Abstract
Osteosarcoma (OS) is the most frequent pediatric malignant bone tumor that has a high propensity for metastases. Through osteoblast-specific alteration of p53 status, we developed a genetically engineered mouse model of localized and metastatic OS to gain an understanding into the molecular pathogenesis of OS. Microarray analysis of both localized tumors and metastatic tumors identified the downregulation of the naked cuticle homolog 2 (NKD2) gene, a negative regulator of Wnt signaling. Overexpression of NKD2 in metastatic human and mouse OS cells significantly decreases cell proliferation, migration and invasion ability in vitro and drastically diminishes OS tumor growth and metastasis in vivo, whereas downregulation enhances migratory and invasive potential. Evaluation of NKD2-overexpressing tumors revealed upregulation of tumor-suppressor genes and downregulation of molecules involved in blood vessel formation and cell migration. Furthermore, assessment of primary human OS revealed downregulation of NKD2 in metastatic and recurrent OS. Finally, we provide biological evidence that use of small-molecule inhibitors targeting the Wnt pathway can have therapeutic efficacy in decreasing metastatic properties in OS. Our studies provide compelling evidence that downregulation of NKD2 expression and alterations in associated regulated pathways have a significant role in driving OS tumor growth and metastasis.
Similar content being viewed by others
Introduction
Osteosarcoma (OS) is the most frequent malignant bone tumor in children and adolescents and has a relatively high propensity for metastases1, 2 to distal sites such as the lungs, which is associated with extremely poor long-term survival rates of 10–30%.3, 4, 5 Thus identification of genes and molecular signaling pathways underlying OS metastasis may provide novel insights and therapeutic targets.
In both familial OS and sporadic OS, the TP53 gene is frequently mutated, including somatic structural variants or single-nucleotide variants and insertions/deletions as well as germline single-nucleotide variants/insertions/deletions in >90% of OS cases.6 In addition, human tumors frequently exhibit p53 missense mutations, such as the ‘hotspot’ R175H, which accounts for approximately 6% of the identified missense mutations7 and 4% of TP53 missense mutations observed in human OS.8 These missense mutations confer both dominant-negative and gain-of-function activities beyond mere loss of tumor-suppressor function.9
TP53 heterozygous knockout mice develop a high percentage of non-metastatic OS,10, 11 whereas mice with a germline R172H mutation (corresponding to the human R175H mutation) develop tumors with high metastatic potential.12 With these insights, we have established two osteosarcoma-susceptible lines of genetically engineered mice (GEMM) utilizing an osteoblast- specific Cre allele crossed with floxed p53 or LSL-p53 R172H alleles to generate localized or metastatic OS. Because of the tremendous propensity for p53 alterations in OS, and the presence of hotspot mutations, we believe that these GEMMs most authentically represent human OS development and progression. Subsequently, we analyzed global differential gene expression between localized primary and highly metastatic tumor tissues.
Our analysis of metastatic tumors revealed alterations in the expression for components of the Wnt signaling pathway, specifically we identified the downregulation of naked cuticle homolog 2 (NKD2) in metastatic OS, which is involved in the degradation of Dvl proteins and downstream regulation of β-catenin. We examined NKD2 expression in localized OS, primary OS that had associated metastases (referred now as primary metastatic OS) and pulmonary metastatic OS tumors. Through both in vitro and in vivo studies, we have identified and characterized NKD2 as a novel suppressor of OS tumor growth and metastasis in both mouse and human OS. Re-expression of NKD2 in metastatic OS cells correlated with downregulation of signaling pathways that drive cell motility, angiogenesis and growth signaling. Finally, we demonstrate that the use of small-molecule Wnt inhibitors decreases OS proliferation and invasive potential. Overall, our data identify NKD2 as a novel regulator of OS metastasis and suggest that implementation of Wnt signaling inhibitors for clinical use may represent a novel treatment strategy.
Results
Generation and characterization of mouse non-metastatic and metastatic osteosarcoma models
For our non-metastatic OS model, we crossed Col2.3-Cre transgenic mice with osteoblast-specific Cre expression in mice with a germline p53 allele flanked by LoxP recombination sites (‘p53 floxed’)[18,19] (Figure 1a). Progeny of this cross with one Col2.3-Cre allele and one or two floxed p53 alleles exhibit deletion of one or both wild-type p53 genes, respectively, in bone tissues. For our metastatic model, we crossed the Col2.3-Cre mice to mice with a germline R172H p53 allele preceded by a Lox-Stop-Lox (LSL) cassette in the p53 promoter region (LSL-p53R172H).13 In the bi-allelic Col2.3:LSL-p53R172H progeny, activation of the mutant p53 allele occurs only in osteoblasts (Figure 1a). Four distinct Col2.3-Cre transgenic genotypes were then monitored over 2 years for tumor incidence: F/+ (floxed p53 allele/wt p53 allele), F/F (two floxed p53 alleles), R/+ (LSL-p53R172H allele/wt p53 allele), and R/F (LSL-p53R172H allele/floxed p53 allele). Mice homozygous for the LSL-p53R172H (R/R) were rarely observed, possibly due to embryonic selection against this genotype.
Design and characterization of a novel genetically engineered mouse model of metastatic osteosarcoma. (a) Schematic representing osteosarcoma-susceptible mice. Top panel shows the Col2.3-Cre; floxed p53 mice that delete one or both p53 alleles in osteoblasts, while the bottom panel shows the Col2.3-Cre; LSL-p53R172H mice that express a gain-of-function mutant p53. (b) Kaplan–Meier plots for each of four different genotypes of mice. ‘F’=floxed p53, ‘R’=LSL-p53R172H and ‘+’=WTp53 allele. F/+ (N=77); F/F (N=55); R/+ (N=38); R/F (N=28). (c) Frequency (%) of metastatic osteosarcomas varies according to p53 genotype with statistical analyses (as described in Materials and methods). (d) Anatomical distribution of primary tumor location for osteosarcomas of all p53 genotypes. (e) Tumor histopathology demonstrating primary osteoblastic osteosarcoma (upper left panel) and distal lung metastatic lesions (upper right panel). Spectral karyotype of a primary osteosarcoma (lower panel).
Comparison of osteosarcoma incidences in the various Cre/p53 allele genotypes showed that Cre+F/+ mice exhibited a median onset of OS at 49 weeks of age compared with a median onset of 37 weeks of age for Cre+F/F mice (Figure 1b). Kaplan–Meier analyses showed the differences between these two groups to be highly significant (P<0.0001). Similarly, Cre+R/+ mice had a median osteosarcoma incidence of 52 weeks compared with Cre+R/F mice with a median incidence of 40 weeks (P=0.05; Figure 1b). At the time of necroscopy, a thorough pathological analysis of all the major organs was performed in search of metastatic lesions. Lungs were the most frequent site of metastatic lesion. We observed that 31% of Cre+F/+ mice exhibited metastatic lesions compared with 55% for Cre+R/+ mice (Figure 1c). Interestingly Cre+F/F mice also showed enhanced metastatic potential (54%) compared with Cre+F/+ mice. Not surprisingly, the Cre+R/F mice also had high rates of metastasis (57%).
The sites of osteosarcoma development were variable, with the spine showing the highest frequency of lesions (48%), followed by the forelimbs and hindlimbs (31%) (Figure 1d). Other less frequent sites included the ribs (12%), pelvis (7%) and jaw/cranium (2%). The sites of OS development were not affected by p53 genotype. Examination of primary and metastatic tumors by hematoxylin and eosin (H&E) staining revealed highly cellular tumors consisting of basophilic malignant cells, with evidence of osteoid production consistent with osteoblastic osteosarcoma (Figure 1e, upper panels). Furthermore, we performed spectral karyotyping of several of the osteosarcomas derived from these mice that consistently revealed high levels of genomic instability (Figure 1e), with complex karyotypes displaying chromosomal rearrangements, similar to that observed in human OS.
NKD2 expression is downregulated in metastatic tumors and cell lines
To identify genes and pathways that regulate OS metastasis, we performed gene expression microarray analysis comparing six non-metastatic OS with those of 12 primary metastatic OS tumors. In addition, seven lung metastatic lesions gene expression profiles were compared with those of the primary tumor from which they originated. Differentially expressed genes were selected for further analysis when expression differences were greater than twofold and P-values were <0.05, as determined by two-tailed t-test. Gene ontology analysis demonstrated significant alterations between the non-metastatic and primary metastatic OS bone tumors. Table 1 highlights some categories and representative genes differentially expressed.
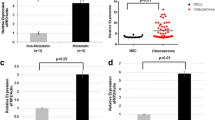
Besides changes in expression in ribosomal/translational regulating genes, extracellular matrix (ECM) proteins and inflammatory-related genes, we also noted that among the most differentially regulated genes were those that regulate Wnt signaling (Table 1). We determined that both human and mouse metastatic OS cells have enhanced Wnt signaling activity compared with local OS, as assessed by TOPFlash/FOPFlash Wnt reporter assay (Figure 2a). To validate the microarray results, we performed real-time reverse transcriptase PCR (RT–PCR) on RNA from an additional nine localized and primary metastatic tumors. In particular, RT–PCR results showed that Nkd2, a known inhibitor of Wnt signaling,14 to be one of the most significantly downregulated genes in metastases relative to primary tumors (Figure 2b). In addition, two other Wnt inhibitory genes associated with inhibition of canonical Wnt signaling, Wnt5a and Apcdd1, were also downregulated in mouse OS tumors associated with metastases (Figure 2b). To further corroborate the relevance of our mouse model, recent transcriptome analysis of human OS cell lines with varying phenotypic properties reported both NKD2 and APCDD1 as being significantly downregulated in highly tumorigenic (NKD2) or in highly invasive and proliferative (APCDD1) OS cell lines.15 Our studies focus on delineating the functional role of NKD2 in OS tumor development and metastasis, which has not previously been implicated in OS.
Enhanced Wnt signaling activity and decreased NKD2 expression in metastatic OS tumors and cell lines. (a) TOPFlash/FOPFlash assay of low and high metastatic human sublines HOS and 143B as well as murine local (n=3) and metastatic (n=4) OS cell lines. (b) Quantitative PCR showing relative expression of endogenous Wnt inhibitors Nkd2, Wnt5a and Apcdd1 from localized (LT; n=9) and primary metastatic (PM; n=9) mouse tumors. (c) Relative Nkd2 expression in paired mouse primary (PM) and distal lung (ML) metastatic tumors. (d) Western blotting analysis of NKD2 protein expression in murine local, primary metastatic (PM) OS and distal lung (ML) cell lines. Actin used as a loading control. (e) Relative expression of NKD2 in local (LT), primary metastatic (PM) and recurrent human osteosarcoma tumors. (f) Kaplan scan for NKD2 derived from clinically annotated human osteosarcoma gene expression database (Kuijjer: mixed osteosarcoma http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) High expression (blue line) and low expression (red line).
We further examined the expression of Nkd2 RNA by real-time RT–PCR in three pairs of primary metastatic tumors and their metastatic lung lesions and again found that Nkd2 expression is decreased in pulmonary metastases compared with primary tumors (Figure 2c). Analysis of NKD2 protein expression in OS cell lines developed from the GEMM showed that NKD2 was downregulated in both primary metastatic cell lines and pulmonary metastatic cell lines when compared with non-metastatic cell lines (Figure 2d).
Furthermore, we examined the relative expression of NKD2 in primary human tumor specimens consisting of 10 non-metastatic, 7 primary metastatic and 7 recurrent lesions. As shown in Figure 2e, our analysis revealed that NKD2 expression is generally downregulated in metastatic tumors. When aggregated with NKD2 expression from patients with recurrent metastatic disease, the downregulation of NKD2 was statistically significant (P=0.02). Finally, a Kaplan scan plot of the R2 genomic analysis and visualization platform (Kuijjer: mixed osteosarcoma; http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) osteosarcoma database revealed that downregulation of NKD2 in human OS tumors is associated with a significantly worse overall survival (Figure 2f). Taken together, these results indicate that decreased expression of NKD2 is associated with highly aggressive OS states.
NKD2 overexpression suppresses cell proliferation, migration and invasion in vitro
To study the effects of NKD2 on OS cell behavior, we expressed NKD2 in a highly metastatic mouse OS cell line, RF379L (designated Nkd2/RF379L), derived from a mouse lung OS metastatic lesion (Figure 3a). To examine the modulation of canonical Wnt activity by NKD2, TOPFlash reporter activity was reduced by ~50% in Nkd2/379L cells compared with control cells (Supplementary Figure S1a). In addition, Nkd2/RF379L cells showed decreased expression of Wnt target genes, including Mmp9 and Ctgf (Supplementary Figure S1b), and decreased proliferation rates compared with vector control RF379L cells (Figure 3b). Cell cycle analysis revealed significant changes upon NKD2 overexpression, with a 10% increase in G0/G1 phase cell populations (P<0.01; data not shown) compared with control cells.
Ectopic NKD2 expression inhibits cell growth, migration and invasion, and inhibits primary tumor growth and pulmonary metastasis in orthotopic OS mouse model. (a) Western blot of NKD2 expression in RF379L cells overexpressing NKD2. Actin is used as a loading control. The bar graph on the right shows the corresponding mRNA levels. (b) Colorimetric in vitro proliferation assay of Nkd2/RF379L and control cells. Duplicates performed±s.e. (c) Transwell cell invasion assay for control and Nkd2/RF379L cells. Crystal violet staining for control and Nkd2-expressing cells are shown. (d) Images of scratch wounds at 0 and 18 h. Gap measurements were quantified for Nkd2/RF379L and control cells. Average of three independent experiments±s.e. is shown in the bar graph. (e) 2.5 × 105 Nkd2/RF379L cells or empty vector cells were injected intratibially into nude mice (n=5/group). Two-dimensional tumor measurements were determined, and volumes were calculated twice a week (left panel). Control tumors were harvested at Days 34 and 40 for the NKD2 group, and tumor weights were measured (right panel). Error bars were generated based upon analysis of five mice in each group. (f) Gross lung pathology of mice from Nkd2/RF379L or control empty vector. The green arrows indicate macroscopic pulmonary metastatic lesions (left panel). H&E-stained lung sections from the control (middle upper) and NKD2-expressing groups (middle lower). Red arrows highlight evident microscopic lesions. Bar graph (right panel) quantifying the average number of pulmonary nodules in each group. Error bars were generated from five mice in each group.
We next performed a transwell invasion and scratch assays to analyze the role of NKD2 on OS invasive and migratory potential. As shown, Nkd2/RF379L cells exhibited greater than 80% decreased invasive potential compared to controls (in Figure 3c), and less migratory ability when compared with vector-transfected control cells (Figure 3d).
NKD2 overexpression inhibits primary tumor growth and pulmonary metastasis in vivo
To examine NKD2 in vivo effects, control or Nkd2/RF379L cells were orthotopically injected and tumor growth evaluated. Figure 3e (left panel) shows that all the mice injected with Nkd2/RF379L cells exhibited a slower rate of tumor growth compared with control cells. At the time of killing, the average weight of Nkd2/RF379L tumors was ~90% less than that of vector control tumors (P<0.01) (Figure 3e, right panel).
We next wanted to evaluate the effects of NKD2 on tumor metastasis. As shown in Figure 3f (left panel), all the mice in the control group had gross evidence of pulmonary metastatic lesions, whereas no lung nodules were visible in the NKD2 group. Lungs were sectioned and stained with H&E (Figure 3f, middle panel). In the control mice, large metastases in the lungs of all five animals was detected, while the Nkd2/RF379L animals were free of overt metastatic lesions. The average number of pulmonary nodules on H&E-stained slides in the control group was 9.2, whereas that in the NKD2-overexpression group was 0.2 (Figure 3f, right panel). To reduce the potential effects of tumor size on metastasis, we performed intratibial injections with four times the number of Nkd2/RF379L cells versus control cells and harvested the tumors when approximately the same primary tumor size (Supplementary Figure S2a). Even then, mice bearing tumors with Nkd2 overexpression exhibited decreased tumor growth rates and significantly fewer lung metastases compared with the control mice (Supplementary Figures S2b and c). To determine whether overexpression of NKD2 in a human OS line has similar cellular phenotypes, we overexpressed NKD2 in the highly metastatic human OS cell line, 143B (NKD2/143B).16, 17
After establishment of stable 143B NKD2-overexpressing cells (Supplementary Figure S1c, right panel), we assessed proliferation and invasiveness properties in vitro and found that overexpressed NKD2 inhibits all of these behaviors compared with empty vector-transfected 143B cells (Figures 4a and b, respectively). Next, we intratibially injected human metastatic vector control 143B or NKD2/143B cells into nude mice. As shown in Figure 4c, stable NKD2/143B cells exhibit a significant suppression of tumor growth compared with 143B control cells. In addition, we determined that NKD2/143B developed fewer pulmonary lesions compared with the control group (Figure 4d).
NKD2 inhibits cell growth, invasion and metastatic potential of human metastatic osteosarcoma in vitro and in vivo. (a) In vitro proliferation assay of NKD2/143B versus control vector cells. (b) Transwell assay for 143B control and NKD2-overexpressing cells. Crystal violet staining images (left panels) and quantification of the relative cell invasion (right) are shown. (c) 0.5 × 105 NKD2/143B or an empty vector cells were injected intratibially into nude mice. Measured tumor weights after killing are shown in the bar graph. Error bars are generated from five mouse tumors. (d) Images of lungs from the control (upper left panel) or NKD2 group (lower left panel). Green arrows highlight metastatic disease. H&E images (middle panels) and quantification (right panel) of metastatic lesions within lung sections (lower panels). Error bars generated from five animals in each group.
NKD2 upregulates ECM proteins and Dickkopf-related protein 3 (DKK3) expression and downregulates expression of cell motility and angiogenesis factors
To explore the molecular pathways that NKD2 regulates to restrict tumor growth and metastasis, we used microarray analysis to compare gene expression patterns in the intratibially transplanted RF379L control tumors and Nkd2/RF379L tumors. Table 2 highlights some of the differential curated gene sets and ontology categories (using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) and Gene Set Enrichment Analysis (http://www.broad.mit.edu/gsea/msigdb/annotate.jsp), which includes upregulation of ECM and focal adhesion genes and downregulation of genes involved in cell proliferation and motility in NKD2-expressing tumors. We also noted that the highly metastatic control tumors had alterations in genes associated with enhanced chemotherapy resistance and stem cell properties.
Supplementary Table S1 lists specific genes differentially expressed between the two sets of tumors. Interestingly, Fbln1, an ECM component, was downregulated in the primary metastatic OS tumors (Table 1), and this reduced expression was subsequently confirmed by RT–PCR (Supplementary Figure S3a). Several genes involved in cell growth, motility and tumor angiogenesis were found to be downregulated upon NKD2 overexpression (Supplementary Table S1, Supplementary Figure S3b). DKK3 is a specific inhibitor of canonical Wnt signaling whose tumor-suppressive activity is reduced in several human cancers.18, 19, 20, 21 In addition, it has previously been reported to inhibit invasion and motility of human OS cells by modulating the Wnt-ß-catenin pathway.22 In our microarray studies, we found that Dkk3 expression was increased approximately 14-fold in the Nkd2-overexpressing tumor group (P=0.002). Upregulation of Dkk3 by NKD2 was confirmed by RT–PCR (Supplementary Figure S3a). Interestingly, transient knockdown of DKK3 (Supplementary Figure S3c) in the NKD2-overexpressing OS cells caused an increase in invasive potential (Supplementary Figure S3d), suggesting that DKK3 is a critical mediator of NKD2-driven suppression of metastasis. However, the precise mechanism by which NKD2 regulates DKK3 is unknown and warrants further investigations.
NKD2 downregulation in osteosarcoma cells enhances cell proliferation and migration in vitro
To investigate the effects of reducing NKD2 levels in non-metastatic OS cells, we generated stable knockdown of NKD2 by shRNA in two independent non-metastatic mouse OS cell lines, RF43 (Figure 5a left panel) and RF575 (Figure 5a, right panel). This led to enhanced proliferation (Figure 5b), migratory and invasive potential (Figures 5c and d). These results corroborate the NKD2 overexpression effects and signify the importance of NKD2 expression in osteosarcoma development and progression.
NKD2 depletion in non-metastatic OS cell lines enhances cell proliferation, migration and invasion. (a) Quantitative PCR showing relative NKD2 expression in non-metastatic murine osteosarcoma cell lines transfected with control or shNkd2 plasmids, RF43 (left panel) and RF575 (right panel). Western blotting of NKD2 knockdown in RF43 is shown. (b) Colorimetric in vitro proliferation assay of NKD2-depleted non-metastatic OS cell lines, RF43 (left panel) and RF575 (right panel) and control cells. Duplicate wells for each condition and ±s.e. are shown. (c) Scratch assay assessing migratory ability for NKD2 knockdown osteosarcoma cells compared with control non-metastatic mouse OS cells in RF43 cells. (d) Transwell cell invasion assay of control and knockdown NKD2 OS cells. Crystal violet staining of control and NKD2 knockdown cells. Below is the relative quantification of invasive potential.
Wnt signaling inhibition leads to altered tumor proliferation and invasive potential
We utilized a highly specific Porcupine enzyme inhibitor, Wnt-C59, which prevents the normal maturation of the Wnt ligands and function of the signaling pathway, to investigate targeting Wnt activity in metastatic OS. As shown in Figure 6a, treatment of multiple metastatic OS cell lines (lower three panels) with Wnt-C59 reduces tumor cell proliferation and invasive potential (Figure 6b). Interestingly, treatment of non-metastatic mouse OS cell lines showed decreased sensitivity to Wnt-C59 than did the metastatic cells (Figure 6a, top panel). Similar results were seen for the treatment of human OS cells with Wnt-C59 (Supplementary Figure S4a). These functional alterations are associated with a concomitant decrease in Wnt signaling activity as determined by TOPFlash/FOPFlash (Figure 6c). Finally, we tested another small molecule Wnt inhibitor (IWR-1, Sigma-Aldrich), which antagonizes Wnt signaling via stabilization of Axin. This also led to a decrease in proliferation of mouse metastatic OS cells in vitro (Supplementary Figure S4b). Finally, we provide evidence that the Wnt-antagonists do not significantly inhibit invasive potential of low Wnt activity, non-metastatic OS cells (Supplementary Figure S4c). Overall, these results suggest the potential use of small-molecule Wnt inhibitors as novel therapeutic options in the future treatment of metastatic OS with dysregulated Wnt signaling.
Small-molecule targeting of the Wnt signaling pathway preferentially decreases metastatic OS proliferation and invasive properties. (a) In vitro proliferation assay in the presence of Porcupine inhibitor, Wnt-C59 for non-metastatic (top panel) and metastatic OS cells (bottom three panels). (b) Transwell assay for metastatic mouse OS cell lines in the presence and absence of Wnt-C59. Quantification of invasive potential for OS cells treated with and without Wnt-C59. (c) TOPFlash/FOPFlash Wnt reporter assay in the presence and absence of Wnt-C59. *P<0.05; **P<0.01.
Discussion
There are very few studies using immunocompetent model systems to reliably investigate the metastatic mechanisms in mesenchymal malignancies, such as sarcomas.23, 24 We believe recent whole-genome sequencing analysis of human OS tumors provides strong credence to our model in which we specifically alter p53 status in osteoblast cells to devise a mouse model system that is truly representative of the genomic and biological phenotypes seen in the human disease. Furthermore, we have engineered a model with a high incidence of developing macroscopic metastatic disease that allows for comprehensive molecular and biological studies. We have been able to identify several unknown gene expression alterations in metastatic OS that we corroborated in the human disease.
The Naked cuticle (Nkd) family of proteins has been associated with the regulation of the Wnt and transforming growth factor (TGF)-α pathways.25, 26, 27 The NKD family (including NKD1 and NKD2) were predicted to be candidate tumor suppressors28 and hypermethylated in astrocytic gliomas.29 Analysis of NKD1 expression in metastatic mouse and human OS tumors did not reveal any significant differences in expression compared with localized tumors (Supplementary Figures S5a and b). Therefore, we have focused on the identification of NKD2 as a novel gene regulating OS tumor proliferation and metastasis.
Although little is known about the role of NKD2 in OS, it should be noted that this gene was recently reported to be significantly downregulated in human OS cell lines with higher propensity for tumorigenicity.15 In addition, we provide evidence that low expression of NKD2 is associated with a decreased overall survival. Thus this only further corroborates our GEMM data, and our novel functional investigations into the regulation and role of NKD2 in OS development and metastasis demonstrate the need to study NKD2-dependent mechanisms. In addition, the Wnt signaling pathway itself has been implicated in cancer metastasis.30 Besides an important role in bone differentiation and proliferation,31, 32 the Wnt/β-catenin signaling pathway has recently been identified as a potential target in both human and mouse osteosarcoma models. The current evidence that Wnt signaling is directly involved in driving OS metastatic potential remains relatively fragmentary and inconclusive, with very few functional studies providing definitive evidence.26, 33
Using overexpression and silencing experiments, we showed that NKD2 inhibits OS cell proliferation and invasiveness in vitro. Although we recognize that the re-expression of NKD2 may not be fully physiological, we believe that these studies do significantly demonstrate a biological phenotype regulated by NKD2. Besides the use of multiple cell line studies, the reciprocal, complementary knockdown (shNKD2) experiments performed confirm the biological importance and role of NKD2 in helping regulate osteosarcoma metastasis. In addition, we have assessed the subcellular localization of endogenous and overexpressed NKD2 in our metastatic cell lines (Supplementary Figure S1c; left and middle panels). Using immunofluorescence, we noted higher endogenous expression in the low metastatic HOS cell line34, 35, 36, 37 compared with the highly metastatic 143B cell line. Furthermore, upon overexpression of FLAG-tagged NKD2 we noted increased membrane-associated NKD2 expression (Supplementary Figure S1c; right panel) and overall levels more comparable to the low metastatic HOS cells.
In vivo overexpression of NKD2 in mouse and human OS cells dramatically inhibited both tumor growth and metastatic potential. When we attempted to silence NKD2 expression in non-metastatic OS cells in xenograft models, we obtained only modest enhancement of tumorigenic and metastatic potential (data not shown). This suggests that decreased levels of NKD2 may be necessary but not sufficient, requiring downregulation of other Wnt signaling inhibitors for full metastatic potential. Consistent with this, our microarray comparisons of non-metastatic and metastatic OS reveal high levels of other Wnt inhibitory molecules (for example, Apcdd1, Wnt5a) in the non-metastatic tumors.
Furthermore, direct analysis of downstream Wnt signaling through activation of Wnt-responsive promoters and Wnt-target genes demonstrated increased Wnt activity in metastatic tumors. Finally, use of newly available and highly specific Wnt small-molecule inhibitors showed significant reductions in metastatic OS cell proliferation and invasiveness. The mechanisms by which Wnt signaling enhances metastasis remain unclear, though the genetic comparison of tumors with and without overexpressed NKD2 provide some valuable insights, including alterations in cell adhesion genes, cell motility, angiogenesis and ECM genes, such as FBLN1. FBLN1 has been associated with several human cancers, including hepatocellular carcinoma and prostate cancer.38, 39, 40
Besides demonstrating its role in the regulation of the Wnt signaling cascade, we also briefly investigated the effects of NKD2 expression levels on the TGF-α pathway by studying downstream effectors of TGF-α-EGFR (epidermal growth factor receptor) signaling. Specifically, we assessed for changes in the activation of the extracellular signal–regulated kinase (ERK)/mitogen-activated protein kinase and phosphatidylinositol 3′-kinase-Akt pathways. Our studies did not reveal any changes in Akt activation (data not shown); however, interestingly it did reveal that NKD2 expression levels altered activation of ERK, via changes in phospho-ERK. Although knockdown of NKD2 mildly enhanced ERK activity (Supplementary Figure S1d, RF43 data), overexpression of NKD2 in human and mouse OS cell lines caused a decrease in ERK activation, (Supplementary Figure S1d, 143B and 379L data). Whether the ERK signaling changes are direct or indirect effects of NKD2 in osteosarcoma cells still needs to be investigated. In epithelial cells, NKD2 has been reported to enhance TGF-α transportation to the basolateral membrane of polarized cells;41, 42 however, the exact influence of NKD2 on TGF-α-mediated signaling in mesenchymal cells has not been investigated. Further studies will address the mechanisms by which NKD2 regulates both TGF-α and Wnt signaling and their contributions towards understanding OS tumor development and metastasis. These results insinuate that inhibition of both Wnt and ERK activation through dual combination therapy could have additional therapeutic benefit for treating human OS.
Here we provide strong evidence for the role of NKD2 downregulation, in concert with enhanced Wnt signaling, as key events in driving metastatic potential. Further exploration of NKD2-regulated pathways for OS tumor growth and metastasis may be helpful to identify biomarkers and therapeutic targets for NKD2-deficient OS.
Materials and methods
Mice
Col2.3-Cre mice contain a rat Col1a1 2.3-kb rat promoter driving a Cre transgene in osteoblast lineages.43 These mice were backcrossed into a C57BL/6 background and further crossed to either floxed p53 mice or LSL-p53R172H mice. All research with mice was conducted in compliance with the Baylor Animal Protocol Committee (Baylor College of Medicine Animal Protocol AN336) and AAALAC recommendations as published in The Guide for the Care and Use of Laboratory Animals (NRC1996). All animals had comprehensive necroscopies upon time of euthanasia with complete dissection of tumor as well as evaluation of all major organs, with special attention given to the lungs and liver and other bones. Portions of all tumor specimens as well as all that of the lungs and liver were sent to pathology for histological analysis as well as evaluation of distal organs for evidence of metastatic disease.
High molecular weight genomic DNA was prepared from 5 mm tail tips or 50 mg tissue and prepared as previously described.44 All PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). The presence of the Col2.3-Cre transgene was verified using the primers Cre-F: 5′-TTACTGACCGTACACCAAAATTTGCCTGC-3′ and Cre-R: 5′-CCTGGCAGCGATCGCTATTTTCCATGAGTG-3′, which produce a 450-bp product following PCR. Genotyping for the floxed p53 allele was performed using the primers FLOX-F:5′-CACAAAAAACAGGTTAAACCCAG-3′ and FLOX-R: 5′-GCTTGTCCCAACTCTAAGAAGC-3′. Using these primers, wild-type p53 alleles produce a 431-bp fragment, while the floxed p53 allele produces a 584-bp fragment following PCR. Successful excision of exons 2–10 by Cre recombinase was verified with PCR using primers Int1FOR (5′-CACAAAAAACAGGTTAAACCCAG) and Int10REV (5′-GAAGACAGAAAAGGGGAGGG-3′), which produce a 612-bp fragment with floxed p53 excision. Finally, the presence of the LSL-p53R172H allele was verified using the primers LSL-F (5′-CCATGGCTTGAGTAAGTCTGC-3′) and LSL-R (5′-CTTGGAGACATAGCCACACTG-3′) that produces a 299-bp fragment. Recombination of the LSL allele was confirmed by using primers LSL-rec1 (5′-CTTGGAGACATAGCCACACTG-3′ and LSL-rec2 (5′-AGCCTGCCTAGCTTCCTCAGG-3′). Excision of the LSL cassette produces a 330-bp fragment.
Cell culture, reagents and plasmids
Metastatic mouse OS cell lines (RF379L, CR175L, RF229, RF1044, RF892, FC456 primary and FC456L) were isolated from either primary OS tumors and/or lung lesions in highly metastatic p53+/R172H or p53 null OS mouse models. Non-metastatic mouse OS cell lines (C2984, RF575, RF1026 and RF43) were isolated from p53+/− or singly floxed p53+/F-Col2.3 mice. All cell lines used for functional assays were extensively characterized for their migratory, invasive and metastatic potentials both in vitro and in vivo prior to genetic alteration of NKD2 status.
Cell lines 1066 and 3441 were established from lung metastasis obtained from Texas Children’s Cancer Center Research Tissue Support Service. The human osteosarcoma cell lines, HOS, 143B, SaOs2, LM7 and MNNG were purchased from ATCC (Manassas, VA, USA). Cells were maintained in complete Dulbecco’s modified Eagle’s media containing 10% fetal bovine serum (GIBCO, Life Technology, Grand Island, NY USA). The mouse NKD2 overexpression vector pCMV6-NKD2 was obtained from ORIGENE (Rockville, MD, USA). The shNKD2 vectors were purchased from the Baylor College of Medicine Genome Wide shRNA Screening Core. The canonical Wnt signaling inhibitors Wnt-C59 and IWR-1 were purchased from Cellagen Technology (San Diego, CA, USA) and Sigma-Aldrich (St Louis, MO, USA), respectively. For Wnt-C59 proliferation experiments, cells were treated with 1 μM of reagent on Day 0. For invasion assays, cells were pretreated with 1 μM 24 h prior to initiating assay. IWR-1 (10 μM) was added on Day 0 of proliferation studies.
Human osteosarcoma specimens
Primary local and primary metastatic bone tumor total RNA were provided by Dr Ching Lau. Recurrent bone and lung osteosarcoma tumors were provided by the UT MD Anderson Sarcoma Research Laboratory.
RNA isolation and real-time RT–PCR
Total RNA from cell lines and tumor tissues was isolated using the Trizol Reagents (Ambion, Life Technology), and cDNA templates were generated via RT–PCR using cDNA Synthesis Supermix (Quanta, Gaithersburg, MD, USA). The cDNA products were used for real-time PCR templates. Gene expression was monitored using real-time primer pairs with SYBR Green detection (Applied Biosystems, Life Technology) and analyzed using SteOnePlus real-time PCR machine (AB Biosystems, Life Technology). PCR conditions were as follows: 10 mins at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Relative fold change in mRNA expression compared with control was calculated using the ΔΔCt method. All samples were normalized to β-actin expression levels. Primer sequences are listed in Supplementary Table S2.
Microarray profiling
Briefly, RNA samples were isolated from localized tumors, primary tumors from metastatic mice and metastatic lung lesions for target preparation. Total RNA was amplified and purified using the Illumina TotalPrep RNA Amplification Kit (Ambion, Cat. no. IL1791) following the manufacturer’s instructions. An aliquot of 750 ng of amplified products was loaded onto Illumina Sentrix Beadchip Array Mouse Ref8.v2 arrays and hybridized at 58 °C in an Illumina Hybridization Oven (Illumina, San Diego, CA, USA; Cat. no. 198361) for 17 h, washed and incubated with straptavidin-Cy3 to detect biotin-labeled cRNA on the arrays. Arrays were dried and scanned with the BeadArray Reader (Illumina). Data were analyzed using the Genome Studio software (Illumina).
The microarray data were generated by the University of Texas Medical School Genomics Core Facility. Array data are available at the Gene Expression Omnibus (GEO, accession GSE43281).
Gene ontology analysis
Differentially expressed genes with expression differences greater than two-fold and P-values, as determined by two-tailed t test, <0.05 were subjected to gene ontology analysis by using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) and Gene Set Enrichment Analysis (http://www.broad.mit.edu/gsea/msigdb/annotate.jsp).
Western blotting
Cells lysates were fractionated by 4–12% Bis-Tris Gel (Novex, Life Technology) and transferred to polyvinylidene difluoride membrane. The membrane was then blocked by 5% nonfat milk in PBST (3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, 0.05% Tween 20, pH 7.4) for 60 min. The membranes were then incubated with the primary antibody. Membranes were washed three times for 5 min and incubated with a 1:1000 dilution of horseradish peroxidase-conjugated secondary antibodies for 60 min. Blots were washed with PBST three times and developed with the enhanced chemiluminescence system (GE Healthcare, Pittsburgh, PA, USA). The antibodies used include: anti-NKD2 antibody (Protein Tech Group, Chicago, IL, USA, Cat. no. 16699-1-AP), phospo-ERK1/2 (Cell Signaling, Danvers, MA, USA, Cat. no. 4370p), total ERK (Cell Signaling, Cat. no. 4695p), anti-beta actin (NovusBio, Littleton, CO, USA, Cat. no. 600-503), anti-Flag (Cell Signaling, Cat. no. 2368p).
In vitro cell proliferation assay
To assess cell proliferation, 1 × 103 cells/well were seeded in a 96-well plate in 100 μl cell culture medium. Cells were cultured in a 5% CO2 incubator at 37 °C. At the indicated time points, viable cells were counted using the Cell Counting Kit-8 (Dojindo Laboratories, Rockville, MD, USA) following the manufacturer’s protocol. The relative proliferation rates were determined by measuring the absorbance at 450 nm using a Perkin Elmer Victor2 microplate reader (Perkin Elmer, Waltham, MA, USA). Individual time points were run in triplicates. Each experiment was performed at least twice, with representative results shown.
Cell migration assay
Cell lines, both control and NKD2 altered, were cultured to confluence in six-well plates. Using a sterile 200-μl pipette tip, a scratch was drawn on the monolayer of the cells. The cells were then washed once with phosphate-buffered saline. The width of the scratch wounds was measured with the NIH Image J (NIH, Bethesda, MD, USA).45 The relative migration at an indicated time point was calculated by comparing the wound width to the original one at the initial time point. Assays were performed at least three times.
In vitro invasion assay
The in vitro invasion assay was performed using 24-well, 8-μm pore-sized invasion chamber system (Greiner Bio-one, Monroe, NC, USA) and 0.5 μg/ml collagen (Advanced Biomatrix, Carlsbad, CA, USA) monolayer. Prior to loading the cells onto the upper chamber, the lower chamber was filled with medium containing 10% fetal bovine serum. Cells were starved overnight and added to serum-free medium in the upper chamber at a concentration of 1 × 105 cells/well followed by incubation for 12–18 h in a humidified 5% CO2 atmosphere at 37 °C. After incubation, the cells on the upper surface of the well were removed completely by a Q-tip. The wells were fixed in methanol and stained with 15% crystal violet (Alfa Aesar, Ward Hill, MA, USA). The number of cells migrating to the lower surface of the filters was counted in five fields under × 20 magnification, and the mean of the number of cells in each field was calculated.
Luciferase reporter gene assay
Reporter gene assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The FOPFlash plasmid containing mutated TCF-binding sites is used as a negative control. Renilla luciferase was used for normalization purposes. After 24 h of seeding, cells were transfected with luciferase reporter constructs (TOPFlash or FOPFlash, respectively) and 1 nM Renilla gene for normalization using Lipofectamine 2000 (Invitrogen, Life Technology). After 24 h of transfection, cells were lysed in 1 × Passive Lysis Buffer (Promega) and collected for luciferase and Renilla activity assays. TOPFlash/FOPFlash ratios were used to calculate the relative Wnt signaling.
Immunofluorescence Assay
Control cells and NKD2-overexpressed cells were fixed with 4% paraformaldehyde in phosphate-buffered saline. Fixed cells were then permeabilized by 0.25% TritonX-100 and incubated with a polyclonal rabbit antibody against either NKD2 or Flag. The primary antibodies used were a rabbit polyclonal antibody for Flag-tag (1:1,000 dilution; Cell Signaling, Cat. no. 2369), a rabbit polyclonal antibody against human NKD2 (1:200 dilution, Abcam, Cat. no. ab170804) and Alexa 488-conjugated anti-rabbit IgG (Invitrogen). DNA was stained with 4′,6-diamidino-2-phenylindole. Stained cells were mounted in ProLong Gold anti-fade mounting media (Invitrogen). All images were collected using the DeltaVision Inverted Deconvolution Microscope (Integrated Microscopy Core, BCM, Houston, TX, USA).
In vivo tumorigenesis and metastasis
Cells stably overexpressing NKD2 or a control vector were harvested during log phase growth and re-suspended in 0.1 ml cold phosphate-buffered saline. In all, 200 000 cells/injection were mixed with the same volume of matrigel and intratibially injected into nude mice (5 mice/group). Tumor size was determined every 3–4 days by obtaining two-dimensional measurements. The tumor volume was calculated by the formula ½ab2, where a is the long axis and b is the short axis of the tumor. Primary tumors and lungs were harvested for further evaluation. Lung metastatic nodules were visualized on 5 μm paraffin-embedded sections with H&E staining.
Statistical analysis
A two-tailed Student’s t-test was used to compare the NKD2 stably expressed (or knockdown) or drug-treated cell lines with control cell lines in proliferation assays, invasion assays and cell cycle analyses. Proportional tests were performed to compare the frequency of metastatic OS between F/+ and the other groups. The s.e. for each group was generated by the formula: s.e.(p)=sqrt (p(1−p)/n); where n represents the number of total mice in each group, and p represents the proportion of metastatic OS in each group. Gene array data were quantile normalized; differential gene expression was assessed by two-sided t-test and fold change (using log-transformed data). Kaplan scan analysis was performed as previously reported.46 Briefly, for each gene or other numerical characteristic, R2 calculates the optimal cutoff expression level dividing the patients into a good and bad prognosis cohort. Samples within a data set are sorted according to the expression of the investigated gene and divided into two groups based on a cutoff expression value.
Accession codes
References
Ottaviani G, Jaffe N . The epidemiology of osteosarcoma. Cancer Treat Res 2009; 152: 3–13.
Wang LL . Biology of osteogenic sarcoma. Cancer J 2005; 11: 294–305.
Bacci G, Briccoli A, Rocca M, Ferrari S, Donati D, Longhi A et al. Neoadjuvant chemotherapy for osteosarcoma of the extremities with metastases at presentation: recent experience at the Rizzoli Institute in 57 patients treated with cisplatin, doxorubicin, and a high dose of methotrexate and ifosfamide. Ann Oncol 2003; 14: 1126–1134.
Kager L, Zoubek A, Potschger U, Kastner U, Flege S, Kempf-Bielack B et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J Clin Oncol 2003; 21: 2011–2018.
Meyers PA, Heller G, Healey JH, Huvos A, Applewhite A, Sun M et al. Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J Clin Oncol 1993; 11: 449–453.
Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 2014; 7: 104–112.
Kansara M, Thomas DM . Molecular pathogenesis of osteosarcoma. DNA Cell Biol 2007; 26: 1–18.
Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007; 28: 622–629.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr ., Butel JS et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992; 356: 215–221.
Donehower LA, Lozano G . 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer 2009; 9: 831–841.
Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci USA 2000; 97: 4174–4179.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004; 119: 847–860.
Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res 2007; 9: R57.
Lauvrak SU, Munthe E, Kresse SH, Stratford EW, Namlos HM, Meza-Zepeda LA et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J Cancer 2013; 109: 2228–2236.
Baranwal S, Alahari SK . miRNA control of tumor cell invasion and metastasis. Int J Cancer 2009; 126: 1283–1290.
Luu HH, Kang Q, Park JK, Si W, Luo Q, Jiang W et al. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin Exp Metastasis 2005; 22: 319–329.
Edamura K, Nasu Y, Takaishi M, Kobayashi T, Abarzua F, Sakaguchi M et al. Adenovirus-mediated REIC/Dkk-3 gene transfer inhibits tumor growth and metastasis in an orthotopic prostate cancer model. Cancer Gene Ther 2007; 14: 765–772.
Hsieh SY, Hsieh PS, Chiu CT, Chen WY . Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene 2004; 23: 9183–9189.
Mizobuchi Y, Matsuzaki K, Kuwayama K, Kitazato K, Mure H, Kageji T et al. REIC/Dkk-3 induces cell death in human malignant glioma. Neuro-oncol 2008; 10: 244–253.
Veeck J, Bektas N, Hartmann A, Kristiansen G, Heindrichs U, Knuchel R et al. Wnt signalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res 2008; 10: R82.
Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA et al. Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res 2004; 64: 2734–2739.
Miretti S, Roato I, Taulli R, Ponzetto C, Cilli M, Olivero M et al. A mouse model of pulmonary metastasis from spontaneous osteosarcoma monitored in vivo by Luciferase imaging. PLoS ONE 2008; 3: e1828.
Hong SH, Briggs J, Newman R, Hoffman K, Mendoza A, LeRoith D et al. Murine osteosarcoma primary tumour growth and metastatic progression is maintained after marked suppression of serum insulin-like growth factor I. Int J Cancer 2009; 124: 2042–2049.
Hu T, Li C, Cao Z, Van Raay TJ, Smith JG, Willert K et al. Myristoylated Naked2 antagonizes Wnt-beta-catenin activity by degrading Dishevelled-1 at the plasma membrane. J Biol Chem 2010; 285: 13561–13568.
Rousset R, Mack JA, Wharton KA Jr ., Axelrod JD, Cadigan KM, Fish MP et al. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev 2001; 15: 658–671.
Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C et al. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci USA 2001; 98: 3802–3807.
Guo Y, Zi X, Koontz Z, Kim A, Xie J, Gorlick R et al. Blocking Wnt/LRP5 signaling by a soluble receptor modulates the epithelial to mesenchymal transition and suppresses met and metalloproteinases in osteosarcoma Saos-2 cells. J Orthop Res 2007; 25: 964–971.
Gotze S, Wolter M, Reifenberger G, Muller O, Sievers S . Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int J Cancer 2010; 126: 2584–2593.
Nguyen DX, Massague J . Genetic determinants of cancer metastasis. Nat Rev 2007; 8: 341–352.
Baron R, Rawadi G . Wnt signaling and the regulation of bone mass. Curr Osteoporos Rep 2007; 5: 73–80.
Johnson AS, Couto CG, Weghorst CM . Mutation of the p53 tumor suppressor gene in spontaneously occurring osteosarcomas of the dog. Carcinogenesis 1998; 19: 213–217.
Flores RJ, Li Y, Yu A, Shen J, Rao PH, Lau SS et al. A systems biology approach reveals common metastatic pathways in osteosarcoma. BMC Syst Biol 2012; 6: 50.
Brennecke P, Arlt MJ, Campanile C, Husmann K, Gvozdenovic A, Apuzzo T et al. CXCR4 antibody treatment suppresses metastatic spread to the lung of intratibial human osteosarcoma xenografts in mice. Clin Exp Metastasis 2014; 31: 339–349.
Husmann K, Arlt MJ, Muff R, Langsam B, Bertz J, Born W et al. Matrix Metalloproteinase 1 promotes tumor formation and lung metastasis in an intratibial injection osteosarcoma mouse model. Biochim Biophys Acta 2013; 1832: 347–354.
Kimura R, Ishikawa C, Rokkaku T, Janknecht R, Mori N . Phosphorylated c-Jun and Fra-1 induce matrix metalloproteinase-1 and thereby regulate invasion activity of 143B osteosarcoma cells. Biochim Biophys Acta 2011; 1813: 1543–1553.
Muff R, Ram Kumar RM, Botter SM, Born W, Fuchs B . Genes regulated in metastatic osteosarcoma: evaluation by microarray analysis in four human and two mouse cell line systems. Sarcoma 2012; 2012: 937506.
Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K et al. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol 2008; 294: G39–G49.
Kanda M, Nomoto S, Okamura Y, Hayashi M, Hishida M, Fujii T et al. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog 2011; 50: 571–579.
Wlazlinski A, Engers R, Hoffmann MJ, Hader C, Jung V, Muller M et al. Downregulation of several fibulin genes in prostate cancer. Prostate 2007; 67: 1770–1780.
Li C, Franklin JL, Graves-Deal R, Jerome WG, Cao Z, Coffey RJ . Myristoylated Naked2 escorts transforming growth factor alpha to the basolateral plasma membrane of polarized epithelial cells. Proc Natl Acad Sci USA 2004; 101: 5571–5576.
Li C, Hao M, Cao Z, Ding W, Graves-Deal R, Hu J et al. Naked2 acts as a cargo recognition and targeting protein to ensure proper delivery and fusion of TGF-alpha containing exocytic vesicles at the lower lateral membrane of polarized MDCK cells. Mol Biol Cell 2007; 18: 3081–3093.
Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol 2004; 48: 645–653.
Dumble ML, Donehower LA, Lu X . Generation and characterization of p53 mutant mice. Methods Mol Biol 2003; 234: 29–49.
Schneider CA, Rasband WS, Eliceiri KW . NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483: 589–593.
Acknowledgements
We thank Yiqun Zhang for technical assistance and Dr Neha Parikh for her assistance with figure layout and design. This work was supported by The Cancer Prevention and Research Institute of Texas (to SZ, T-KM, CL, PR, JH, LK, CJC, LAD, JTY), WWW.W Foundation, Inc. (QuadW) (to JTY), The St Baldrick’s Foundation (to JTY), the Sarcoma Foundation of America (to LAD), NIH R01 CA123238 (to AMCB) and NCI/NIH grant P30 CA125123 (to CJC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, S., Kurenbekova, L., Gao, Y. et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene 34, 5069–5079 (2015). https://doi.org/10.1038/onc.2014.429
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.429
- Springer Nature Limited
This article is cited by
-
NKD2 Trigger NF-κB Signaling Pathway and Facilitates Thyroid Cancer Cell Proliferation
Molecular Biotechnology (2023)
-
NKD2 is correlated with the occurrence, progression and prognosis of thyroid carcinoma
European Journal of Medical Research (2022)
-
Osteosarcoma
Nature Reviews Disease Primers (2022)
-
Expression pattern and clinical significance of β-catenin gene and protein in patients with primary malignant and benign bone tumors
Scientific Reports (2022)
-
Identification of key genes as predictive biomarkers for osteosarcoma metastasis using translational bioinformatics
Cancer Cell International (2021)