under the allele model (t = 1.82; P = 0.087 > 0.05), suggesting that there was no publication bias.
Discussion
Colorectal cancer, the third most common cause of cancer death in the world, has 150,000 new cases and 50,000 deaths in the USA annually. In North America and Europe, the incidence rate is approximately 30–50/100,000. For colorectal cancer, one of the most common cancers, early diagnosis is critical. The susceptibility of colorectal cancer contains many factors. Studies have pointed that individual risks for colorectal cancer depend on genetic factors. Data has indicated an association of the descent productivity of DNA repair and the increased susceptibility of colorectal cancer [36]. It is widely recognized that mismatch repair pathway is an etiological factor of individual risk to colorectal cancer [37]. Many study data indicated an association of hOGG1 polymorphisms which were involved in oxidative DNA lesions repair with the risk occurrence of colorectal cancer patients [38]. Researchers found that hOGG1 polymorphisms may affect DNA repair capacity which suggested its role in colorectal cancer pathogenesis [33].
Relations of hOGG1 polymorphisms with cancer susceptibility have been certified by several analyses. As shown above, at present, studies suggest that hOGG1 polymorphisms might modify the susceptibility to colorectal cancer among the total population (allele model and homozygote model). Meanwhile, in the subgroup analysis of ethnicity, increased cancer risk was observed among Caucasians under the heterogeneity, recessive, and dominant models. The difference may be induced by ethnic differences in genetic backgrounds and the environment where they lived. Different socioeconomic status could also affect colorectal cancer risk. The most important etiological factors of sporadic colorectal tumors are inflammation, fat metabolism, tobacco smoking, and consumption of meat and alcohol [12, 39, 40]. This might explain the racial disparities. The difference may be caused by the limited number of included studies. Insufficient statistical power for assessment might be caused by small sample sizes, too. Thus, we should interpret the results carefully.
We used the fixed-effect model in the recessive model for the overall data. Because we observed between-study heterogeneity in the allele, dominant, and homozygote models for the overall data, we used the random-effect model in these models. In every model, we divided the data into subgroups with regard to ethnicity. Then, heterogeneity was removed in Asians, but not in the overall and in the Caucasian populations, which suggested that the heterogeneities may be multifactorial. Other factors, such as living environment, age, gender, and lifestyle, might induce the heterogeneities. At the same time, publication bias, as an important factor, should be considered in the meta-analysis. Funnel plot evaluation and Begg’s linear regression tests were used to assess the possible publication bias. We found no evident bias, so the potential publication bias might cause little influence on the results.
There were several limitations in our meta-analysis. First, in this meta-analysis, the included 18 studies regarded only Caucasians and Asians, but not other races. Data about other ethnicities, for example, African, should be noticed in the future. Second, because we could not obtain sufficient data from the present publications, in this study, subgroup analyses regarding living environment, age, gender, lifestyle, and other factors have not been expressed. So, selection bias might exist. Meanwhile, in further studies, gene–gene and gene–environment interactions should also be noted. Further well-designed studies with more rigorous matching criteria and large sample sizes are required to assess the relationship between hOGG1 polymorphisms and colorectal cancer risk. Despite the limitations, the overall data indicated a significant association of hOGG1 polymorphisms on colorectal cancer risk (allele model OR = 1.14, 95%CI 1.02–1.27; homozygotemodel OR = 1.32, 95 %CI 0.92–1.92; recessive model OR = 1.12, 95 %CI 1.00–1.26; dominant model OR = 1.15, 95 %CI 1.00–1.32). Furthermore, in the subgroup analysis by ethnicity, increased cancer risk was observed among Caucasians under the allele, heterogeneity, recessive, and dominant models (allele model OR = 1.23, 95 %CI = 1.05–1.44; homozygote model OR = 1.49, 95 %CI 1.05–2.12; recessive model OR = 1.40, 95 %CI 1.16–1.69; dominant model OR = 1.21, 95 %CI = 1.12–1.45). In summary, the present meta-analysis suggested that hOGG1 Ser326Cys polymorphism might modify the susceptibility to colorectal cancer among the total population, especially among Caucasians. In summary, the data of the present studies suggest that hOGG1 polymorphisms might modify the susceptibility to colorectal cancer among the total population, especially among Caucasians.
References
Fujita T. Colorectal cancer. Lancet. 2010;376:331.
O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–5.
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47.
Johnson IT, Lund EK. Review article: nutrition, obesity and colorectal cancer. Aliment Pharmacol Ther. 2007;26:161–81.
Perse M. Oxidative stress in the pathogenesis of colorectal cancer: cause or consequence? Biomed Res Int. 2013;2013:725710.
Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, et al. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radic Biol Med. 2004;37:1449–54.
Michaels ML, Miller JH. The go system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol. 1992;174:6321–5.
Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodg. Nature. 1991;349:431–4.
Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, et al. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell. 1999;10:1637–52.
Shinmura K, Kohno T, Takeuchi-Sasaki M, Maeda M, Segawa T, Kamo T, et al. Expression of the OGG1-type 1a (nuclear form) protein in cancerous and non-cancerous human cells. Int J Oncol. 2000;16:701–7.
Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H, et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–25.
Brevik A, Joshi AD, Corral R, Onland-Moret NC, Siegmund KD, Le Marchand L, et al. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomarkers Prev. 2010;19:3167–73.
Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–302.
Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomark. 2010;14:559–64.
Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39:527–34.
Przybylowska K, Kabzinski J, Sygut A, Dziki L, Dziki A, Majsterek I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutat Res. 2013;745–746:6–15.
Sameer AS, Nissar S, Abdullah S, Chowdri NA, Siddiqi MA. DNA repair gene 8-oxoguanine DNA glycosylase Ser326Cys polymorphism and colorectal cancer risk in a Kashmiri population. DNA Cell Biol. 2012;31:541–6.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21:1559–73.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48.
Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res. 2004;129:39–44.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101.
Curtin K, Samowitz WS, Wolff RK, Ulrich CM, Caan BJ, Potter JD, et al. Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3384–8.
Hansen R, Saebo M, Skjelbred CF, Nexo BA, Hagen PC, Bock G, et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229:85–91.
Hansen RD, Krath BN, Frederiksen K, Tjonneland A, Overvad K, Roswall N, et al. GPX1 Pro(198)Leu polymorphism, erythrocyte GPX activity, interaction with alcohol consumption and smoking, and risk of colorectal cancer. Mutat Res. 2009;664:13–9.
Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–8.
Obtulowicz T, Swoboda M, Speina E, Gackowski D, Rozalski R, Siomek A, et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis. 2010;25:463–71.
Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–53.
Sliwinski T, Krupa R, Wisniewska-Jarosinska M, Pawlowska E, Lech J, Chojnacki J, et al. Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J Exp Med. 2009;218:185–91.
Jin MJ, Liu B, Zhang SS, Zhang YJ, Xu M, Ma XY, et al. [Application of multifactor dimensionality reduction on the interactions between gene-gene, gene-environment and the risk sporadic colorectal cancer in Chinese population]. Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29:535–9.
Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49.
Kim JI, Park YJ, Kim KH, Song BJ, Lee MS, Kim CN, et al. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World J Gastroenterol. 2003;9:956–60.
Park HW, Kim IJ, Kang HC, Jang SG, Ahn SA, Lee JS, et al. The hOGG1 Ser326Cys polymorphism is not associated with colorectal cancer risk. J Epidemiol. 2007;17:156–60.
Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM, Koh WP, et al. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese health study. Cancer Epidemiol Biomarkers Prev. 2007;16:2363–72.
Jiricny J, Marra G. DNA repair defects in colon cancer. Curr Opin Genet Dev. 2003;13:61–9.
Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–9.
Wei B, Zhou Y, Xu Z, Xi B, Cheng H, Ruan J, et al. The effect of hOGG1 Ser326Cys polymorphism on cancer risk: evidence from a meta-analysis. PLoS One. 2011;6:e27545.
Levi F, Pasche C, La Vecchia C, Lucchini F, Franceschi S. Food groups and colorectal cancer risk. Br J Cancer. 1999;79:1283–7.
Wang J, Joshi AD, Corral R, Siegmund KD, Marchand LL, Martinez ME, et al. Carcinogenmetabolism genes, redmeat and poultry intake, and colorectal cancer risk. Int J Cancer. 2012;130:1898–907.
Acknowledgments
This work was funded by the Natural Science Foundation of China (grant no. 11274216).
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, M., MO, R. Association of hOGG1 Ser326Cys polymorphism with colorectal cancer risk: an updated meta-analysis including 5235 cases and 8438 controls. Tumor Biol. 35, 12627–12633 (2014). https://doi.org/10.1007/s13277-014-2586-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2586-5