Abstract
Mitotic arrest-deficient 2 (MAD2) is one of the essential mitotic spindle checkpoint regulators, and it can protect cells from aberrant chromosome segregation. The Mad2 gene is very rarely mutated in many kinds of human cancer, but aberrantly reduced expression of MAD2 has been correlated with defective mitotic checkpoints in several human cancers. We have previously found that the MAD2 expression level is also shown to be associated with the multidrug resistance of tumour cells. In this study, we constructed a small interfering RNA (siRNA) eukaryotic expression vector of MAD2 and downregulated MAD2 expression in the gastric cancer cell line SGC7901 by transfection of MAD2-siRNA. SGC7901 cells stably transfected with the MAD2-siRNA exhibited significantly increased expression of phosphorylated survivin protein and enhanced drug resistance. Furthermore, MAD2-siRNA suppressed the proliferation of SGC7901 cells and inhibited tumour formation in athymic nude mice. This study clearly reveals that downregulation of MAD2 could regulate the cell cycle, increase proliferation, and improve the drug resistance of gastric cancer cells by regulating the activation of phosphorylated survivin. It also suggests both that MAD2 might play an important role in the development of human gastric cancer and that silencing the MAD2 gene may help to deal with the multidrug resistance of gastric cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitotic arrest-deficient 2 (MAD2) is an important component of the spindle assembly checkpoint, and by counteracting the effect of CDC20, MAD2 acts as an inhibitor of APC. When the spindle-assembly checkpoint is activated, MAD2 forms a ternary complex with CDC20 and APC to prevent the activation of APC and thereby arrests cells at prometaphase [1]. Chromosomal instability and the formation of tetraploid or aneuploid cells will occur if there are any errors in MAD2, which results in chromosomal missegregation and mitotic catastrophes [2–5]. The mutation of the Mad2 gene is very rare in many kinds of human cancer [6–9], but deletion or downregulation of MAD2 has also been reported in several human cancers such as lung cancer [10], breast cancer [11], nasopharyngeal cancer [12], soft-tissue sarcoma [13, 14], and ovarian carcinoma [15].
Previously in our laboratory, we indicated that the status of MAD2 is very important for determining the resistance of gastric cancer cells to anticancer drugs [16], and we found an alternative splicing variant of MAD2 named Mad2β, which is only expressed in the drug-resistant gastric cancer cell lines of SGC7901/VCR and SGC7901/ADR [17]. Mad2β could suppress the expression levels of MAD2 and eventually result in a drug-resistant phonotype of the gastric cancer cells SGC7901 by inhibiting apoptosis [18]. We also used small interfering RNA (siRNA) techniques to downregulate the expression of MAD2 in gastric cancer cells and found that the defect of the spindle checkpoint could promote the drug resistance of gastric cancer cells and reverse anticancer drug-induced apoptosis by upregulating Bcl-2 and interfering with the mitochondrial apoptosis pathway [19]. The aim of the present study was to find out (1) whether the downregulation of MAD2 is related to the development and malignant phenotypes of gastric cancer and (2) whether there is another role of MAD2 in the multidrug resistance of gastric cancer.
We found that MAD2 could regulate the cell cycle of cancer cells in the present study. In gastric cancer, the induction of cell-cycle arrest was partly responsible for the antitumour function of MAD2. Our study indicated that MAD2 may be used as a therapeutic target of gastric cancer and that it was a potential tumour regulator. Furthermore, it was also indicated that MAD2 might be a novel negative modifier in multidrug resistance and that it may provide a pivotal mechanism for exerting the efficiency of chemotherapy drugs in gastric cancer treatment.
Materials and methods
Cell culture
The human gastric cancer cell line SGC7901 was cultivated in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% heat-inactivated foetal calf serum, in a 37°C humidified incubator with a mixture of 95% air and 5% CO2 (Forma Scientific).
Small interfering RNA construction and transfection
Two different MAD2-specific siRNA duplexes were designed homologous to the MAD2 mRNA consensus sequence. The first siRNA molecule was MAD2-1 (S: 5′-tttgcagtcttgtcacactcaaacattgagtgtgacaagactgctttt-3′, AS: 5′-ctagaaaaagcagtcttgtccacactcaatgtttgagtgtgacaagactg-3′), whose target sequence was homologous to nt 311–329 of the MAD2 mRNA sequence; the second siRNA molecule, MAD2-2 (S: 5′-tttgcagtcttgtcacactcaaacatcagacagatcacagctagtttt-3′, AS: 5′-ctagaaaaagcagtcttgtccacactcaatgttcagacagatcacagcta-3′), was homologous to nt 401–423 of the MAD2-psilencer3.1 (Ambion, Austin, TX); these were used for the construction of the human MAD2-siRNA vectors (MAD2-siRNA1 and MAD2siRNA2), according to the manufacturer’s protocol.
According to the manufacturer’s instruction, the siRNA plasmids were transfected into SGC7901 cells using Lipofectamine2000 reagent (Invitrogen, Carlsbad, USA). The gastric cancer cells transfected with MAD2siRNA1 and MAD2siRNA2 were named as SGC7901/MAD2-siRNA1 and SGC7901/MAD2-siRNA2, respectively.
Briefly, SGC7901 cells were seeded in six-well plates and cultured in drug-free medium until reaching 70% to 90% confluence, and these cells were not transfected with 1-mg plasmids. Using Lipofectamine2000 reagent, 2 μg of psilencer3.1-MAD2siRNA plasmids was transfected into SGC7901 cells. To stabilise the transfection, G418 (400 mg/ml) was added into cells after 24 hours of transfection. Mixed clones were screened by Western blot analysis and expanded for an additional 6 weeks. The cells transfected with psilencer3.1 vector alone served as the negative control.
RNA extraction and semiquantitative RT-PCR
The RNA of each cell line was extracted using Trizol (Life Technologies, USA) according to the manufacturer’s protocol. DNase was used to decrease the contamination of genomic DNA. The quantity and purity of the RNA prepared from each sample were determined by UV absorbance spectroscopy. The reverse transcription reaction was performed using the First-Strand cDNA Synthesis Kit (Fermentas, USA) in a final volume of 20 μm. After incubation at 42°C for 60 minutes, the reverse transcription reaction was terminated by heating at 70°C for 10 minutes. The newly synthesised cDNA was amplified by PCR. The MAD2 primer was S: 5-AAGGTGAAGGTCGGAGTCAA-3, AS: 5-TGTGGTCATGAGTCCTTCCA-3. Amplification cycles were: 95°C for 2 minutes; then 36 cycles at 94°C for 30 seconds, 51°C for 45 seconds, and 72°C for 50 seconds; followed by 72°C for 10 minutes. β-Actin primer (S: 5-AGCGGGAAATCGTGCGTG-3; AS: 5-CAGGGTACATGGTGGTGCC-3) was used as an internal control. Aliquots of PCR product were electrophoresed on 1% agarose gels, and PCR fragments were visualised by ethidium bromide staining.
Western blot analysis
The Western blotting was done as described [19]. In brief, total proteins were prepared and then quantified by the Bradford method. A measure of 80 μg of lysates was electrophoresed in 12% SDS-PAGE and blotted on a nitrocellulose membrane (Immoblin-P, Millipore, Bedford, MA). Membranes were blocked with 5% fat-free milk powder at room temperature for 2 hours and incubated overnight with polyclonal antibody specific for rabbit anti-MAD2 (diluted 1:200, Santa Cruz Biotech, Santa Cruz, CA, USA) and monoclonal antibody specific for mouse anti-survivin (diluted 1:300, Santa Cruz Biotech) or anti-β-actin antibody (diluted 1:2000, Sigma Chemical Co. CA, USA) at 4°C. After three washes for 15 minutes in PBS-T, the membrane was incubated with the HRP-conjugated goat antimouse IgG antibody (Wuhan, Hubei, China) for 1 hour at room temperature. The membrane was washed again in PBS-T; enhanced chemiluminescence (Amersham Life Science, Piscataway, NJ) was added, and the colour development was monitored for the development of colour. Each experiment was performed in triplicate. All examined gene expression levels were quantitatively analysed and expressed as ratios to β-actin.
Immunofluorescent staining
Cells were plated onto 24-well plates (2 × 104 cell/well) containing 12-mm coverslips (IWAKI, Tokyo, Japan) and subsequently treated with cisplatin (0.5 µg/ml) for 10 minutes. At 24 hours after treatment, cells were fixed in 4% paraformaldehyde in PBS and then washed thrice in PBS and permeabilised in 0.1% Triton X-100 for 5 minutes. After blocking in 1% BSA for 20 minutes, cells were incubated with rabbit anti-MAD2 polyclonal antibody (1:200; Santa Cruz Biotech) for 2 hours, followed by fluorescein-conjugated goat antimouse IgG secondary antibody (1:50) for 45 minutes. The fluorescent signal was visualised with a Zeiss Axiophot microscope (Carl Zeiss, Inc., Thornwood, NY) and photographed using a Spot RT digital camera and associated software (Diagnostic Instrument, Sterling heights, MI). At least 500 cells were counted from three random fields under 400 magnification for each experiment, and the percentage of cells containing >10 fluorescent foci was calculated. Each experiment was repeated at least thrice.
DAPI counterstaining
To observe the different apoptosis morphologies in the cell lines 4 hour after treatment, the cellular DNA was counterstained with 4V,6-diamidino-2-phenylindole (DAPI). DAPI counterstaining allows for determination of a nuclear morphology indicator based on nuclear size and roundness. It is found that the nuclear area factor is an early indicator of cell death (significant after 4 hours post treatment). DAPI is a blue fluorescent probe that fluoresces brightly when it is selectively bound to the minor groove of double-stranded DNA, where its fluorescence is approximately 20-fold greater than in the nonbound state. This selectivity for DNA, along with the cell permeability, allows the staining of nuclei with little background from the cytoplasm, making DAPI the classic nuclear counterstain for assessing apoptosis under immunofluorescence microscopy.
Flow cytometry analysis of cell apoptosis
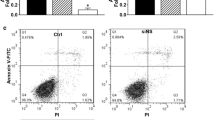
The apoptosis of cells was evaluated by measuring the exposure of phosphatidylserine on the cell membranes using Apoptosis Detection Kits (BD Pharmingen). Cell pellets were resuspended in a staining solution containing propidium iodide (PI, 50 Ag/ml) and Annexin V-FITC (25 Ag/ml) for 15 minutes at room temperature in the dark. The cells were assessed by FACS equipped with the Cell Quest software (BD Pharmingen).
MTT assay
Cells were seeded on a 96-well plate (Corning) at 3 × 104 cells/well in DMEM containing 10% FCS. Each sample had four replicates. The medium was replaced every other day. Viable cells were counted by the MTT assay after 2, 4, 6, and 8 days. Briefly, cells were incubated with 50 μl of 0.2% MTT for 4 hours at 37°C in the 5% CO2 incubator and then lysed in 150 μl of DMSO. The absorbance at 490 nm was determined using the 96-well plate reader (Dynex Technologies).
Tumour growth in nude mice
The logarithmically growing cells were trypsinised and resuspended in D'Hanks solution, and 5 × 106 cells (0.2 ml) were injected subcutaneously into the left flank of 4-week-old female BALB/c nude mice (at least three mice per group). The chemotherapy drugs were injected into the caudal vein after xenografts became palpable. The tumour volume was calculated as (length×width2)×0.5.
Intracellular adriamycin concentration analysis
The amount of intracellular adriamycin was determined by flow cytometry. Briefly, the cells were seeded into six-well plates (1 × 106 cells/well) and treated with adriamycin at the final concentration of 5 μg/ml. For the detection of adriamycin accumulation, the cells were harvested 1 hour after drug treatment, whereas for the detection of adriamycin retention, cells were cultured in drug-free DMEM for another 1 hour before harvest. Then, cells were washed with PBS, and the mean fluorescence intensity was detected using flow cytometry (FACScan, BD Biosciences) with an excitation wavelength of 488 nm and emission wavelength of 575 nm. The experiment was independently done thrice. Finally, the adriamycin-releasing index of the gastric cancer cells was calculated using the following formula: releasing index = (accumulation value-retention value)/accumulation value.
Immunoprecipitation
Cell lysates for immunoprecipitation were prepared using a mild lysis method [20, 21]. The concentrations of cell lysates were determined using the Bradford method (BioRad), and equal amounts of aliquots were incubated with the indicated antibodies at 4°C for 15 hours [22]. For further Western blot analysis, the immunoprecipitates were washed four times with lysis buffer and subjected to typical gel electrophoresis and immunoblotting procedures [23]. The protein bands were finally detected by enhanced chemiluminescence (ECL, Amersham, Amersham Life Science).
Statistical analysis
Each data point represented here was the mean ± SE of at least three independent experiments, and each of them was performed in triplicate unless otherwise indicated. The statistical significance of the flow cytometric assay results was analysed by the two-sided Student’s t-test and the chi-squared test. Differences were considered to be statistically significant when P < 0.05 (indicated by the asterisk in the table and figures). All statistical analyses were performed using SPSS 13.0 software.
Results
Downregulation of MAD2 expression in cancer cells induced by siRNA
MAD2 was found to be highly downregulated in SGC7901 cells by a streptavidin magnetic beads-mediated subtraction cloning method in our previous research [18]. The psilencer3.1-MAD2-siRNA plasmid was designed to decrease the expression of MAD2 in the human gastric carcinoma cell line SGC7901. Two subclones of the MAD2-siRNA transfectants SGC7901/MAD2-siRNA and SGC7901/MAD2-siRNA2 were generated. The efficacy of siRNA transfection was evaluated by reverse transcriptase-polymerase chain reaction (RT-PCR), Western blotting, and immunofluorescent staining (Figs. 1, 2, and 3).
Inhibition of MAD2 expression in gastric cancer cells. Four cell lines were evaluated: SGC7901/MAD2-siRNA1 (transfected with MAD2-siRNA1; lane 1), SGC7901/MAD2-siRNA2 (transfected with MAD2-siRNA2; lane 2), human gastric cancer cells SGC7901 (lane 3), and SGC7901/psilencer (transfected with psilencer3.1; lane 4). a The RT-PCR assay showed that the mRNA of MAD2 in MAD2-siRNA transfectants was decreased compared with that in control cells. b Western blotting showed that the MAD2 protein expression was inhibited significantly upon MAD2-siRNA-transfected cells
Relative protein (a) and mRNA (b) expression levels of MAD2 to their respective β-actin were analysed by one-way analysis of variance. The expression levels of MAD2 in SGC7901/MAD2-siRNA1 or SGC7901/MAD2-siRNA2 cells were lower compared with the controls. *P < 0.05 vs. SGC7901 and SGC7901/psilencer. There were no differences in the expression levels of MAD2 between SGC7901/MAD2-siRNA1 and SGC7901/MAD2-siRNA2 cells. # P > 0.05 vs. SGC7901/MAD2-siRNA1
As shown in Fig. 1a, the siRNA reduced the levels of the endogenous MAD2 mRNA in siRNA-transfected cells as compared with control cells, whereas it showed no effect on β-actin. A Western blot showed that both MAD2-siRNA1 and MAD2-siRNA2 decreased the MAD2 protein expression level almost equally in SGC7901 cells (Fig. 1b). SGC7901/MAD2-siRNA2 cells were selected for subsequent studies. Immunofluorescent staining showed that the MAD2 protein expression was inhibited significantly in MAD2-siRNA-transfected cells (Fig. 3).
Downregulation of MAD2 decreased the apoptosis induced by cisplatin
We examined the apoptosis rate in cells with normal (before siRNA treatment) or low expression levels of MAD2 (after siRNA treatment) at 24 hours after exposure to 0.5 µg/ml cisplatin. As shown in Fig. 4 and supplemental Fig. 1, a low percentage of MAD2 protein-expressing cells showed a combination of a punctuate staining pattern after exposure to cisplatin by DAPI staining as compared to controls. Flow cytometry analysis showed the apoptosis ratio in MAD2-siRNA transfectants to be 3.4%. In contrast, an increased number of apoptotic cells were observed in control cells. The apoptosis ratios of SGC7901 and SGC7901/psilencer3.1 induced by cisplatin were 23.4% and 19%, respectively. These results indicate that the downregulation of MAD2 decreased the apoptosis induced by cisplatin; however, the mechanism is not yet clear.
Elevation of apoptosis in cells by DAPI staining. Nuclei were stained with DAPI (blue). Images were photographed at the same exposure time under a ×20 objective with a SPOT camera. Note that a high percentage of MAD2-siRNA-expressing cells showed a combination of a punctate staining pattern after exposure tocisplatin (0.5 µg/ml). a SGC7901/MAD2-siRNA1 (transfected with MAD2-siRNA1), b SGC7901/MAD2-siRNA2 (transfected with MAD2-siRNA2), c human gastric cancer cells SGC7901, and d SGC7901/psilencer
Suppression of endogenous MAD2 enhances tumour drug resistance
To examine the effect of the downregulation of MAD2 on chemotherapeutic sensitivities, IC50 values of vincristine, paclitaxel, adriamycin, VP-16, and cisplatin in SGC7901/MAD2-siRNA cells were determined by MTT assay. As shown in supplemental Fig. 2, compared with SGC7901 and SGC7901/psilencer3.1 cells, SGC7901/MAD2-siRNA cells exhibited significantly increased IC50 values for vincristine, paclitaxel, adriamycin, and cisplatin (P < 0.05).
Furthermore, an in vivo subcutaneous tumour formation assay was adopted to examine the chemotherapeutic sensitivities of MAD2-siRNA cells. Compared with control cells transfected with empty vector, the injection of SGC7901/MAD2-siRNA cells led to a significantly increased tumour size (P < 0.05; supplemental Fig. 3). We also determined MAD2 protein expression in the MAD2-siRNA transfected tissues of nude mice analysed using Western blot (supplemental Fig. 4). Both in vitro and in vivo assays suggested that the downregulation of MAD2 expression significantly decreased the sensitivity of gastric cancer cells to microtubule inhibitors and DNA damaging agents. This evidence is in agreement with Sudo T’s study [24].
Depression of MAD2 decreases the intracellular accumulation of adriamycin in gastric cancer
To further elucidate whether the downregulation of MAD2 is involved in the multidrug resistance of gastric cancer cells, the intracellular accumulation and releasing of adriamycin were explored by flow cytometry. As shown in supplemental Fig. 5, the accumulation of adriamycin in SGC7901/MAD2-siRNA cells was markedly decreased compared with that in SGC7901/psilencer cells and SGC7901 cells (P < 0.05). Consistent with this, the adriamycin-releasing index of SGC7901/MAD2-siRNA cells was significantly higher than that of control cells (P < 0.05). These data further confirmed that the downregulation of MAD2 expression played an important role in the drug resistance of gastric cancer.
Downregulation of endogenous MAD2 enhances the expression of P-gp, Raf-1 and p-cdc2
Because MAD2 gene silencing could rapidly extrude the ADR from the cytoplasm into the external medium, we found that the expressions of P-gp, Raf-1, and p-cdc2 were increased in the SGC7901/MAD2siRNA cell line compared with that in control cell lines. However, no differences of cyclinB or MRP expression levels were detected among the SGC7901/MAD2-siRNA, SGC7901/psilencer, and SGC7901 cells (Fig. 6).
Survivin protein expression in gastric cancer cell lines
To further study the mechanism of MAD2 effects on apoptosis, the expression of survivin was evaluated in the SGC7901/MAD2-siRNA, SGC7901/psilencer, and SGC7901 cells. No differences in general survivin expression were found among the above cell lines, but the level of phosphorylated survivin was increased in MAD2-defective cells compared with that in control cells (Fig. 5), indicating that MAD2 may affect apoptosis by regulating the activity of survivin. Furthermore, as shown in Fig. 6, MAD2 protein could bind to survivin protein in an immunoprecipitation assay.
Western blot analysis of the parental cells. β-actin was used as the loading control (p-survivin: phosphorylated-survivin) (lane 1, SGC7901/siMAD2; lane 2, SGC7901/ADR; lane 3, SGC7901; lane 4, SGC7901/psilencer). It showed that the level of phosphorylated-survivin is lower in the MAD2-downregulated gastric cancer cell line (SGC7901/siMAD2) and in the drug-resistant (SGC7901/ADR) cancer cell line compared to the parent cell line (SGC7901) and the control cell line (SGC7901/psilencer)
Western blot analysis of the MAD2 levels in the survivin immunoprecipitates from gastric cancer cells; β-actin was used as the loading control (lane 1, SGC7901/siMad2; lane 2, SGC7901/ADR; lane 3, SGC7901; lane 4, SGC7901/psilencer). The results of immunoprecipitation showed that the MAD2 protein was bound to survivin
Discussion
Mitotic catastrophe is an important mechanism capable of inducing the death of cancer cells by antineoplastic agents that damage DNA and activate the apoptosis pathway. Defects in the G1 and G2 checkpoints of the cell cycle are apparent in most cancer cells and allow the cells to enter mitosis with DNA damage [25, 26]. However, several studies showed that DNA damage-induced mitotic arrest can also occur without DNA damage checkpoints by spindle checkpoints [27–29]. Although more of these mechanisms of DNA damage have not been elucidated, it has been suggested that extensive DNA damage could activate the spindle checkpoint and may lead to the impairment of kinetochores [27, 28, 30].
MAD2 is one of the essential players in the spindle checkpoint’s regulation of anaphase [31, 32], and it localises selectively to unattached kinetochores in mitosis metaphase, blocks the activity of the anaphase-promoting complex, and then disappears until all chromosomes of cells are accurately attached to the spindle [33]. The downregulation of MAD2 is responsible for mitotic checkpoint abrogation and chromosomal instability in human cancers [11–15]. Wang et al. [34] reported that MAD2-induced sensitisation to VCR is associated with Raf/Bcl-2 phosphorylation and mitotic arrest in nasopharyngeal carcinoma. Recently, it has been suggested that the overexpression of MAD2 confers sensitivity to DNA-damaging anticancer drug-induced apoptosis and that MAD2-induced mitotic arrest mediates the DNA damage response [35, 36].
Our previous study indicated that the decrease of MAD2 or increase of Mad2β expression could inhibit anticancer drug-induced apoptosis by upregulating Bcl-2 and interfering with the mitochondrial apoptosis pathway. Furthermore, our present results of the cell cycle suggest that in gastric cancer cells with G2 phase checkpoint and spindle checkpoint defects, chemotherapeutic drugs could still cause cells to arrest at the G2/M phase; the spindle checkpoint could not only identify abnormities caused by spindle microtubule inhibitors’ VCR but also detect G2 phase defects during the DNA damage, which might ultimately cause cell-cycle arrest [37].
DNA damage and spindle microtubules' disturbance are able to activate the spindle checkpoint process to block further damages to gastric cancer cells. The spindle checkpoint is able to determine not only the ultimate fate of spindle microtubule injury in gastric cancer cells but also the fate of DNA damage in gastric cancer cells. In this study, through in vitro and in vivo sensitivity tests, it was confirmed that inhibiting the expression of MAD2 might cause SGC7901 spindle checkpoint defects, leading to the multidrug resistance of gastric cancer cells.
Our study suggested that MAD2, as the cell-cycle checkpoint in the final phase of the mitotic spindle checkpoint, played a very important role in drug resistance. It controlled the ability of gastric cancer cells to enter the next cell cycle of the final clearance. For the role of chemotherapy drugs in gastric cancer cells, the spindle checkpoint of the “state” (defected or normal) to determine their ultimate fate is the key, proceeding either “into death” or “into the next cell cycle to continue to survive.”
It has been demonstrated that P-gp, encoded by the human MDR1 gene, plays a key role in tumour drug resistance [38]. Given the capacity for enhancing apoptosis and downregulating P-gp expression, the upregulation of MAD2 protein is expected to reverse gastric cancer drug resistance. The data presented by the drug sensitivity assay in this study strongly support the opinion that downregulating MAD2 could enhance the resistant phenotype of gastric cancer cells not only through reducing apoptosis but also through upregulating the expression of P-gp, Raf-1, and p-cdc2 [39]. It was supposed that MAD2 inhibited the activity of cdc2 while subsequently upregulating the expressions of Raf-1 and P-gp.
The results of immunoprecipitation showed that the MAD2 protein was bound to survivin in mitosis prior to the formation of the medium-term combination of the complex. It was suggested that MAD2 might inhibit the activity of survivin by binding with the phosphorylated site of survivin.
Therefore, the key spindle checkpoint element MAD2, together with survivin, may be involved in the spindle checkpoint-mediated apoptosis. However, whether MAD2 binds to the survivin phosphorylation site to inhibit the activity of survivin is still not clear. The specific molecular mechanism of the MAD2-mediated anti-apoptotic effect during the mitosis period is also not clear. What is the relationship between survivin and MAD2? The answer to these questions may be found in further studies.
In summary, our study suggested that two pathways were involved in MAD2-mediated regulation of drug resistance in gastric cancer: the promotion of P-gp expression and the inactivation of the apoptosis protein survivin.
Abbreviations
- MAD2:
-
mitotic arrest-deficient 2
- VCR:
-
vincristine
- ADR:
-
Adriamycin
- 5-FU:
-
5-fluorouracil
- CDDP:
-
cisplatin
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS:
-
phosphate-buffered saline
- MDR:
-
multi-drug resistance
- P-gp:
-
P-glycoprotein
- MRP:
-
multi-drug resistance-associated protein
- PI:
-
propidium iodide
- siRNA:
-
small interfering RNA
References
Fang G, Yu H, Kirschne MW. Control of mitotic transitions by the anaphase-promoting complex phil. Trans R Soc Lond. 1999;B354:1583–90.
Hisaoka M, Matsuyama A, Hashimoto H. Aberrant MAD2 expression in soft-tissue sarcoma. Pathol Int. 2008;58:329–33.
Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101(6):635–45.
Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haploin sufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9.
Pennisi E. Cell division gatekeepers identified. Science. 1998;279:477–8.
Hernando E, Orlow I, Liberal V, Nohales G, Benezra R, Cordon-Cardo C. Molecular analyses of the mitotic checkpoint components hsMAD2, hBUB1 and hBUB3 in human cancer. Int J Cancer. 2001;95(4):223–7.
Gemma A, Hosoya Y, Seike M, Uematsu K, Kurimoto F, Hibino S, et al. Genomic structure of the human MAD2 gene and mutation analysis in human lung and breast cancers. Lung Cancer. 2001;32(3):289–95.
Imai Y, Shiratori Y, Kato N, Inoue T, Omata MJ. Mutational inactivation of mitotic checkpoint genes, hsMAD2 and hBUB1, is rare in sporadic digestive tract cancers. Cancer Res. 1999;90(8):837–40.
Percy MJ, Myrie KA, Neeley CK, Azim JN, Ethier SP, Petty EM. Expression and mutational analyses of the human MAD2L1 gene in breast cancer cells. Genes, Chromosomes Cancer. 2000;29(4):356–62.
Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–8.
Takahashi T, Haruki N, Nomoto S, Masuda A, Saji S, Osada H, et al. Identification of frequent impairment of the mitotic checkpoint and molecular analysis of the mitotic checkpoint genes,hsMAD2 and p55CDC, in human lung cancers. Oncogene. 1999;18:4295–300.
Wang X, Jin D-Y, Wong YC, Cheung ALM, Chun ACS, Lo AKF. Correlation of defective mitotic checkpoint with aberrantly reduced expression of MAD2 protein in nasopharyngeal carcinoma cells. Carcinogenesis. 2000;21:2293–7.
Hisaoka M, Matsuyama A, Hashimoto H. Aberrant MAD2 expression in soft-tissue sarcoma. Pathol Int. 2008;58(6):329–33.
Ruddy DA, Gorbatcheva B, Yarbrough G, Schlegel R, Monahan JE. No somatic mutations detected in the Mad2 gene in 658 human tumors. Mutat Res. 2008;641(1-2):61–3.
Wang X, Jin D-Y, Ng RWM, Feng H, Wong YC, Cheung ALM, et al. Significance of MAD2 expression to mitotic checkpoint control in ovarian cancer cells. Cancer Res. 2002;62:1662–8.
Zhao YQ, You H, Liu F, An HZ, Shi YQ, Yu Q et al (2002) Differentially expressed gene profiles between multidrug resistant gastric adenocarcinoma cells and their parental cells. Cancer Lett. 185211–8.
Yin F, Hu WH, Qiao TD, Fan DM. Multidrug resistant effect of alternative splicing form of MAD2 gene-MAD2beta on human gastric cancer cell. Chin J Oncol. 2004;26:201–4.
Yin F, Du Y, Hu W, Qiao T, Ding J, Wu K, et al. Mad2β, an alternative variant of Mad2 reduce mitotic arrest and apoptosis of gastric cancer cells induced by adriamycin. Life Sci. 2006;78:1277–86.
Pan Y, Bi F, Liu N, Xue Y, Yao X, Zheng Y, et al. Expression of seven main Rho family members in gastric carcinoma. Biochem Biophys Res Commun. 2004;315(3):686–91.
Smits VA, Klompmarker R, Arnaudm L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000;2:672–6.
Huang T-S, Duyster J, Wang JYJ. Biological response to phorbolester determined by alternative G1 pathway. Proc Natl Acad Sci USA. 1995;92:4793–7.
Huang T-S, Shu C-H, Yang WK, Whang-Peng J. Activation of CDC 25 phosphatase and CDC 2 kinase involved in GL331 induced apoptosis. Cancer Res. 1997;57:2974–8.
Huang T-S, Yang WK, Whang-Peng J. GL331-induced disruption of cyclin B1/CDC2 complex and inhibition of CDC2 kinase activity. Apoptosis. 1996;1:213–7.
Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004;64(7):2502–8.
Schwartz GK. Development of cell cycle active drugs for the treatment of gastrointestinal cancers: a new approach to cancer therapy. J Clin Oncol. 2005;20:4499–508.
Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59(4):928–42.
Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics. 2002;161:521–34.
Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–806.
Tulub AA, Stefanov VE. Cisplatin stops tubulin assembly into microtubules. A new insight into the mechanism of antitumor activity of platinum complexes. Int J Biol Macromol. 2001;28:191–8.
Chow JP, Siu WY, Fung TK, Chan WM, Lau A, Arooz T, et al. DNA damage during the spindle-assembly checkpoint degrades CDC25A, inhibits cyclin-CDC2 complexes, and reverses cells to interphase. Mol Biol Cell. 2003;14(10):3989–4002.
Musacchio, Hardwick KG (2002) The spindle checkpoint: structural insights into dynamic signaling. Nat. Rev. Mol. Cell. Biol. 3731–41.
Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores from epigenetics to mitotic checkpoint signaling. Cell 112407–21.
Glotzer M. Mitosis: don’t get mad, get even. Curr Biol. 1996;6:1592–4.
Wang X, Jin D-Y, Wong HL, Feng H, Wong Y-C, Tsao SW. MAD2-induced sensitisation to vincristine is associated with mitotic arrest and Raf/Bcl-2 phosphorylation in nasopharyngeal carcinoma cells. Oncogene. 2003;22:109–16.
Fung MKL, Han H-Y, Leung SCL, Cheung HW, Cheung ALM, Wong Y-C, et al. MAD2 interacts with DNA repair proteins and negatively regulates DNA damage repair. J Mol Biol. 2008;381(1):24–34.
Zhang P, Cong B, Yuan H, Chen L, Lv Y, Bai C, et al. Overexpression of spindlin1 induces metaphase arrest and chromosomal instability. J Cell Physiol. 2008;217(2):400–8.
Du Y, Yin F, Liu C, Hu S, Wang J, Xie H, et al. Depression of MAD2 inhibits apoptosis of gastric cancer cells by upregulating Bcl-2 and interfering mitochondrion pathway. Biochem Biophys Res Commun. 2006;345(3):1092–8.
Fujita T, Washio K, Takabatake D, Takahashi H, Yoshitomi S, Tsukuda K, et al. Proteasome inhibitors can alter the signaling pathways and attenuate the P-glycoprotein-mediated multidrug resistance. Int J Cancer. 2005;117:670–82.
Zhao YQ, You H, Liu F, An HZ, Shi YQ, Yu Q, et al. Differentially expressed gene profiles between multidrug resistant gastric adenocarcinoma cells and their parental cells. Cancer Lett. 2002;185:211–8.
Acknowledgments
We are grateful to Dr. Bin Guo for his proofreading of the manuscript. We thank technician Yunxin Cao for excellent technical assistance. This study was supported in part by grants from the Chinese National Foundation of National Sciences (C03031905, 30973422, 30600551, and 30530780).
Author information
Authors and Affiliations
Corresponding author
Additional information
Li Wang, Fang Yin and Yulei Du contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Effects of MAD2-siRNA on the apoptosis of gastric cancer cells treated with cisplatin for 24 hours. The apoptosis rate is increased in the SGC7901/MAD2-siRNA cell line compared with control and blank vector groups. (JPEG 121 kb)
Supplementary Fig. 2
Effects of MAD2-siRNA on cellular sensitivity to chemodrugs. The value shown is the mean of three determinations. IC50 values of the cells described as above in vitro. a Cell number was evaluated by the absorbance at 490 nm in an MTT assay at the indicated time. Representative experiment of three, with similar results. b Soft agar clone-forming assay of the cells were performed as described above. The data represent means ± SD of three independent experiments. (JPEG 201 kb)
Supplementary Fig. 3
Transplanted tumours in the BALB/c nu/nu mice after chemotherapy. a ADR-A; b ADR-B; c ADR-C (ADR-A: the ADR therapy group that was injected into the SGC7901/MAD2-siRNA gastric carcinoma cells subcutaneously; ADR-B: the ADR therapy group that was injected into the SGC7901/psilencer gastric carcinoma cells subcutaneously; ADR-C: the ADR therapy group that was injected into the SGC7901 gastric carcinoma cells subcutaneously). The tumour sizes of the SGC7901/MAD2-siRNA group grew more slowly than the other two groups after drug therapy. (JPEG 118 kb)
Supplementary Fig. 4
MAD2 protein expression in the transfected tissues of nude mice analysed by Western blot (ADR-A: the ADR therapy group that was subcutaneously injected into SGC7901/MAD2-siRNA gastric carcinoma cells; ADR-B: the ADR therapy group that was subcutaneously injected into SGC7901/psilencer gastric carcinoma cells; ADR-C: the ADR therapy group that was subcutaneously injected into SGC7901 gastric carcinoma cells; VCR-A: the VCR therapy group that was subcutaneously injected into SGC7901/MAD2-siRNA gastric carcinoma cells; VCR-B: the VCR therapy group that was subcutaneously injected into SGC7901/psilencer gastric carcinoma cells; VCR-C: the VCR therapy group that was subcutaneously injected into SGC7901 gastric carcinoma cells). The Western blot shows the levels of MAD2 protein in the transplanted gastric tumour that was subcutaneously injected into SGC7901/MAD2-siRNA carcinoma cells; the levels were very low compared with the other groups. (JPEG 36 kb)
Supplementary Fig. 5
The effect of the MAD2-siRNA on drug cellular adriamycin accumulation was analysed by using FCM. The drug fluorescence intensity is expressed as the mean of fluorescence that could be calculated from the flow cytometric profiles. After being incubated with 1 or 5 mg/L of adriamycin for 1 hour, the positive fluorescence rates of SGC7901/MAD2-siRNA cells were significantly lower than that of SGC7901 and that of SGC7901/psilencer cells. The adriamycin-releasing ratio of the three groups of cancer cells. A Intracellular accumulation fluorescence intensity of adriamycin in gastric cancer cells; R Intracellular retention fluorescence intensity of adriamycin in gastric cancer cells (*P < 0.05: blank vector vs. SGC7901/MAD2-siRNA). (JPEG 264 kb)
Supplementary Fig. 6
Western blot analysis of P-gp, MRP, Raf-1, p-cdc2, and cyclin B in three cell lines. β-actin was used as a loading control (lane 1, SGC7901; lane 2, SGC7901/psilencer; lane 3, SGC7901/MAD2-siRNA). It showed that the downregulation of MAD2 could raise the levels of P-gp, Raf-1, and p-cdc2, but it did not influence the level of CyclinB and MRP. (JPEG 51 kb)
Rights and permissions
About this article
Cite this article
Wang, L., Yin, F., Du, Y. et al. Depression of MAD2 inhibits apoptosis and increases proliferation and multidrug resistance in gastric cancer cells by regulating the activation of phosphorylated survivin. Tumor Biol. 31, 225–232 (2010). https://doi.org/10.1007/s13277-010-0036-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-010-0036-6