Abstract
An increasing body of evidence has shown that the amino acid changes at position 1298 might eliminate methylenetetrahydrofolate reductase (MTHFR) enzyme activity, leading to insufficient folic acid and subsequent human chromosome breakage. Epidemiological studies have linked MTHFR single-nucleotide polymorphism (SNP) rs1801131 to myeloid leukemia risk, with considerable discrepancy in their results. We therefore were prompted to clarify this issue by use of a meta-analysis. The search terms were used to cover the possible reports in the MEDLINE, Web of Knowledge, and China National Knowledge Infrastructure (CNKI) databases. Odds ratios were estimated to assess the association of SNP rs1801131 with myeloid leukemia risk. Statistical heterogeneity was detected using the Q-statistic and I 2 metric. Subgroup analysis was performed by ethnicity, histological subtype, and Hardy-Weinberg equilibrium (HWE). This meta-analysis of eight publications with a total of 1,114 cases and 3,227 controls revealed no global association. Nor did the subgroup analysis according to histological subtype and HWE show any significant associations. However, Asian individuals who harbored the CC genotype were found to have 1.66-fold higher risk of myeloid leukemia (odds ratio, 1.66; 95 % confidence interval, 1.10 to 2.49; P h = 0.342; I 2 = 0.114). Our meta-analysis has presented evidence supporting a possible association between the CC genotype of MTHFR SNP rs1801131 and myeloid leukemia in Asian populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leukemia commonly diagnosed in childhood is a complex and heterogeneous disease arising as a consequence of the irreversible genetic lesions in initially normal hematopoietic cells [1]. Acute myeloid leukemia (AML) characterized by gross chromosomal abnormalities, particularly translocations of inversions [2, 3], represents one major type of myeloid leukemia. Chronic myeloid leukemia (CML), another primary form of myeloid leukemia, is a mature hematologic malignancy with two notable properties: reciprocal translocation and subsequent BCR-ABL fusion transcript [4]. Most of the etiology of AML and CML remains unknown, though exposure to benzene, chemotherapy, and ionizing radiation that may cause chromosomal breakage are being considered as etiological factors [5]. There has been extensive research on the role of inherited susceptibility due to epidemiological data on an association between methylenetetrahydrofolate reductase (MTHFR) gene single-nucleotide polymorphisms (SNPs) and increased risk of leukemia in both children and adults [6].
MTHFR is a mediator of homocysteine levels and folate, an essential component in the process of nucleotide synthesis. Lack of folate results in misincorporation of uracil into human DNA during replication, and this may cause DNA and chromosome breakage consequently [7, 8], while sufficient folate maintains cellular differentiation in a normal condition, for example, the replication of hematopoietic cells. Effective metabolism of folic acid requires the enzyme MTHFR that converts 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, the primary form of serum folate [7]. Mutation in the MTHFR gene could diminish the enzyme activity and decrease plasma folate levels and thereby initiates many common disorders [9].
The gene coding the MTHFR enzyme is located at the short (p) arm of chromosome 1 at position 36.3 [10]. Genetic polymorphisms in the MTHFR gene have been reported to reduce enzyme activities, thus influencing folate metabolism [11]. A common SNP (rs1801131, Glu429Ala) with an amino acid change from glutamate to alanine (A → C) has been identified in exon 7 of the MTHFR gene. This polymorphism is related to lower enzyme activity, and homozygotes have around 60 % of activity in lymphocytes compared to controls [12]. Studies of genetic association in the past decade have extensively examined the effects of SNP rs1801131 on myeloid leukemia risk [13–16]. However, replications of the reported association have been mixed, indicating that it is of importance to conduct a study with strong statistical power to identify the high-risk individuals. In the present study, we performed a meta-analysis to quantitatively assess the association between SNP rs1801131 and myeloid leukemia risk.
Materials and methods
Search strategy and selection criteria
Using the keywords polymorphism, polymorphisms, methylenetetrahydrofolate reductase, MTHFR, and myeloid leukemia, we carried out electronic searches in the MEDLINE, Web of Knowledge, and China National Knowledge Infrastructure (CNKI) databases to cover the relevant reports published prior to Jan. 31, 2014. The computer-based searches were supplemented by manual searches of the references cited in original articles. We used no language restrictions. Two investigators screened all articles to evaluate their eligibility. The case–control studies were considered eligible if the authors estimated risk of myeloid leukemia associated with the presence of SNP rs1801131 and provided detailed genotype data that could assist to calculate odds ratios.
Data extraction
Data were independently extracted by the same two investigators and then checked by the third investigator. Discrepancies were resolved through discussion. Information including the first author’s last name, year of publication, study country, ethnicity and age of the populations studied, subtypes of myeloid leukemia (AML or CML), minor allele frequency (MAF) of cases and controls, and genotype numbers was extracted from each study.
Statistical analysis
The association between SNP rs1801131 and myeloid leukemia risk (odds ratios) was examined assuming diverse genetic models (homozygous, heterozygous, dominant, recessive, and allele model). Heterogeneity was determined using the Q-statistic. p < .05 indicated heterogeneous results. In addition, I 2 metric that takes values between 0 and 100 % was used to quantify the heterogeneity across studies, with higher values indicating larger heterogeneity (no heterogeneity 0–25 %, moderate heterogeneity 25–50 %, large heterogeneity 50–75 %, and extreme heterogeneity 75–100 %) [17]. When substantial heterogeneity was suggested, the random effects (RE) model (DerSimonian and Laird) was preferably used to estimate the pooled odds ratios and vice versa [18, 19]. Funnel plots described by Begg and the liner regression test described by Egger were performed to test the publication bias [20, 21]. Departure from Hardy-Weinberg equilibrium (HWE) was checked using Pearson’s Χ 2 test. Sensitivity analysis was performed by deleting the studies with HWE deviation to examine their effects on the overall estimates. Stratified analysis was also performed according to histological subtype, ethnicity, and HWE. All statistical data were analyzed using Stata software (version 12.0, StataCorp LP, College Station, TX, USA), with p values < .05 being considered statistically significant.
Results
Eligible studies
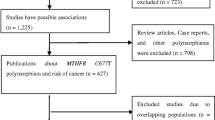
As shown in Fig. 1, the electronic searches yielded a total of 81 relevant reports. Seventy-five articles that failed to meet the pre-described inclusion criteria were excluded after screening the titles, abstracts, and full-texts. Through manual search, we identified two additional articles. This meta-analysis therefore included eight articles involving 10 subpopulations [13–16, 22–25], as two articles provided more than one independent population [15, 16]. Of the studies published between 1999 and 2014, six studies focused on AML and the remainders on CML. Both adults and children were combined in this analysis. As for ethnicity, Asian was investigated in most of the studies, accounting for 60 %. MAF distribution between cases and controls was similar in each study. All studies, except for Lordelo et al. and Zheng et al., were all consistent with HWE (Table 1).
Meta-analysis results
Meta-analysis of the association between SNP rs1801131 and myeloid leukemia risk included 10 studies with a total of 1,114 cases and 3,227 controls. As shown in Table 2, the genotypes of SNP rs1801131 showed no global association with risk of myeloid leukemia. Likewise, we did not find any statistical evidence indicating a significant association when stratifying the pooled data according to histological subtype and HWE. However, it is interesting that in the subgroup of Asian populations, the recessive model provided an odds ratio of 1.66 (95 % confidence interval, 1.10 to 2.49; P h = 0.342; I 2 = 0.114) (Fig. 2, Table 2), suggesting that the Asian individuals carrying the CC genotype were at higher risk of myeloid leukemia relative to the individuals carrying both AC and AA genotypes.
Sensitivity analysis by deleting the studies inconsistent with HWE showed no substantial influence on the overall results (figure not shown). The funnel plots and Egger’s test did not detect any significant publication bias in this meta-analysis. Figure 3 shows the funnel plots of the recessive model (p = 0.452).
Discussion
As a result of the availability of SNPs and advances in genotyping methods, there has been a sharp rise in the number of published genetic association studies (GAS) in recent years. Most of these GAS, however, are characterized by a relative sample size, nonhomogeneous populations, and distinct ethnic groups and usually underestimate the true associations [26]. Meta-analysis as a quantitative method has strong detection power in determining the genetic association of functionally important SNPs and common diseases. Herein, we chose to perform a meta-analysis in an effort to derive a precise estimation of the association between SNP rs1801131 and myeloid leukemia risk. In the analysis of 4,342 unique subjects from eight publications, we found that the presence of SNP rs1801131 was not correlated with the development of myeloid leukemia. A similar story was revealed in stratified analysis according to histological subtype and HWE. However, it is interesting that further subgroup analysis by ethnicity showed statistical evidence of a significant association between the carriage of CC genotype and increased risk of myeloid leukemia in Asians. Due to the limited number of Asian subjects analyzed (585 patients and 2,157 controls), these findings merit further investigation.
Since the genotypes of SNP rs1801131 were shown to not have any associations with adult AML and folate inadequacy may not play a critical role in the progression of the common leukemia [13], SNP rs1801131 has been of special interest for the studies of molecular epidemiology. A subsequent replication of 496 individuals representing Caucasian ethnicity reported that the AC genotype was linked to an approximately 3-fold increased risk of childhood AML [14]. Such inconsistent evidence can similarly be seen in the Asian studies. Hur et al. genotyped 396 individuals and found a significant decrease in risk of CML [15], while a completely different result suggesting an increased risk of CML was implicated in a larger replication study [16]. These small-sampled GAS of individuals representing distinct ethnicities may be underpowered to detect the real associations, befuddling later researchers consequently. With a view of this, we performed a meta-analysis and determined that SNP rs1801131 was associated with significantly increased risk of myeloid leukemia among Asians, suggesting that this SNP may be a potential biomarker for myeloid leukemia.
Nevertheless, accumulating evidence has well documented high incidence of myeloid leukemia in both children and adults, with either Caucasian or Asian ancestry [27–29]. Inconsistent with the earlier reports, our meta-analysis showed a significant association only in Asian populations. There are several plausible explanations. Although amino acid changes in the SNP rs1801131 at position 1298 possibly cause elimination of the MTHFR enzyme activity and subsequent insufficient folic acid [12, 30], this insufficiency has been suggested to confer no substantial effects on the development of myeloid leukemia [13]. Moreover, a recent meta-analysis clarified whether SNP rs1801131 played a major role in acute lymphoblastic leukemia (ALL), with statistical evidence of no effect modification of ALL risk either in adults or in children [31]. Yan et al. carried out a quantitative assessment almost at the same time and presented evidence supporting that SNP rs1801131 did not have an effect on childhood ALL risk [32]. AML, CML, and ALL are major forms of leukemia [33], and the SNP rs1801131 is very likely to confer similar effects on these common diseases. According to the data aforementioned, we can presume that SNP rs1801131 may not represent a risk factor for AML and CML. Considering the sample inadequacy, the findings of our analysis should be treated as preliminary and are worthy of further validation in future larger studies.
In addition to the small sample size, two other limitations should be addressed. First, publication bias arises when small studies are included and the forthcoming publications are not identified. In this analysis, we selected studies without a limitation on a minimum of sample size and failed to identify the unpublished studies, making publication bias possible. But, the overall results were not affected according to the analytical tools. Second, genetic variation has been thought to be a cause of many leukemia subtypes; AML and CML nevertheless are multifactorial diseases, and it is likely that gene-to-gene and gene-to-environment interactions are important components in the process of leukemia. Lack of original data did not permit further analysis on common confounders, such as sex, dietary patterns, and exposure to environmental carcinogens.
To the best of our knowledge, this is the first meta-analysis on the association of SNP rs1801131 with myeloid leukemia risk. The results revealed that the Asians harboring the CC genotype had higher risk to develop myeloid leukemia. To validate this finding, future studies with a very large number of subjects are required.
References
Bloomfield CD, Caligiury M. Molecular biology of leukemias. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 2389–404.
Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519.
Pui CH. Childhood leukaemias. 2nd ed. Cambridge: Cambridge University Press; 2006.
Gabert J et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17(12):2318–57.
Smith MT, Zhang L. Biomarkers of leukemia risk: benzene as a model. Environ Health Perspect. 1998;106 Suppl 4:937–46.
Robien K, Ulrich CM. 5,10-Methylenetetrahydrofolate reductase polymorphisms and leukemia risk: a HuGE minireview. Am J Epidemiol. 2003;157(7):571–82.
Blount BC et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94(7):3290–5.
Duthie SJ et al. Impact of folate deficiency on DNA stability. J Nutr. 2002;132(8 Suppl):2444S–9S.
Franco RF et al. A second mutation in the methylenetetrahydrofolate reductase gene and the risk of venous thrombotic disease. Br J Haematol. 1999;105(2):556–9.
Goyette P et al. Human methylenetetrahydrofolate reductase: isolation of cDNA mapping and mutation identification. Nat Genet. 1994;7(4):551.
Frosst P et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111–3.
Weisberg I et al. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64(3):169–72.
Skibola CF et al. Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc Natl Acad Sci U S A. 1999;96(22):12810–5.
da Costa Ramos FJ et al. Association between the MTHFR A1298C polymorphism and increased risk of acute myeloid leukemia in Brazilian children. Leuk Lymphoma. 2006;47(10):2070–5.
Hur M et al. Methylenetetrahydrofolate reductase A1298C genotypes are associated with the risks of acute lymphoblastic leukaemia and chronic myelogenous leukaemia in the Korean population. Clin Lab Haematol. 2006;28(3):154–9.
Moon HW et al. MTHFR 677CC/1298CC genotypes are highly associated with chronic myelogenous leukemia: a case–control study in Korea. Leuk Res. 2007;31(9):1213–7.
Higgins JP et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Egger M et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Lightfoot TJ et al. Genetic variation in the folate metabolic pathway and risk of childhood leukemia. Blood. 2010;115(19):3923–9.
Lordelo GS et al. Association between methylenetetrahydrofolate reductase and glutathione S-transferase M1 gene polymorphisms and chronic myeloid leukemia in a Brazilian population. Genet Mol Res. 2012;11(2):1013–26.
Zheng MM et al. Association of single nucleotide polymorphism of methylenetetrahydrofolate reductase gene with susceptibility to acute leukemia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013;30(4):451–5.
Khorshied MM et al. Methylene tetrahydrofolate reductase (MTHFR) gene polymorphisms in chronic myeloid leukemia: an Egyptian study. Med Oncol. 2014;31(1):794.
Zintzaras E, Lau J. Trends in meta-analysis of genetic association studies. J Hum Genet. 2008;53(1):1–9.
Cheson BD. The chronic lymphocytic leukemias. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 2447–65.
Kantarjian HM, Faderl S, Talpaz M. Chronic myelogenous leukemia. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 2433–47.
Weinstein HJ, Tarbell N. Leukemias and lymphomas of childhood. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 2235–56.
van der Put NM et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62(5):1044–51.
Wang H et al. Methylenetetrahydrofolate reductase polymorphism C677T is a protective factor for pediatric acute lymphoblastic leukemia in the Chinese population: a meta-analysis. Genet Test Mol Biomark. 2012;16(12):1401–7.
Yan J et al. A meta-analysis of MTHFR C677T and A1298C polymorphisms and risk of acute lymphoblastic leukemia in children. Pediatr Blood Cancer. 2012;58(4):513–8.
Cole P, Rodu B. Descriptive epidemiology: cancer statistics. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: principles and practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 228–41.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, S., Liu, Y. & Chen, J. MTHFR gene polymorphism and risk of myeloid leukemia: a meta-analysis. Tumor Biol. 35, 8913–8919 (2014). https://doi.org/10.1007/s13277-014-2082-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2082-y