Abstract
Activation of the wingless-type (Wnt) signaling pathway is common in various human cancers including colorectal cancer (CRC). Wnt inhibitory factor-1 (WIF-1) is a secreted antagonist that can bind Wnt ligands and therefore inhibits the Wnt signaling pathway. In this study, we aimed to analyze the expression of two members of Wnt signaling (WIF-1 and Wnt5a) in Tunisian patients with sporadic CRC. WIF-1 was frequently methylated in tumor tissues (87.95 %) compared to normal mucosa (39.54 %) and correlated with distant metastasis and vascular invasion (P = 0.001 and 0.037, respectively). The unmethylated profile of the WIF-1 promoter conferred a benefit to patients in terms of overall survival (P log rank = 0.024). In addition, in the group of patients with methylated WIF-1 promoter, the overall survival rate was significantly prolonged for those with small tumor size (<5 cm) and absence of distant metastasis (P log rank = 0.007 and 0.036, respectively). Aberrant CpG methylation of the WIF-1 promoter leads to transcriptional silencing of this tumor suppressor gene in tumor tissues (P = 0.001). Furthermore, we showed that the level of Wnt5a mRNA was significantly lower in tumor compared to normal tissues (P = 0.031) and lower still in those showing more aggressive behavior (presence of lymph nodes and advanced TNM stage). Our finding supports that WIF-1 is frequently methylated and that Wnt5a acts as a tumor suppressor gene in CRC. Loss of WIF-1 and Wnt5a functions results in more aggressive behavior of the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) remains the most prevalent gastrointestinal cancer [1]. In Tunisia, the incidence of CRC was 2.5–4.5/100,000 [2]. During the last decade, the prognosis for CRC has been significantly improved because of advances in the treatment; however, only 30–40 % of patients are diagnosed at an early stage [3].
In colorectal carcinogenesis, progressive accumulation of genetic and epigenetic alterations contributes to malignant transformation of normal colonic mucosa to neoplasia [4, 5]. Two canonical genetic instability pathways appear to drive tumors in a majority of CRC patients, namely chromosomal instability (CIN) and microsatellite instability (MSI), through a well-established “adenoma-carcinoma” transition sequence [6]. Moreover, epigenetic changes including CpG island methylation and global hypomethylation have been reported to occur early in colorectal carcinogenesis [7, 8]. According to the number of methylated promoters of a well-defined panel of genes, tumors were classified as CIMP+ or CIMP− [9]. Aberrant methylation of CpG islands in the promoter region is responsible for transcriptional silencing of tumor suppressor genes and genes involved in cell cycle control, DNA repair, etc. [10, 11].
Compelling evidence has implicated the wingless-type (Wnt) signaling pathway in the pathogenesis of CRC [12–14]. Wnt ligands are secreted proteins that bind to transmembrane receptors of the frizzled (Fz) family. During normal developmental processes, Wnt signaling plays essential roles in the regulation of cell proliferation, patterning, and fate determination [15]. The binding of Wnt ligands to Fz leads to dephosphorylation and stabilization of β-catenin, which translocates into the nucleus and interacts with members of the T cell factor/lymphocyte enhancer factor (TCF/LEF) family of transcription factors to stimulate the expression of target genes [15]. Aberrant expression of Wnt signaling has been reported in various human cancers, including breast [16], melanoma [17], non-small cell lung cancer [18], leukemia [19], and nasopharyngeal carcinoma [20].
Constitutive activation of the Wnt signaling pathway can be induced by the alteration of the Wnt pathway members, like the overexpression of β-catenin and disheveled [18–21] or the silencing of the Wnt antagonists such as Wnt inhibitory factor-1 (WIF-1) and SRFPs [22, 23].
Wnt5a is one of the ligands in the Wnt family. It has been shown to stimulate intracellular Ca2+ release and activation of protein kinase C (i.e., non-canonical Wnt pathway) [24]. In addition, Wnt5a is involved in the canonical Wnt signaling pathway (i.e., β-catenin-mediated pathway) by either activating or antagonizing it [25]. Therefore, the role of Wnt-5a in tumorigenesis is still ambiguous. Indeed, Wnt-5a was initially proposed as a proto-oncogene because its expression was increased in the lungs and breasts [26, 27]; however, in other tumors, including brain and thyroid cancers, Wnt-5a has been shown to inhibit cell proliferation and acts as tumor suppressor [28, 29]. In CRC, Wnt5a expression has been associated with favorable outcome in early stage tumors [30], while loss of Wnt5a expression is associated with liver metastases [31]. Ying et al. [32] showed that Wnt5a is expressed in normal tissues but frequently silenced by methylation in CRC cell lines and primary tumors. Restoration of Wnt5a expression in silenced cells antagonizes Wnt signaling by promoting intracellular β-catenin degradation and inhibits the clonogenicity of CRC cells [32]. Recently, it was reported that expression of Wnt5a was decreased in the highly metastatic human colon cancer cell line compared with the non-metastatic human colon and that Wnt5a silencing might result from transcriptional regulation of the gene by histone modifications [33]. Among the Wnt signaling components implicated in oncogenesis, Wnt5a is particularly interesting since it acts both as an oncoprotein and a tumor suppressor. To explain the opposing roles of Wnt5a, Bauer et al. [34], showed the presence of two isoforms of Wnt5a: an amino-terminally truncated isoform exhibits tumor-promoting activities, while the full-length Wnt5a protein exhibits tumor-suppressive function. The authors showed that expression of these two Wnt5a isoforms is altered in breast and cervical carcinomas, as well as in neuroblastoma tumors.
This study was conducted to elucidate whether the WIF-1 and Wnt5a genes are associated with the pathogenesis of CRC. For this purpose, we analyzed the epigenetic alteration of the WIF-1 promoter and the expression of the Wnt5a gene in Tunisian patients and their association with major clinical parameters.
Material and methods
Patients’ characteristics
A total of 83 primary sporadic adenocarcinomas were collected between January 2003 and December 2007, from patients who underwent radical surgical resection at the Department of Digestive Surgery of Habib Bourguiba University Hospital (Sfax, Tunisia). All patients gave informed consent prior to specimen collection according to institutional guidelines. None of the patients had preoperative or postoperative chemotherapy. Tissues were also taken from the neighboring non-cancerous mucosa (at least 10 cm away from the tumorous lesions) of 43 patients, and used as controls. At the time of surgery, the age of patients ranged from 25 to 85 years (mean 63.5 years) and the sex ratio was 1:1.37 (female to male ratio). The histological subtypes were classified according to the World Health Organization criteria. The carcinomas were staged according to the TNM (tumor, lymph node, and metastases) classification adopted by the American Joint Committee on Cancer.
DNA extraction and methylation-specific PCR
Genomic DNA was isolated from tissue sections for sporadic CRC patients by phenol/chloroform extraction, and the quantity was checked with a spectrophotometer. DNA samples were stored at −20 °C for further use.
For methylation-specific PCR (MSP) assays, DNA samples (1–2 μg) were treated by sodium bisulfite which converts the unmethylated cytosine to uracil using the EZ Methylation Kit according to the manufacturer’s recommendations (Zymo Research). The bisulfite-treated DNA was amplified using specific primers for methylated and unmethylated alleles. The sequences of the primers, annealing temperature, and product size are listed in Table 1.
For the WIF-1 gene, modified DNA samples were used for MSP assay as described previously [35]. We used nested MSP as follows: in the first round, 2 μL of bisulfite-treated DNA was amplified using the external primers, which recognize the bisulfite-modified template but do not discriminate between methylated and unmethylated alleles (Table 1). The cycling conditions consisted of an initial denaturation step at 95 °C for 5 min, followed by 40 cycles (30 s at 94 °C, 45 s at 50 °C, and 45 s at 72 °C) and a final extension at 72 °C for 10 min. An aliquot (1 μL) of the PCR product was subjected to the second round of PCR using primers selective for the methylated or unmethylated alleles (Table 1). The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles (30 s at 94 °C, 30 s at optimal annealing temperature, and 30 s at 72 °C) and a final extension at 72 °C for 10 min. The reactions were performed in a total volume of 25 μL, containing 0.2 μM of each primer, 200 μM dNTP, 1× PCR buffer, and 1 unit of Dream Taq DNA polymerase (Fermentas). The amplified products were analyzed by electrophoresis on 2 % agarose gel, stained with ethidium bromide and visualized under ultraviolet light.
RNA extraction and reverse transcription-PCR
Total RNA was isolated from fresh frozen tissues using TRIzol reagent [36]. First-strand cDNA synthesis was performed on total RNA, previously treated with DNaseI (Amersham-Biosciences), using 2 μg of RNA, 0.5 μg oligo dT, 2 mM dNTP, 0.5 unit/μL RNase inhibitor (Amersham-Biosciences), 4 μL of 5× RT buffer, and 200 units of MMLV reverse transcriptase (Fermentas). The reaction mixture was incubated at 37 °C for 1 h, followed by 70 °C for 10 min. The cDNA (2 μL) was used as a template in PCR using specific primers for WIF-1 and GAPDH (Table 1). The PCR products were analyzed on 2 % agarose gel, stained with ethidium bromide and visualized under UV light.
Real-time PCR
The real-time PCR was performed using the CFX96™ Real-Time PCR system (Bio-Rad) on 21 colon cancer samples and 4 controls, for Wnt5a (target gene) and GAPDH (control gene). The PCR was carried out with SsoFast™ EvaGreen® Supermix (Bio-Rad) using 1 μL of cDNA in a 25-μL final reaction volume. The thermal cycling conditions were as follows: 2 min at 95 °C and 50 cycles of 20 s at 95 °C and 30 s at 58 °C for Wnt5a and 60 °C for GAPDH. Each sample was repeated in duplicate, and the mean value was calculated. The Wnt5a and GAPDH primers were listed in Table 1.
Statistical analysis
Statistical analyses were performed using the SPSS 17 statistical software for Windows. The two-sided χ 2 test was used to determine associations between the methylation status of the gene promoter and various clinicopathological features. The correlation with overall survival was performed using Kaplan-Meier survival plots, and the significance was tested using the log rank test.
Based on the formula shown below, analysis of the Wnt5a gene expression ratio in tumor samples was performed with the REST-2009 software [37]. GraphPad Prism software (GraphPad Software, La Jolla, CA) was used for analysis and pairwise fixed reallocation randomization tests were performed with 2,000 iterations to assess the significance by the REST software. A P value less than 0.05 was considered statistically significant.
where E is the efficiency, ref is the reference gene (GAPDH), and target is the Wnt5a gene
Results
Methylation profile of WIF-1 gene promoter and correlation with clinicopathological parameters
To investigate the epigenetic changes in WIF-1, we have analyzed by MSP the promoter methylation status in 83 CRC samples and showed that the methylated allele was detected in 87.95 %. A representative example of the MSP results was presented in Fig. 1. Associations between gene promoter methylation of WIF-1 and clinicopathological features were summarized in Table 2. There was a significant correlation of WIF-1 promoter methylation with distant metastasis (P = 0.039) and vascular invasion (P = 0.029).
Representative example of MSP for WIF-1 gene promoter in tumors (a) and normal mucosa (b). T1–T6 tumor samples showing the methylated (T6), hemi-methylated (T1, T2, T4), and unmethylated (T3, T5) patterns; N1–N6 normal mucosa samples showing the unmethylated (N1, N3) and hemi-methylated (N2, N4, N5, N6); H 2 O negative control for MP, L 100-bp DNA ladder (Fermentas)
For DNA samples of normal colorectal mucosa adjacent to the tumor, 26 out of the 43 analyzed cases were unmethylated (60.46 %), while 15 cases were hemi-methylated and only two samples (4.65 %) showed complete methylation (Fig. 1).
The CpG methylation profile of the WIF-1 promoter in tumors versus control tissues was statistically significant (p = 3.883e−08).
Association of WIF-1 promoter methylation with patient survival
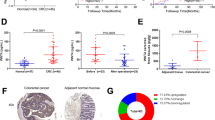
In our series, the survival time was available for only 40 patients among 83 and the follow-up time ranged from 6 to 2,300 days. The Kaplan-Meier plot illustrated a significant reduced overall survival time for the patients with methylated WIF-1 status compared to those with the unmethylated WIF-1 status (P log rank = 0.024, Fig. 2a). Moreover, patients with the methylated WIF-1 pattern combined with large tumor size (≥5 cm) or with advanced metastatic state exhibited a significant reduced overall survival rate compared to those without such poor prognostic features (P log rank = 0.007 and P log rank = 0.036, respectively, Fig. 2b, c).
Correlation of WIF-1 methylated promoter and mRNA expression
In an attempt to validate the effect of aberrant methylation on the expression of WIF-1, we have performed reverse transcription-PCR (RT-PCR) analysis on 23 available sporadic tumor specimens with unmethylated or hemi-methylated profiles using GAPDH as the internal control. Out data showed a significant correlation between the MSP and RT-PCR (P = 0.001). In fact, while the WIF-1 messenger RNA (mRNA) was observed only in 9.5 % of samples with hemi-methylated profile, it has been detected in all samples with the unmethylated profile. Although the number of samples analyzed is small, our result suggests that the CpG methylation is responsible for WIF-1 silencing in tumor tissues (Figs. 3 and 4).
Representative results of MSP and RT-PCR for WIF-1 in CRC. T6 tumor sample with methylated pattern and absence of WIF-1 mRNA. T5 and T4 tumor samples with unmethylated pattern and presence of WIF-1 mRNA, T3 and T2 tumor samples with hemi-methylated status showing weak level of WIF-1 mRNA. Lanes M and U correspond to methylated and unmethylated DNAs, respectively. L 100-bp DNA ladder (Fermentas). GAPDH was used as an endogenous control for RT-PCR
Wnt5a mRNA level in colorectal tumors
We have examined by real-time PCR the expression of Wnt5a in 21 CRC tumors and 4 normal tissues using the GAPDH gene as an internal control. The Wnt5a mRNA was detected at a high level in all the normal tissues, whereas its expression was significantly reduced in most of the tumors. The median Wnt5a expression ratio of the normal tissues was 1.027, whereas in all the tumors it was 0.242. Thus, the expression level of Wnt5a is downregulated in tumors compared to paired controls by a mean factor of 0.131 (P = 0.011, Fig. 5a). Finally, we have examined the Wnt5a expression ratio in relation to clinicopathological parameters. We found that CRC tissues that were with advanced TNM stage (III + IV) or with lymph node invasion (N1) expressed a reduced level of Wnt5a than colorectal cancer tissues without such poor prognostic features (P = 0.014 and P = 0.036, respectively, Fig. 5b, c).
Expression of Wnt5a in sporadic colon cancer patients in comparison to controls (a) and in patient samples according to the TNM status (b) and presence of lymph nodes (c). Each dot plot represents median-centered log2 expression of Wnt5a mRNA from an individual sample. The middle line indicates the median Wnt5a expression level by the group. p indicates the statistical significance of the differences in the collective Wnt5a expression between the two groups, as calculated using Mann-Whitney test
Discussion
Activation of the Wnt signaling pathway occurred in various human tumors including CRC through both genetic and epigenetic mechanisms [12]. The family members of the Wnt pathway have been divided into canonical and non-canonical. The canonical Wnt ligands promote cell growth and proliferation by controlling cytoplasmic degradation and subsequently nuclear accumulation of β-catenin [12]. The non-canonical Wnt ligands were involved in regulating cell motility and polarity through a variety of intracellular mediators including calmodulin-dependent protein kinase II, protein kinase C, and c-Jun N-terminal kinase [38]. Aberrant activation of the Wnt signaling pathway has been firstly described in colorectal carcinoma [8]. Moreover, the secreted Wnt antagonists namely secreted frizzled-related proteins (SFRP1-5), Dickkopf (DKK1-4), and Wnt inhibitory factor-1 (WIF-1) are important mediators of both types of Wnt signaling. These glycoproteins are often silenced by promoter CpG island hypermethylation in CRC as well as various tumors [22, 23, 39, 40].
We showed that WIF-1 is often methylated in CRC since 87.95 % of patients displayed the methylated profile which is in line with previous studies [28, 41]. Lee et al. [41] showed that among seven studied promoters, WIF-1 methylation was the highest (74 %) in a large series of CRC patients.
Furthermore, we have demonstrated that expression of WIF-1 is correlated with the promoter CpG methylation pattern. Indeed, WIF-1 mRNA was detected in 100 % of samples with unmethylated status whereas only 9.5 % of cases exhibiting the methylated profile expressed the WIF-1 mRNA. This suggests that CpG methylation is responsible for WIF-1 silencing in tumors; nevertheless, it should be confirmed on a larger cohort. Our result is consistent with that of Taniguchi et al. [39] showing that the methylation status was correlated with WIF-1 mRNA expression. Similarly, other studies investigated nasopharyngeal [20], breast [42], and lung [43] cancers, suggesting that the WIF-1 gene is subject to DNA hypermethylation-mediated downregulation in different tumor types.
In a univariate analysis, the WIF-1 methylation pattern was significantly correlated with aggressive behavior such as presence of distant metastasis and vascular invasion. With regard to patient survival, Kaplan-Meier plots showed that the overall survival time was significantly reduced for CRC patients with the WIF-1 methylated pattern. In addition, if we considered the group of patients with the methylated profile, we found that the overall survival was significantly prolonged for those with small tumor size (<5 cm) and absence of distant metastasis (P log rank = 0.007 and 0.036, respectively). In the study of Taniguchi et al., no significant association was found between WIF-1 downregulation and clinicopathological characteristics [39].
Finally, we have examined the expression of the Wnt5a gene in CRC patients. Interestingly, in tumor specimens, the level of Wnt5a mRNA was significantly lower than that of normal tissues, in particular in cases with more aggressive behavior such as advanced TNM, and lymph node involvement. This finding supports previous studies showing that Wnt5a acts as a tumor suppressor gene in CRC [30]. It was reported that Wnt5a is frequently silenced by methylation in CRC cell lines and primary tumors but expressed in normal colon tissues [44, 45]. The restoration of Wnt5a antagonizes Wnt signaling by promoting intracellular β-catenin degradation. Similar findings have been described in breast cancer since loss of Wnt5a resulted in stabilization of nuclear β-catenin and expression of target genes, promoting tumor growth [45]. Furthermore, it was recently reported in Tunisian patients with sporadic breast cancer that the level of Wnt5a mRNA was significantly lower in tumor tissues compared to normal tissues, in particular in cases with more aggressive behavior such as advanced TNM, reduced survival, and triple negative status [46].
In conclusion, we showed that loss of WIF-1 and Wnt5a expression is related to the aggressiveness of sporadic CRC in Tunisian patients and thus could be useful as biomarker for prognosis prediction.
References
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2005;55:74–108.
Hsairi M, Fakhfakh R, Ben Abdallah M, Jlidi R, Sellami A, Zheni S, et al. Assessment of cancer in Tunisia. Tunis Med. 2002;80:57–64.
Smith RA, Cokkinides V, Eyre HJ. American Cancer Society guidelines for the early detection of cancer. CA Cancer J Clin. 2006;56:11–25.
Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–65.
Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–11.
Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7.
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6.
Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Med. 2004;23:29–39.
Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93.
Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, et al. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–54.
Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Amouri A, Frikha F, et al. Aberrant methylation of hMLH1 and p16INK4a in Tunisian patients with sporadic colorectal adenocarcinoma. Biosci Rep. 2011;31(4):257–64.
Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51.
Shimizu Y, Ikeda S, Fujimori M, Kodama S, Nakahara M, Okajima M, et al. Frequent alterations in the Wnt signaling pathway in colorectal cancer with microsatellite instability. Genes Chromosome Cancer. 2002;3(1):73–8.
Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25(57):7531–7.
Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–330.
Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56.
Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–88.
Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22:7218–21.
Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2004;101:3118–23.
Fendri A, Khabir A, Hadri-Guiga B, Sellami-Boudawara T, Daoud J, Frikha M, et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter is common and occurs in advanced stage of Tunisian nasopharyngeal carcinoma. Cancer Invest. 2010;28:896–903.
Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90.
Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22.
Aguilera O, Fraga MF, Paz BEMF, Herranz M, Espada J, García JM, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–21.
Slusarski DC, Yang-Snyder J, Busa WB, Moon RT. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol. 1997;182:114–20.
Umbhauer M, Djiane A, Goisset C, Penzo-Méndez A, Riou JF, Boucaut JC, et al. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signalling. EMBO J. 2000;19:4944–54.
Huang C-L, Liu D, Nakano J, Ishikawa S, Kontani K, Yokomise H, et al. Wnt5a expression is associated with the tumor proliferation and the stromal vascular endothelial growth factor: an expression in non-small-cell lung cancer. J Clin Oncol. 2005;23:8765–73.
Lejeune S, Huguet EL, Hamby A, Poulsom R, Harris AL. Wnt5a cloning, expression, and up-regulation in human primary breast cancers. Clin Cancer Res. 1995;1:215–22.
Blanc E, Roux GL, Bénard J, Raguénez G. Low expression of Wnt-5a gene is associated with high-risk neuroblastoma. Oncogene. 2005;24:1277–83.
Kremenevskaja N, von Wasielewski R, Rao AS, Schöfl C, Andersson T, Brabant G. Wnt 5a has tumor suppressor activity in thyroid carcinoma. Oncogene. 2005;24:2144–54.
Dejmek J, Dejmek A, Safholm A, Sjolander A, Andersson T. Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res. 2005;65(20):9142–6.
Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455(6):485–94.
Ying J, Li H, Yu Ju Ng KM, Poon FF, Wong SC, et al. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/b-Catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008;14:55–61.
Li Q, Chen H. Silencing of Wnt5a during colon cancer metastasis involves histone modifications. Epigenetics. 2012;7(6):551–8.
Bauer M, Bénard J, Gaasterland T, Willert K, Cappellen D. WNT5A encodes two isoforms with distinct functions in cancers. PLoS One. 2013;18:8(11).
Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6.
Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9.
Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36.
Kuhl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signalling pathway takes shape. Trends Genet. 2000;16:279–83.
Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–52.
Maehata T, Taniguchi H, Yamamoto H, Nosho K, Adachi Y, Miyamoto N, et al. Transcriptional silencing of Dickkopf gene family by CpG island hypermethylation in human gastrointestinal cancer. World J Gastroenterol. 2008;14:2702–14.
Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–91.
Ai L, Tao Q, Zhong S, Fields CR, Kim W-J, Lee MW, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–8.
Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–20.
Rawson JB, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, et al. Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. Br J Cancer. 2011;104(12):1906–12.
Roarty K, Baxley SE, Crowley MR, Frost AR, Serra R. Loss of TGF-beta or Wnt5a results in an increase in Wnt/beta-catenin activity and redirects mammary tumour phenotype. Breast Cancer Res. 2009;11:R19.
Trifa F, Karray-Chouayekh S, Jmal E Jmaa ZB, Khabir A, Sellami-Boudawara T, Frikha M, Daoud J, Mokdad-Gargouri R. Loss of WIF-1 and Wnt5a expression is related to aggressiveness of sporadic breast cancer in Tunisian patients. Tumor Biol. 2013
Acknowledgments
This work was supported by a grant of the Tunisian Ministry of High Education and Scientific Research.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abdelmaksoud-Dammak, R., Miladi-Abdennadher, I., Saadallah-Kallel, A. et al. Downregulation of WIF-1 and Wnt5a in patients with colorectal carcinoma: clinical significance. Tumor Biol. 35, 7975–7982 (2014). https://doi.org/10.1007/s13277-014-2015-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2015-9