Abstract
Background
A large proportion of eukaryote nuclear genomes is composed of repetitive DNA. Tracing the dynamics of repetitive elements in the genomes of related taxa can reveal important information about their phylogenic relationships as well as traits that have become distinct to a lineage.
Objective
Study the genomic abundance and chromosomal location of repetitive DNA in Capsicum annuum L. to understand the repeat dynamics.
Method
We quantified repeated DNA content in the C. annuum genome using the RepeatExplorer pipeline.
Results
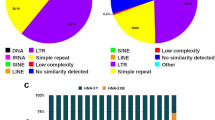
About 42% of the C. annuum genome dataset comprised repetitive elements. Of these, 0.011, 0.98, 3.09, and 0.024% represented high and low confidence satellite repeats, putative long-terminal repeats (LTRs), and rDNA sequences, respectively. One novel high confidence 167-bp satellite repeat with a genomic proportion of 0.011%, Ca167TR, was identified. Furthermore, FISH with Ca167TR on metaphase chromosomes of C. annuum revealed signals in the subtelomeric regions of the short and long arms of chromosome 3 and 4, respectively.
Conclusion
Further understanding of the origin and associated functions of Ca167TR and other repeats in Capsicum will give us insights into the genomic relationships and functions of the genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Capsicum annuum belongs to the Solanaceae family. There are five domesticated and approximately 25 wild species in the economically important genus Capsicum (Cheema and Pant 2013). The fruits of Capsicum species are used as spices, and C. annuum is among the most consumed vegetable crops worldwide (Kwon and Kim 2009). Capsicum peppers were introduced to Asia from South America during the sixteenth century via trade routes (Castro-Concha et al. 2014). Currently, the largest producer is China (Paran et al. 2007). Moreover, there are two distinct groups present in the genus: species that have 12 pairs of chromosomes (2n = 24), and species that have 13 pairs of chromosomes (2n = 26) (Rohami et al. 2010). C. annuum is diploid with a genome size of 3.26 Gb (Qin et al. 2014).
Recent advances in genomics have shown that large proportion of eukaryotic genomes are repetitive elements (REs). These sequences could be up to 90% of the genome size (Mehrotra and Goyal 2014; Pertea 2012). The REs can be classified into two broad classes: satellite sequences, which include micro-satellites and mini-satellites, and dispersed repeats like transposable elements (TEs) (Wang et al. 2012). REs play very important roles in species evolution because they are involved in several processes, such as the movement, pairing, recombination, and arrangement of chromosomes, regulation of genes expressions, and genic responses to environmental stimuli (Mehrotra and Goyal 2014). The TEs play key roles in adaptation, diversification, and speciation (Kalendar et al. 2000; Schrader et al. 2014). One TE, the long terminal repeat (LTR) retrotransposon represents one of the largest portions of DNA component (Liu et al. 2018), such as the proportions of LTR are greater than 75%, 41.35%, and 68.68% in maize (Schnable et al. 2009), wheat (Jia et al. 2013), and coix (Cai et al. 2014), respectively. In general, LTR retrotransposons are distributed along the chromosomes and found in the centromere region (Li et al. 2017), playing significant roles in gene regulation and evolution (Galindo-González et al. 2017).
The latest genome assembly of C. annuum showed that 80.9% comprised repeats. Among those REs, the majority was the long terminal repeat (LTR) which represents approximately 70.3% of the genome, and the most of the LTR were Gypsy elements (Kim et al. 2014; Qin et al. 2014). In a previous study, an unknown type of repeat CaLUR with a long unit length (18–24 kb) was identified and randomly distributed in C. annuum pachytene chromosomes. However its functional role was unclear (Park et al. 2012).
Ribosomal DNAs are essential genetic elements involved in ribosome function (Richard et al. 2008). They include two major subfamilies: the 5S and 45S (18S–5.8S–25S) rDNAs (Galian et al. 2012). It has been observed that the copy number and chromosome distribution of rDNAs could be changed rapidly (Muratović et al. 2010). Because of this property, rDNAs have been used as molecular markers for genome mapping (Witsenboer et al. 1997) and cytogenetic research (Gupta and Varshney 2000) in different plants.
Characterizing the types of repeats in a genome and understanding their chromosomal distribution and potential function will allow the improvement of crop breeding programs. A rapid and efficient approach to identifying major genomic REs is through the RepeatExplorer pipeline (Novák et al. 2013), using low-coverage whole-genome sequencing (WGS) datasets obtained either through de novo sequencing or from public repositories. Here, we performed a genome-wide analysis of major repeats in C. annuum using RepeatExplorer and performed FISH on one novel satellite DNA.
Materials and methods
Plant samples
Seeds of C. annuum L. (IT032384) were collected from the National Agrobiodiversity Center, Republic of Korea. The seeds were germinated in a plastic cup filled with horticulture-grade soil. Root tips were harvested when the seedlings were about 5 cm tall. The root tips were pre-treated with 2 mM 8-hydroxyquinoline solution for 5 h at 18 °C. The samples were then fixed in Carnoy’s fixative (3:1 v/v absolute ethanol: glacial acetic) overnight. Finally, the samples were stored in 70% alcohol at 4 °C for later use.
Genomic DNA extraction
Two grams of young seedling leaves of C. annuum were collected for DNA extraction. The genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) protocol described by Allen et al. (2006). The concentration of the obtained genomic DNA was measured using Colibri Microvolume Spectrometer (Titertek Berthold, Pforzheim, Germany).
Identification of satellite DNA
The C. annuum WGS reads (SRR653476) were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=BGI_Zunla_1). Read quality trimming, sampling of 0.05 × of the C. annuum genome, and read cluster were carried out using the Tandem repeat analyzer (TAREAN) (Novák et al. 2013) workflow in RepeatExplorer (Novák et al. 2017).
Probe preparation
The pre-labeled oligo nucleotide probes (PLOPs) for 5S and 45S rDNAs and Arabidopsis-type telomeric repeats were designed by Waminal et al. (2018) and provided by the Bioneer Corporation (South Korea). The probe for Ca167TR was prepared by PCR amplification using primers (Table 1) designed using Primer3Plus (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi). A TaKaRa Ex Taq® DNA Polymerase kit (TaKaRa RR001B, Kusatsu, Japan) was used for PCR amplification of Ca167TR. The PCR mixture contained 150 ng of template DNA, 2 μM each of forward and reverse primers, 0.2 mM dNTP mixture (TaKaRa), 2.5 units of Ex Taq polymerase (TaKaRa), 5 μl of 10 × Ex Taq buffer (TaKaRa), and nuclease-free water (Sigma-Aldrich RNBG3073, Darmstadt, Germany) up to a final volume of 50 μl. The PCR amplification was carried out as follows: initial denaturation at 95 °C for 5 min; followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 20 s, and extension at 72 °C for 15 s; and a final extension for 5 min at 72 °C. The PCR amplicons were labeled with biotin-16-dUTP according to the manufacturer’s protocol (Biotin-Nick Translation Mix, Roche).
Slide preparation
Chromosome preparation was carried out according to a previously descibed method (Waminal et al. 2011) with some modifications. Briefly, 2 mm long root tips were dissected and digested in 1:2 (%) ratio of pectolyase and cellulase for 90 min at 37 °C. The root tips were then washed with distilled water. The meristematic tissue was pipetted into the Carnoy’s solution (3:1 v/v absolute ethanol:glacial acetic acid), squashed, and simultaneously vortexed for 15 s. The suspension was then centrifuged at 5000 rpm for 5 min, and the supernatant was decanted carefully. The protoplast was re-suspended in acetic acid:ethanol (9:1 v/v) solution. The final suspensions were mounted on a 70 °C pre-warmed glass slide in a humidity chamber and air dried at room temperature (23–25 °C).
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) with PLOPs was based on the method described by Waminal et al. (2018). Briefly, a total 40-µl FISH mixture of 25 ng each of 5S and 45S rDNA and telomeric repeat PLOP probes, 50% formamide, 10% dextran sulfate, and 2 × SSC was prepared for each slide. The FISH mixture was placed on a chromosomal DNA slide and denatured at 80 °C on the slide heater for 5 min. Then, the hybridization was proceeded at room temperature in a humid chamber for 30 min in the dark. The steps of stringent washes were: 2 × SSC at room temperature for 10 min, 0.1 × SSC at 42 °C for 25 min, and 2 × SSC at room temperature for 5 min. Then, the slides were dehydrated through an ethanol series (70%, 90%, and 100%) at room temperature for 3 min each and dried in the dark. Finally, the slide was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Roche 70217321, Hoffmann-La Roche Ltd., Basel, Switzerland) at a ratio of 1:100 DAPI (100 µg/ml stock) to Vectashield (Vector H1000, Vector Laboratories, Burlingame, CA, USA). Slides were observed under an Olympus BX53 fluorescence microscope (Olympus Co., Tokyo, Japan) with a built-in CCD camera (CoolSNAP, Photometrics, Tucson, AZ, USA), using an oil lens (100 × magnification).
Chromosomes were washed and reprobed with Ca167TR following the method described by Lim et al. (2005). In order to remove the oil and coverslip, the slides were washed in 70% ethanol and 2 × SSC for 5 min and 10 min, respectively. Then, the slides were immersed in 4 × SSC containing 0.2% Tween-20 for 1 h, incubated in 2 × SSC containing 70% formamide for 5 min at 80 °C, and dried in the air for re-probing. The biotin-labeled Ca167TR probe was detected with streptavidin-Cy3 conjugate (Zymed Lab., USA).
Results and discussion
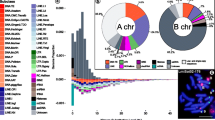
The RepeatExplorer analysis generated eight major repeats, which included one high confidence satellite repeat, three low confidence satellites repeats, three LTRs, and one rDNA (Table 2). We identified one read cluster (CL240) representing a novel high confidence 167-bp satellite repeat with a genomic proportion of 0.011%, which we named Ca167TR (Table 3). We developed a FISH probe of this tandem repeat to test its potential as a cytogenetic marker for pepper.
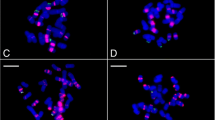
Cytogenetic studies of C. annuum, including the publications of chromosome number, karyotype by fluorescence staining (CMA/DA/DAPI), and FISH with rDNA and telomeric probes have been previously conducted (Hwang et al. 2010; Lippert et al. 1966; Moscone et al. 2007; Pickersgill 1991). Here, we observed a 2n = 24 chromosome number in C. annuum, like other previous studies (Cheema and Pant 2013; Dixit 1931; Huskins and La-Cour 1930). Our FISH analysis on sporophytic metaphase chromosomes showed one and three signals of 5S and 45S rDNA, respectively (Fig. 1), in accordance with results reported by Hwang et al. (2010). One of the 5S rDNA signals hybridized in the subtelomeric region of the short arm of chromosome 9, while one pair of major and two pairs of minor signals of 45S rDNA were detected in the subtelomeric region of the short arm of chromosomes 12 and 4, and the long arm of chromosome 10, respectively. Two pairs of Ca167TR repeat signals were detected in the subtelomeric region of the short arm of chromosome 3 and the long arm of chromosome 4. Telomeric repeat signals could be detected in the telomere region of almost both end of all chromosomes (Fig. 2; Table 4).
FISH metaphase chromosomes of C. annuum L. a 5S rDNA (green), b 45S rDNA (red), c Ca167TR repeat (magenta), d telomere (yellow) signals, and e merged. The red and magenta arrows in b, c and e indicate the weak 45S rDNA and Ca167TR repeat signals, respectively. Scale bar = 10 µm (color figure online)
FISH karyogram of C. annuum L. showing the merged signals (E) of 5S rDNA (green, A), 45S rDNA (red, B), Ca167TR (magenta, C), and Arabidopsis-type telomere (yellow, D). The red and magenta arrows in B, C, and E indicate the weak 45S rDNA and Ca167TR repeat signals, respectively. Scale bar = 10 µm (color figure online)
Hwang et al. (2010) and Romero-da Cruz et al. (2017) performed FISH-based karyotype analyses of C. annuum with rDNAs signals that could reliably distinguish two chromosomes (5S and 45S major signals). With the newly found tandem repeat Ca167TR, aside from chromosome 9 (5S signal) and 12 (45S major signal), we could reliably distinguish three more chromosomes, chromosome 3 (Ca167TR), 4 (45S minor and Ca167TR signals), and 10 (45S minor signal). Thus, Ca167TR is confirmed to be a good cytogenetic marker for C. annuum.
The genomes of higher plants comprise a mass of repetitive elements (REs). These REs have a well-defined impact on chromosome composition, which is the main reason for genome size variation (Garrido-Ramos 2015; Piegu et al. 2006). The genome size of C. annuum was approximately four times larger than that of tomato (760 Mb) (Consortium TG 2012) and potato (727 Mb) (Xu et al. 2011) genomes, mainly due to the large proportion of REs in both heterochromatic and euchromatic regions (Kim et al. 2014). The genome of C. annuum contained 80.9% of repeats (Qin et al. 2014). Usually REs like tandem repeat could be richly detected in telomere and centromere regions (Rao et al. 2010). Similarly, the newly found Ca167TR repeat of C. annuum was detected in the subtelomeric regions (Fig. 2).
Here, we have confirmed the usefulness of Ca167TR as a cytogenetic marker for C. annuum. Further FISH experiments are needed in order to localize other major repeats identified in this study. Additionally, whether these repeats affect the spiciness and stress response of C. annuum by interfering with the capsaicin biosynthetic pathway and the biotic and abiotic stress signalling pathways remains unknown and therefore, are interesting topics for future research.
References
Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325
Cai Z, Liu H, He Q, Pu M, Chen J, Lai J, Li X, Jin W (2014) Differential genome evolution and speciation of Coix lacryma-jobi L. and Coix aquatica Roxb. hybrid guangxi revealed by repetitive sequence analysis and fine karyotyping. BMC Genomics 15:1025–1040
Castro-Concha LA, Tuyub-Che J, Moo-Mukul A, Vazquez-Flota FA, Miranda-Ham ML (2014) Antioxidant capacity and total phenolic content in fruit tissues from accessions of Capsicum chinense Jacq. (Habanero Pepper) at different stages of ripening. Sci World J 2014:809073
Cheema S, Pant M (2013) Karyotype analysis of seven cultivated varieties of Capsicum annuum L. Caryologia 66:70–75
Consortium TG (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Dixit P (1931) A cytological study of Capsicum annuum. Indian J Agric Sci 1:419–433
Galian JA, Rosato M, Rossello JA (2012) Early evolutionary colocalization of the nuclear ribosomal 5S and 45S gene families in seed plants: evidence from the living fossil gymnosperm Ginkgo biloba. Heredity 108:640–646
Galindo-González L, Mhiri C, Deyholos MK, Grandbastien M-A (2017) LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25
Garrido-Ramos MA (2015) Satellite DNA in plants: more than just rubbish. Cytogenet Genome Res 146:153–170
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Huskins C, La-Cour L (1930) Chromosome numbers in Capsicum. Am Nat 64:382–384
Hwang YJ, Lee SN, Song KA, Ryu KB, Ryu KH, Kim HH (2010) Karyotype analyses of genetically modified (GM) and Non-GM hot peppers by conventional staining and FISH method. Hortic Environ Biotechnol 51:525–530
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607
Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278
Kwon J-K, Kim B-D (2009) Localization of 5S and 25S rRNA genes on somatic and meiotic chromosomes in Capsicum species of chili pepper. Mol Cells 27:205–209
Li S-F, Su T, Cheng G-Q, Wang B-X, Li X, Deng C-L, Gao W-J (2017) Chromosome evolution in connection with repetitive sequences and epigenetics in plants. Genes 8:290–308
Lim K-B, De Jong H, Yang T-J, Park J-Y, Kwon S-J, Kim JS, Lim M-H, Kim JA, Jin M, Jin Y-M (2005) Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Mol Cells 19:436–444
Lippert L, Smith P, Bergh B (1966) Cytogenetics of the vegetable crops. Garden pepper, Capsicum sp. Bot Rev 32:24–55
Liu Z, Liu Y, Liu F, Zhang S, Wang X, Lu Q, Wang K, Zhang B, Peng R (2018) Genome-wide survey and comparative analysis of long terminal repeat (LTR) retrotransposon families in four Gossypium species. Sci Rep 8:9399
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genomics Proteomics Bioinform 12:164–171
Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE, Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Hortic 745:137–170
Muratović E, Robin O, Bogunić F, Šoljan D, Siljak-Yakovlev S (2010) Karyotype evolution and speciation of European lilies from Lilium sect. Liriotypus. Taxon 59:165–175
Novák P, Neumann P, Pech J, Steinhaisl J, Macas J (2013) RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29:792–793
Novák P, Ávila Robledillo L, Koblížková A, Vrbová I, Neumann P, Macas J (2017) TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res 45:e111–e111
Paran I, Ben-Chaim A, Kang B-C, Jahn M (2007) Capsicums. In: Kole C (ed) Genome mapping and molecular breeding in plants, vol 5. Vegetables. Springer, Berlin, pp 209–226
Park M, Park J, Kim S, Kwon JK, Park HM, Bae IH, Yang TJ, Lee YH, Kang BC, Choi D (2012) Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. Plant J 69:1018–1029
Pertea M (2012) The human transcriptome: an unfinished story. Genes 3:344–360
Pickersgill B (1991) Cytogenetics and evolution of Capsicum L. Chromosome Eng Plants Genet Breed Evol 33:139–160
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci 111:5135–5140
Rao SR, Trivedi S, Emmanuel D, Merita K, Hynniewta M (2010) DNA repetitive sequences-types, distribution and function: a review. J Cell Mol Biol 7:1–11
Richard G-F, Kerrest A, Dujon B (2008) Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72:686–727
Rohami M, Mohammadi A, Khosroshahli M, Ahmadi H, Darandeh N (2010) Karyotype analysis of several ecotypes of Capsicum annuum L. in Iran. Not Bot Horti Agrobot Cluj-Napoca 38:177–180
Romero-da Cruz MV, Urdampilleta JD, Forni Martins ER, Moscone EA (2017) Cytogenetic markers for the characterization of Capsicum annuum L. cultivars. Plant Biosyst 151:84–91
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K, Weichselgartner T, Kemena C, Stökl J, Schultner E (2014) Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun 5:5495
Waminal NE, Kim NS, Kim HH (2011) Dual-color FISH karyotype analyses using rDNAs in three Cucurbitaceae species. Genes Genomics 33:521–528
Waminal NE, Pellerin RJ, Kim N-S, Jayakodi M, Park JY, Yang T-J, Kim HH (2018) Rapid and efficient FISH using pre-labeled oligomer probes. Sci Rep 8:8224
Wang D, Su Y, Wang X, Lei H, Yu J (2012) Transposon-derived and satellite-derived repetitive sequences play distinct functional roles in mammalian intron size expansion. Evol Bioinform Online 8:301–319
Witsenboer H, Michelmore R, Vogel J (1997) Identification, genetic localization, and allelic diversity of selectively amplified microsatellite polymorphic loci in lettuce and wild relatives (Lactuca spp.). Genome 40:923–936
Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
Acknowledgements
This paper was supported by the Sahmyook University Research Fund in 2017 (Grant no. RI12017006).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, H.C., Waminal, N.E. & Kim, H.H. In silico mining and FISH mapping of a chromosome-specific satellite DNA in Capsicum annuum L.. Genes Genom 41, 1001–1006 (2019). https://doi.org/10.1007/s13258-019-00832-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00832-8