Abstract
S100A4, a 10–12 kDa calcium-binding protein, plays functional roles in tumor progression and metastasis. The present study aimed to investigate the function of S100A4 in osteosarcoma (OS) metastasis, using a loss-of-function approach. Our previous expression profiling analysis revealed that S100a4 was preferentially expressed in the highly metastatic mouse OS cell line, LM8. Introducing a short hairpin ribonucleic acid (shRNA) targeting S100a4 using a newly established vector containing insulators and transposons, we established stable LM8 subclones with almost 100% silencing of endogenous S100a4 protein. These transfectants showed a significant suppression of cell migration in vitro as well as a marked reduction in their ability to colonize the lung and form pulmonary metastases in vivo following intravenous inoculation, whereas there was no significant change in cell proliferation or cell attachment to fibronectin, laminin, and type I collagen. Expression and phosphorylation of ezrin, an emerging OS metastasis-associated factor, and expression of MMPs, remained the same in S100a4-shRNA clones. In 61 human OS, immunohistochemical analysis showed that lesional cells in 85.2% samples expressed S100A4 protein, and the immunoreactivity was primarily cytoplasmic, but it also showed occasional nuclear localization. Chondroblastic and osteoblastic OS subtypes expressed more S100A4 than fibroblastic subtypes. The causative role of S100A4 in OS lung metastasis shown in the murine xenograft model, together with the high proportion of primary human OS expressing S100A4, suggest that S100A4 protein represents an important potential target for future OS therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor affecting mostly young adults. OS has a high tendency to metastasize to the lung, and a high number of the patients presented in the clinic have already developed metastatic pulmonary disease, which results in a very poor prognosis [1]. Thus, understanding the molecular mechanisms underlying pulmonary metastasis and developing targeted therapies against OS metastasis are extremely important. We have previously employed transplantable mouse OS cell lines as one in vivo metastasis model. The mouse OS cell line, LM8, has been isolated originally from the Dunn OS cell line by in vivo selection as a subline with a high metastatic potential to the lung [2]. Our previous study using gene chip-based global gene expression analysis of differential screening between parental Dunn and LM8 cells revealed several genes predominantly expressed in LM8 cells, which correlate with high metastatic potential [3]. Of these, we focused here on the S100a4 messenger ribonucleic acid (mRNA), which is preferentially expressed in LM8 cells compared to the parental Dunn cell line.
The S100A4 protein belongs to the S100 family of calcium-binding proteins constituting more than 25 members [4]. S100A4 is also referred to as 18A2, mts1, CAPL, PEL-98, 42A, p9Ka, FSP1, calvasculin, and metastasin. S100A4 has been suggested to act as a tumor progression factor, facilitating processes such as cell motility and invasion, promoting angiogenesis with upregulation of matrix metalloproteinases (MMPs), adhesion to extracellular matrix (ECM), epithelial— mesenchymal transition, and modulation of both cell cycle progression and cell survival [5]. Previous gain-of-function experiments in vivo showed that overexpression of S100A4 induced a metastatic phenotype in nonmetastatic rat primary mammary epithelial cells [6]. Furthermore, two independent experiments using S100A4 transgenic mouse lines showed an increase in metastatic potential of breast cancers following crossing with transgenic mice susceptible to mammary tumor formation [7, 8]. Meanwhile, S100A4 loss-of-function studies, employing antisense mRNA and ribozyme techniques and introducing mutations and transient small interfering RNA (siRNA) transfection, reduced the metastatic potential of highly metastatic cell lines [9–11]. Clinically, the expression level of S100A4 has been suggested to be a prognostic molecular marker for the metastatic potential of a wide range of human neoplasms including breast, esophageal squamous, primary gastric, colorectal, pancreatic, bladder, nonsmall cell lung, thyroid, prostate, bladder cancers, malignant melanomas, and most recently OS [5, 12–14 and references therein].
Although S100A4 has been regarded as one of the crucial genes that establish tumor metastasis, the loss-of-function studies in vivo, to date, using siRNA have employed only a thyroid cancer metastasis model [15]. We therefore embarked on investigating the potential role of S100A4 in OS metastasis by knocking down S100a4 in LM8 cells. We obtained stable knockdown clones using a new efficient shRNA-expressing vector, where the shRNA cassette would be protected from the chromosome position effects. We investigated their in vitro parameters that are associated with the metastatic ability of cells including motility, adhesion, expression of MMPs and ezrin [16] as well as their in vivo metastatic potential in mice. Furthermore, expression of S100A4 protein was assessed by immunohistochemistry (IHC) in 61 primary human OS samples. Our study suggests that targeting S100A4 has a potential therapeutic application to prevent OS metastasis.
Materials and methods
Cell culture of Dunn and LM8
The mouse OS cell line Dunn and its highly metastatic cell line LM8 [2] were a kind gift from Dr. T. Ueda (Department of Orthopedic Surgery, Graduate School of Medicine, Osaka University, Osaka, Japan) and were employed for experiments within ten passages. The cells were grown in Eagle’s minimum essential medium (MEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-incubated fetal bovine serum (FBS; CELLect, Lot No. 5766H, MP Biomedicals, Irvine, CA), l-glutamine, antibiotics, and 1% nonessential amino acid (Sigma-Aldrich), as described previously [3]. The human fibrosarcoma cell line, HT1080, was obtained from ATCC and cultured under standard conditions.
High-density oligonucleotide array analysis
Approximately 100 μg of biotin-labeled complementary RNA (cRNA) from LM8 and Dunn cells was hybridized to a high-density oligonucleotide array (GeneChip® Mouse Genome 430 2.0 Array, Affymetrix, Santa Clara, CA), of which probe set is newer than that of the previously employed [3], as described [17]. Following scanning using Hewlett-Packard Scanner (Palo Alto, CA), the intensity of each feature of the array was calculated using Affymetrix Gene Chip software version 3.3. In calculating the average differences in expression, normalization of all probe sets was performed using the MAS5.0 algorithm and the fold change between the hybridization intensity of Dunn and LM8 samples was obtained.
RT-PCR analyses
Multiplex reverse transcription-polymerase chain reaction (RT-PCR) was carried out in 50 μl containing four primers for amplifying both S100a4 and Gapdh transcripts. Quantitative RT-PCR (qRT-PCR) was carried out in a 96-well optical tray using the iCycler iQTM Multi-Color Real Time PCR Detection system (Bio-Rad, Hercules, CA). Standard curves for templates of S100a4 and Gapdh were generated by serial dilution of the control templates. All the samples were amplified in triplicate. The primer sequences are shown in Table S1.
Generation of S100a4 shRNA vectors and transfection
In order to prevent the transcriptional suppression of shRNA expression by the chromosomal position effect during the in vivo metastasis assay, we devised the pInSB-Neo-shS100a4 vector that possesses chicken HS4 insulator cores, which are deoxyribonucleic acid (DNA) sequence elements that can act either to block the extension of a condensed chromatin domain into a transcriptionally active region or to prevent the interaction of a distal enhancer with a promoter (Fig. 1a) [18] (Figs. S1–S3). Furthermore, to enable an efficient genomic integration of the shRNA expression cassette, we utilized the sleeping beauty transposable DNA element flanked by a couple of inverted repeat sequences, which are excised by the sleeping beauty transposase at these repeat sequences and are then inserted again almost at random elsewhere in the genome [19]. The pInSB-Neo-shCont vector, with no insertion of oligonucleotides into the shRNA expression cassette, was utilized as the Mock vector. These two vectors were co-transfected into LM8 cells each with the sleeping beauty transposase expression vector (pCMV-SB; kindly provided from Dr. Hackett, University of Minnesota) using FuGENE® 6 (Roche Diagnostics, Basel, Switzerland). The transfected LM8 transfectants, representating the S100i knockdown clones and Mock controls, were selected in the presence of 400 μg/ml G418 (Invitrogen, Carlsbad, CA).
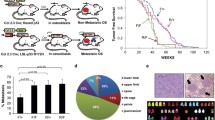
Isolation of cell clones with silenced S100A4 expression. (a) Map of pInSB-Neo-shS100a4, the shRNA-expressing vector against S100a4. 2 × HS4 core and IR/DR indicate the tandem of the 250 bp core HS4 insulator and sleeping beauty transposable elements, respectively. For further details, see Electronic Supplementary Material. (b) Top panel: qRT-PCR analysis of the mRNA expression levels of S100a4 in LM8, Dunn, Mock, and S100i clones. The relative expression levels are normalized to Gapdh and compared to the mean of the S100a4 expression levels in LM8 set to 100%. The asterisk (*) indicates p < 0.05 by one-way ANOVA comparing LM8 and Dunn or Mock and S100i clones. Bottom panel: Western blot analysis of the S100A4 protein as a 16 kDa band in LM8 and Mock cells, and negligible expression in Dunn and S100i clones. Loading was confirmed by actin protein expression. (c) Immunofluorescence staining of S100a4 protein using an anti-S100A4 antibody, under the same exposure times. a LM8, b Mock, c S100i10, d S100i16. Magnification 100×. (d) Growth curve analysis of parental LM8, Mock, and S100i clones. Cells were plated at 5000 cells/35-mm dish in standard medium (day 0), and the number of cells was counted on each day. Data represent the mean ± SD of the number of cells in triplicate wells
Western blot analysis
LM8 cells, Dunn cells, and S100i clones were prepared in lysis buffer [4.8% sodium dodecyl sulfate (SDS), 2 M urea, 8% sucrose, 0.5 M NaCl, 1 mM NaF, 0.1 mM NaVO4, 30 mM β-glycerophosphate, and 1 mM DTT with proteinase inhibitors], and the protein samples were separated by SDS polyacrylamide gel electrophoresis, followed by transferring onto polyvinylidene fluoride membrane (Millipore, Bedford, MA). The membranes were exposed to anti-S100A4 (A5114, Dako, Glostrup, Denmark), anti-ezrin (Sigma-Aldrich), anti-phospho-ERM (Cell Signalling, Boston, MA), anti-MMP14 (113-5B7, Calbiochem, Nottingham, UK), and anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, followed by incubations with a horseradish peroxidase (HRP)-labeled secondary antibody and HRP detection regent (Millipore). The anti-S100A4 antibody (A5114) does not show cross-reactivity with other S100 protein family members, but it has potential cross-reactivity with cytokeratin 8 (~80 kDa) [20] which can easily be distinguished from S100A4 (~10–12 kDa) by size in Western blotting. The ratio of expression level was determined by densitometry, using Image J software (National Institutes of Health, Bethesda, MD) and by normalizing to actin levels.
Immunofluorescence staining of S100a4
LM8 cells and, mock S100i clones were grown in 35-mm2 chamber slides (Becton Dickinson Labware, Franklin Lakes, NJ), fixed with acetone/ethanol and permeabilized with 0.1% Triton-X in phosphate buffered saline (PBS) for 5 min. Following incubation with an anti-S100A4 Rabbit polyclonal antibody (1:600 dilution, Ab-8, Cat. #RB-1804, Lab Vision, Fremont, CA), the cell layer was linked to Alexa Fluor 488-labeled goat anti-rabbit antibody (Invitrogen). Evaluation of immunoreactivity was performed with a fluorescence microscope and photographed using a digital cooled charge-coupled device (CCD) camera (Olympus, Tokyo, Japan) under the same exposure time.
Growth properties of tumor cells in vitro
Cell lines were cultured in 35-mm dishes in standard medium at a starting density of 5 × 103 cells/dish. Cell growth was estimated by counting the total number of cells in each dish every 24 h. Each experiment was performed in triplicate and repeated three times.
In vitro cell migration assay
Cell motility was estimated in a trans-well chamber with 8 μm pore membranes (Becton Dickinson Labware, Bedford, MA), with assays repeated a minimum of three times as previously described [3]. Cells that penetrated through to the other side of the pore filter were fixed and stained with Giemsa solution and counted microscopically using ten random fields at 100× magnification.
In vitro cell attachment assay
The diluted ECMs, fibronectin (Sigma-Aldrich), laminin (Becton Dickinson), type I collagen (Sigma-Aldrich), and bovine serum albumin (BSA), were each coated onto a 96-well plate. Following blocking with 0.2% BSA in PBS, each cell line (5 × 104 cells) in MEM containing 0.1% BSA was placed in each well and incubated for 30 min. The number of attached cells in triplicate wells was compared using a cell counting kit-8 (Dojin Chemicals, Kumamoto, Japan) by measuring the absorbance at 450 nm using a plate reader (Model 550 Microplate Reader, Bio-Rad).
Gelatin zymography
Serum-free conditioned medium from each cell line was assayed for gelatinase activity, using a gelatin zymo electrophoresis kit (Primary Cell, Hokkaido, Japan), according to the manufacturer’s protocols.
Lung metastasis assay in vivo
C3H male mice aged 10–11 weeks (CLEA Japan, Shizuoka, Japan) housed under specific pathogen-free conditions were used to estimate the in vivo lung metastatic potential of S100i clones. Each tumor cell line was suspended in 0.2 ml Hank’s balanced salt solution (HBSS, Invitrogen) and injected subcutaneously either into the back (106 cells) or into the tail vein (2 × 105 cells) on day 0. The mice were sacrificed on day 14, and the lungs were bisected, embedded in paraffin, cut, and stained with hematoxylin and eosin (H&E). Finally, the total number of the metastatic nodules on each slide glass was counted under microscope. The experimental protocol was approved by the Animal Care and Use committee of Tokyo University (1822 T-020).
Ki-67 IHC
IHC staining for murine Ki-67 was carried out using biotin-free polymer kits (ChemMate envision plus, Dako) according to the manufacturer’s protocol. Paraffin sections of lungs from the mouse xenograft model described above were incubated with an anti-mouse Ki-67 (clone TEC-3, Dako, 1:100) antibody that does not recognize human Ki-67.
Analysis of human tumor tissue samples
All the cases were diagnosed as conventional OS at the University of Tokyo Hospital. Tissue microarrays (TMAs) were constructed from formalin-fixed and paraffin-embedded blocks as described elsewhere [21]. All the cases were conventional OSs, and the samples comprised of 44 osteoblastic OS, seven chondroblastic OS, and ten fibroblastic OS, according to the preferential morphology observed following H&E staining. IHC staining on human TMAs was carried out as described above using an anti-S100A4 polyclonal antibody (Lab Vision, 1:600, Ab-8) [22]. Only immunoreactive tumor cells were included in our quantification, and grading of the IHC staining for S100A4 was scored independently by two experienced pathologists using the following categories, representing the proportion of positive immunoreactive tumor cells as follows: -, no immunoreactivity; +, <30%; 2+, between 30% and 67%; and 3+, more than 67% of the tumor cells. This study was approved to use for research by the Ethical Review Board of Tokyo University (no. 1220).
Statistical analysis
Student’s t-test or one-way analysis of variance (ANOVA) test followed by Bonferroni’s post hoc test was performed in order to compare data set between either LM8 and Dunn or Mock and each clone, respectively, using Graphpad Prism v.4 (Graphpad Software, La Jolla, CA). For in vitro growth assays, the data sets were log-transformed prior to drawing a normalized regression line. All p values were two-sided with statistical significance at p < 0.05.
Results
S100a4 mRNA expression is markedly upregulated in LM8 metastatic OS cells
Microarray-based global gene expression analysis in both LM8 and Dunn cells identified hundreds of genes upregulated in LM8. Generally, a signal ratio of >80 and fold increase of >2.0 are considered to be reliable [17]. The data discussed in this publication have been deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information (NCBI) and are accessible through GEO Series accession number GSE26648. Among the ten highest genes listed in Table S2, S100a4 depicted the highest expression level in normalized signal intensity in LM8 in comparison to Dunn. We next investigated the expression ratios of the other S100 family gene members. As listed in Table 1, from 11 family members analyzed, S100a1 gene expression was quite low in LM8 cells compared to S100a4 levels, and the expression levels of S100a6, S100a10, and S100a11 did not differ between LM8 cells and the nonmetastatic parental Dunn cells. In addition, S100A8 and S100A9, which have chemokine activity, can establish a premetastatic niche in the lung by recruiting bone marrow-derived cells [23], and are expressed in osteocartilagenous tissues [24], showed negligible expression in both Dunn and LM8 cells. Taken together, among all the S100 proteins tested, only S100a4 showed a significant increase in LM8 cells compared to the parental Dunn cells that might correlate with its high pulmonary metastatic capacity. Indeed, validation by qRT-PCR analysis demonstrated that LM8 expressed an approximately tenfold higher level of S100a4 transcripts than Dunn, and the protein expression levels as determined by densitometry analysis of the Western blots confirmed the qRT-PCR result (Fig. 1b).
Establishment of stable S100a4 knockdown cell clones (S100i clones)
By introducing the unique shRNA using a transposon insulator-associated vector (Fig. 1a), we have selected two representative knockdown clones out of a total of 24 clones, namely, S100i10 and S100i16, that expressed significantly reduced levels of S100a4 mRNA and S100a4 protein compared to Mock controls, as shown by qRT-PCR and Western blot analysis, respectively (Fig. 1b). The negligible S100a4 protein expression level in each S100i clone was confirmed by immunofluorescence using an anti-S100A4 antibody (Fig. 1c). S100a4 protein showed intensive cytoplasmic localization in both the LM8 cell line and the Mock clone, whereas S100i10 and S100i16 clones showed no detectable protein. These two S100i transfectants were employed for all further analyses.
S100a4 knockdown does not affect cell proliferation in vitro
We next investigated whether or not the knockdown of S100a4 in the S100i clones affected their growth properties. There were no significant differences in the growth rates between LM8, Mock, and S100i clones, as the slopes of the log-transformed data representing the population doubling time were all similar (up to 0.75, p = 0.33; Fig. 1d).
Cell migration in vitro is strongly suppressed in S100i clones
The effects of knockdown of S100a4 on cell migration were analyzed next using transwell assays. The results showed that the cell mobility of S100i clones was dramatically suppressed, with a significant decrease of 92.9% and 67.7% in S100i10 and S100i16 clones, respectively, compared to Mock cells (Fig. 2a).
Metastatic parameter assays in vitro. (a) In vitro migration assay. The number of cells migrating through 8 μm pores on Boyden chamber membranes was determined by averaging ten random fields at 100× magnification. Asterisk (*) indicates p < 0.05 using one-way ANOVA comparing Mock and S100i clones. There were no significant differences in the cell migration ability between Mock and LM8 cells. (b) Adhesion assay to fibronectin (FN), laminin (LN), type i collagen (Col I) and bovine serum albumin (BSA). Cells were plated on the respective ECM molecules as indicated and adhesion was measured as described in the “Materials and methods.” The absorbance at 450 nm indicates the number of attached cells to each matrix. There were no significant differences between all cell lines. (c) Analysis of MMP expression. The transcript levels of i Mmp2 and ii Mmp13 were examined using qRT-PCR analysis. The asterisk (*) indicates p < 0.05 by one-way ANOVA comparing LM8 and Dunn or Mock and S100i clones. iii Western blot analysis for Mmp14. Protein samples from each cell line are indicated. HT1080 human fibrosarcoma cells were employed as a positive control. iv Gelatin zymography using culture supernatants from each cell line as indicated. The activities of the gelatinases proMMP9, proMMP2, and MMP2 were visualized as cleared regions. MMP marker was used as a control. (d) Expression and phosphorylation status of ezrin, radixin, and moesin (ERM) family proteins. The indicated cell lines were cultured and expression of total ezrin and phospho-ERM proteins was examined by Western blot analysis. Actin was used as a loading control
Cell adhesion to ECM is not altered following knockdown of S100a4
We next investigated the adhesive properties of each cell line to different ECM substrates. Plating cells on fibronectin, laminin, type I collagen, or BSA showed similar adhesion properties of all cell lines, although adhesion of S100i clones to fibronectin showed an approximately 30% decrease compared to Mock cells, although these differences were not statistically significant (Fig. 2b).
Expression of MMPs in S100i clones
As MMPs have important roles in metastasis, we examined potential alterations in MMP expression in the S100i clones. We focused on MMP2, MMP13, and MMP14 because these were among the only MMPs that showed a significant change in expression levels between LM8 and Dunn cells using the gene chip analysis (Table S3). qRT-PCR analysis showed that the expression of Mmp2 was no different between LM8, Mock, and S100i clones, and there was negligible expression of Mmp13 transcripts in LM8, Mock, and S100i clones (Fig. 2ci,ii). We further confirmed the levels of MMP2 and MMP9 by gelatin zymography and protein expression of MMP14 by Western blotting. The results demonstrated that both S100i clones expressed proMMP2 at the same levels as LM8 and Mock cells, whereas Dunn cells showed less proMMP2 expression (Fig. 2civ). The expression levels of MMP2 and MMP9 were negligible in all the cells (Fig. 2civ). Despite the low expression levels of MMP14 protein, no differences between the Dunn, LM8, and S100i transfectants were observed (Fig. 2ciii), which was consistent with the array expression data of Mmp14 (Table S3: relative signal levels, 323.1 and 287.1 in LM8 and Dunn, respectively; log2 (Fold change), 0.2).
Expression of ERM family proteins is not affected by knockdown of S100a4
Since ezrin, one of the ezrin–radixin–moesin (ERM) family proteins, has been regarded as one of the causative genes in OS metastasis [25], we next examined the expression levels of ezrin in the investigated cell lines. Western blot analysis revealed that the expression of both total ezrin and phosphorylated ERM proteins was not significantly different between any of the cell lines (Fig. 2d), which was consistent with the array expression data of ezrin (relative signal levels, 121 and 101 in Dunn and LM8, respectively). The amount of phosphorylated ERM was proportional to the basal ezrin. Thus, we conclude that ERM proteins are generally unaffected by the S100a4 knockdown.
S100i clones show reduced lung metastatic activity in vivo
As S100i clones showed a significantly reduced migratory activity in vitro, we next evaluated whether their ability to cause lung metastasis was altered using an in vivo model. Whereas formation of spontaneous metastases following subcutaneous inoculation did not reveal any significant differences in lung colonization between the clones (data not shown), intravenous injection of S100i and Mock clones into the tail vein of mice showed a marked suppression of pulmonary metastases by both S100i clones (Fig. 3a). There were significant differences in the number of metastatic lung nodules between the Mock and the S100i clones, representing 83% and 75% decreases in clones S100i10 and S100i16, respectively (Fig. 3b). Furthermore, IHC analysis employing an anti-S100A4 antibody revealed negligible S100a4 protein expression in lung metastatic nodules formed by S100i cells compared to that of Mock (Fig. 3a). Ki-67 labeling studies revealed that Mock cells as well as the two S100i clones exhibited a similar labeling index in the metastatic nodules of approximately 55% (Fig. 3c). These findings suggest that the suppression of lung metastases from cells with reduced expression of S100a4 is unlikely to be due to differences in the growth properties of the cells in vivo.
In vivo metastasis assay. (a) Microphotographs of lung metastatic nodules which formed 2 weeks following intravenous injection of the a Mock and b S100i16 clones (H&E staining; bar 250 μm). IHC staining of sections of lung nodules from Mock (c) and S100i16 (d) clones using an anti-S100A4 antibody (bar 50 μm). T Metastatic nodules, * bronchus. (b) Histogram showing the number of lung metastatic nodules formed by each clone following intravenous injection. The asterisk (*) indicates p < 0.05 by one-way ANOVA, comparing Mock and S100i clones. The lower panel shows RT-PCR analysis confirming the absence of S100a4 mRNA expression in the S100i clones. n Number of mice used for each clone. (c) Proliferation index of metastatic tumors. The upper panel shows IHC staining with an anti-mouse Ki-67 antibody (bar 100 μm). The bar chart displays the percent Ki-67 positive cells for each cell line. There were no significant differences between the clones (p > 0.168)
Human OS expresses high levels of S100A4
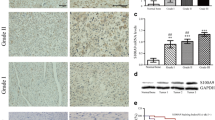
To establish the relevance of our findings to human OS, we examined the expression of S100A4 protein in a total of 61 OS samples. The lesional cells in 85.2% human OS (52/61) showed intensive S100A4 protein expression, as summarized in Table 2. Moreover, 80.8% of positive tumors showed a high number of lesional cells (grade 2 or 3), whereas the remaining 19.2% revealed a fewer number of cells (grade 1). The S100A4 protein was mainly localized to the cytoplasm, while some tumor cells also revealed dense immunoreactive signals in the nucleus (Fig. 4). This nuclear immunolocalization was especially evident in the chondroblastic subtypes. All the histological subtypes including osteoblastic, chondroblastic, and fibroblastic tumors generally showed strong S100A4 expression, although the fibroblastic subtype showed lower positivity than the other two subtypes, where 50% of the samples (5/10) were negative for S100A4 immunoreactivity (Table 2). In addition to tumor cells, inflammatory cells as well as osteoclasts, which are highly chemotactic, were also strongly positive for S100A4 protein (data not shown). From this evidence, we conclude that human OS generally expresses high levels of S100A4 protein, despite minor differences between histological subtypes.
Expression of S100A4 in human OS. Microphotographs showing H&E staining (a–c) and representative IHC staining of conventional human OS samples labeled with an anti-S100A4 antibody (d–f). A, D Typical osteoblastic OS subtype. B, E Typical chondroblastic OS subtype. C, F Typical fibroblastic OS subtype. Bar 200 μm
Discussion
In the present study, we used global gene expression analysis in the highly metastatic OS cell line LM8 and showed that S100a4 is preferentially expressed in LM8 cells and is a key factor in OS pulmonary metastasis. We developed a new shRNA expression vector containing both chicken beta-globin insulator cores [18] and transposon units [26], with little chance of losing the shRNA cassette during our investigation, and we succeeded in nearly 100% stable knockdown of S100a4 protein. Two independent S100i clones demonstrated up to 83% suppression in their ability to form pulmonary metastasis in vivo in comparison to the Mock control cells following intravenous injection, suggesting that S100a4 plays an important role in seeding and colonizing a distant organ in this mouse model. To identify the potential underlying mechanisms, we investigated a number of parameters that are commonly associated with metastasis such as cell migration and adhesion, cytoskeleton-associated proteins, and MMP activity.
S100A4 protein was mainly localized to the cytoplasm in LM8 cells. Several target proteins, such as tropomyosin, the heavy chain of nonmuscle myosin IIA (MHC-IIA or MYC9) and IIB, p37, S100A1, methionine aminopepdidase 2, liprin β1, CCN3, and septin 2, 6, and 7, are known to interact with cytoplasmic S100A4 in a Ca2+-dependent manner [5]. S100A4 protein should exist as a homodimer to interact with these target molecules; inhibition of its dimerization leads to a significantly reduced binding affinity to MHC-IIA and p53, suppressing further tumor metastasis in in vivo models [11]. MHC IIA, one of the filaments comprising nonmuscle myosin II, also plays an important role in cell migration, cell adhesion, and cell motility through its contractile and actin cross-linking properties [27, 28]. Direct binding MHC IIA to S100A4 inhibits MHC IIA phosphorylation by kinases including Rho-associated kinase, casein kinase C and PKC (Protein kinase C) and leads to biological cellular consequences [28, 29]. Thus, it is possible that the suppression of cell motility observed by S100a4 knockdown is associated with the subsequent loss of MHC IIA function.
Cell motility is also regulated by the actin cytoskeleton which is modified and regulated by the ERM family proteins. Phosphorylated ERM proteins link actin filaments and cell surface receptors such as CD44 [30], in which the vast majority of human OS express [14]. Recently, it has been reported that ezrin is responsible for developing OS metastasis, and its expression level is correlated with clinical prognosis [31]. Like S100A4, ezrin plays a key role in cell motility, cellular adhesion, and also modulation of intracellular signaling pathways via activating Rho family members [32]. Regarding LM8 cells, Fukaya et al. [33] reported that ezrin activated the MAPK and Akt pathways. Moreover, ezrin phosphorylation is modified by protein kinase C alpha [34], which is under S100A4 regulation [5]. In our study, nevertheless, introducing shRNA against S100a4 did not affect either the expression levels of ezrin or phosphorylated ERM proteins. Hence, it is likely that ezrin did not contribute significantly to either the suppressed cell migration or the reduced in vivo metastasis observed in the present study.
Inoculation of the two S100i clones into the tail vein revealed a significant suppression in the number of pulmonary metastatic nodules compared to the control clone. The differences in in vivo metastatic potential in the present study could not be explained by alterations in S100A4-dependent tumor cell proliferation because of no differences both in vitro and in vivo. Invading the basement membrane of endothelial cells followed by ECM degradation by MMPs and further adhesion to connective tissue is an essential serial process for escaping from the intravascular space to distant organs. Regarding the invasive potential of OS cells, previous reports have described a correlation between the downregulation of expression of MMP2, MMP3, MMP13, and MMP14 and the suppression of S100A4 expression [5, 35, 36]. In the present study, however, both the S100i knockdown transfectants expressed equivalent amounts of proMmp2/Mmp2 to LM8, and no detectable Mmp9 as originally described [2], which did not allow us to carry out in vitro invasion assay; this was supported by both the gene chip analysis and qRT-PCR studies showing negligible expression levels of both Mmp3 and Mmp13 transcripts in LM8 cells. Western blot analysis for Mmp14 expression revealed the membrane-anchored MMP was weakly expressed in Dunn, LM8, and transfectants with no differences. A recent study has shown that MMPs are not necessarily upregulated during tumor invasion [37]. Rather, a morphological transformation into an ameboid or rounded shape, driven by myosin light chain-Rho kinase-dependent matrix deformation without proteolysis, is responsible for tumor invasion through meshed matrices. In terms of adhesion affinity to ECM, knockdown of S100a4 resulted in no significant changes in adhesion to laminin, type I collagen or BSA, although there was a tendency for S100i clones to attach less onto fibronectin-coated plates. Taken together, our data suggest that the suppression of LM8 pulmonary metastatic potential by knocking down S100a4 was mainly due to attenuation of cell motility, yet the downregulation of adhesion to fibronectin may also play an important role.
In human OS, it was recently been reported that a large proportion of tumor cells expressed high cytoplasmic levels of S100A4 protein [14]. We examined S100A4 expression in human OS in greater detail and showed that there was some variation among different histological subtypes, with fibroblastic subtypes expressing lower S100A4 than the osteoblastic and chondroblastic OS. The variations in S100A4 expression level between OS histological subtypes are consistent with the fact that S100A4 is highly expressed in chondrocytes [38] and immature osteoblasts [39]. The clinical relevance of S100A4 in OS, however, could not be assessed in this study because of the lack of a reliable retrospective cohort of this rare tumor from a single institute in Japan. Nevertheless, our data suggest that the high percentage of S100A4 expression in primary OS samples might serve as a predictor of metastatic potential.
Taken together, we have shown that S100A4 protein is highly expressed in both mouse and human OS, and we provide functional evidence in a mouse metastasis model that it has a very important role in OS metastasis in vivo. While the underlying molecular mechanisms are still not fully defined, further investigation would pave the way for targeting therapy for OS metastasis development.
Abbreviations
- OS:
-
Osteosarcoma
- shRNA:
-
Short hairpin RNA
- ECM:
-
Extracellular matrix
- MMP:
-
Metalloproteinase
- TMA:
-
Tissue microarray
- MHC:
-
Myosin heavy chain
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- IHC:
-
Immunohistochemistry
References
Kempf-Bielack B, Bielack SS, Jurgens H, Branscheid D, Berdel WE, Exner GU, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS). J Clin Oncol. 2005;23:559–68.
Asai T, Ueda T, Itoh K, Yoshioka K, Aoki Y, Mori S, et al. Establishment and characterization of a murine osteosarcoma cell line (LM8) with high metastatic potential to the lung. Int J Cancer. 1998;76:418–22.
Kashima T, Nakamura K, Kawaguchi J, Takanashi M, Ishida T, Aburatani H, et al. Overexpression of cadherins suppresses pulmonary metastasis of osteosarcoma in vivo. Int J Cancer. 2003;104:147–54.
Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–14.
Boye K, Maelandsmo GM. S100A4 and metastasis: a small actor playing many roles. Am J Pathol. 2010;176:528–35.
Davies BR, Barraclough R, Davies MP, Rudland PS. Production of the metastatic phenotype by DNA transfection in a rat mammary model. Cell Biol Int. 1993;17:871–9.
Davies MP, Rudland PS, Robertson L, Parry EW, Jolicoeur P, Barraclough R. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–7.
Ambartsumian NS, Grigorian MS, Larsen IF, Karlstrom O, Sidenius N, Rygaard J, et al. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–30.
Maelandsmo GM, Hovig E, Skrede M, Engebraaten O, Florenes VA, Myklebost O, et al. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res. 1996;56:5490–8.
Grigorian MS, Tulchinsky EM, Zain S, Ebralidze AK, Kramerov DA, Kriajevska MV, et al. The mts1 gene and control of tumor metastasis. Gene. 1993;135:229–38.
Ismail TM, Zhang S, Fernig DG, Gross S, Martin-Fernandez ML, See V, et al. Self-association of calcium-binding protein S100A4 and metastasis. J Biol Chem. 2010;285:914–22.
Ito Y, Yoshida H, Tomoda C, Uruno T, Miya A, Kobayashi K, et al. S100A4 expression is an early event of papillary carcinoma of the thyroid. Oncology. 2004;67:397–402.
Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–80.
Ma X, Yang Y, Wang Y, An G, Lv G. Small interfering RNA-directed knockdown of S100A4 decreases proliferation and invasiveness of osteosarcoma cells. Cancer Lett. 2010;28;299(2):171–81
Shi Y, Zou M, Collison K, Baitei EY, Al-Makhalafi Z, Farid NR, et al. Ribonucleic acid interference targeting S100A4 (Mts1) suppresses tumor growth and metastasis of anaplastic thyroid carcinoma in a mouse model. J Clin Endocrinol Metab. 2006;91:2373–9.
Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–6.
Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–40.
West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–88.
Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10.
Cabezon T, Celis JE, Skibshoj I, Klingelhofer J, Grigorian M, Gromov P, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121:1433–44.
Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–73.
Kwak JM, Lee HJ, Kim SH, Kim HK, Mok YJ, Park YT, et al. Expression of protein S100A4 is a predictor of recurrence in colorectal cancer. World J Gastroenterol. 2010;16:3897–904.
Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–93.
Zreiqat H, Howlett CR, Gronthos S, Hume D, Geczy CL. S100A8/S100A9 and their association with cartilage and bone. J Mol Histol. 2007;38:381–91.
Leonard P, Sharp T, Henderson S, Hewitt D, Pringle J, Sandison A, et al. Gene expression array profile of human osteosarcoma. Br J Cancer. 2003;89:2284–8.
Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, et al. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–22.
Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, et al. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530–9.
Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121:11–8.
Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–76.
Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, et al. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51.
Salas S, Bartoli C, Deville JL, Gaudart J, Fina F, Calisti A, et al. Ezrin and alpha-smooth muscle actin are immunohistochemical prognostic markers in conventional osteosarcomas. Virchows Arch. 2007;451:999–1007.
Khanna C, Khan J, Nguyen P, Prehn J, Caylor J, Yeung C, et al. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res. 2001;61:3750–9.
Fukaya Y, Ishiguro N, Senga T, Ichigotani Y, Sohara Y, Tsutsui M, et al. A role for PI3K-Akt signaling in pulmonary metastatic nodule formation of the osteosarcoma cell line, LM8. Oncol Rep. 2005;14:847–52.
Ren L, Hong SH, Cassavaugh J, Osborne T, Chou AJ, Kim SY, et al. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene. 2009;28:792–802.
Bjornland K, Winberg JO, Odegaard OT, Hovig E, Loennechen T, Aasen AO, et al. S100A4 involvement in metastasis: deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res. 1999;59:4702–8.
Mathisen B, Lindstad RI, Hansen J, El-Gewely SA, Maelandsmo GM, Hovig E, et al. S100A4 regulates membrane induced activation of matrix metalloproteinase-2 in osteosarcoma cells. Clin Exp Metastasis. 2003;20:701–11.
Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–23.
Yammani RR, Carlson CS, Bresnick AR, Loeser RF. Increase in production of matrix metalloproteinase 13 by human articular chondrocytes due to stimulation with S100A4: Role of the receptor for advanced glycation end products. Arthritis Rheum. 2006;54:2901–11.
Duarte WR, Shibata T, Takenaga K, Takahashi E, Kubota K, Ohya K, et al. S100A4: a novel negative regulator of mineralization and osteoblast differentiation. J Bone Miner Res. 2003;18:493–501.
Acknowledgements
The authors would like to thank Dr. Felsenfeld and Dr. Hackett for providing chicken HS4 insulator cores and pCMV-SB, respectively, and Mrs. Ogiwara, Ms. Yamamura, and Ms. Meguro for their technical assistance.
The work described in this report was funded by a grant (no. 16790202) from the Ministry of Education, Culture, Sports, Science & Technology to TGK and a grant from the UK Bone Cancer Research Trust .
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 75 kb)
Table S1
Primers sets used in this study (DOC 81 kb)
Table S2
List of the highest ten genes preferentially expressed in LM8 than in Dunn by oligonucleotide arrays (DOC 137 kb)
Table S3
MMP family expression profiling by oligonucleotide arrays (DOC 143 kb)
Fig. S1
The map of pInSB(R) (JPEG 449 kb)
Fig. S2
The map of pInSB(L) (JPEG 538 kb)
Fig. S3
Construction of pInSB-Neo-shS100a4 (JPEG 489 kb)
Rights and permissions
About this article
Cite this article
Fujiwara, M., Kashima, T.G., Kunita, A. et al. Stable knockdown of S100A4 suppresses cell migration and metastasis of osteosarcoma. Tumor Biol. 32, 611–622 (2011). https://doi.org/10.1007/s13277-011-0160-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-011-0160-y