Abstract
The pig provides meat products for human consumption on a large scale. High-throughput sequencing technology provides a powerful approach for the characterization of the muscle transcriptome of pigs. Despite many studies using high-throughput technologies, the functional complexity of porcine muscle transcriptome is not fully elucidated. Therefore, the aim of this study was to gain insight into the longissimus lumborum muscle transcriptome between two pig breeds with distinct phenotypes, Laiwu pig, a Chinese indigenous pig breed, and Yorkshire pig, a Western breed. RNA-seq was applied to sample transcripts of the longissimus lumborum muscle between the two pig breeds to gain insights into whole-genome transcription profiles. For the Laiwu and Yorkshire libraries, we obtained a total of 40,498,476 and 59,072,892 high quality reads, respectively. Moreover, the resulting data set provided expression patterns for many annotated, predicted and novel pig genes. 178 significantly differentially expressed genes were identified between the Laiwu and Yorkshire pigs, with 98 up-regulated and 80 down-regulated genes in Laiwu pig compared with Yorkshire pig. Gene expression results of the RNA-seq data were validated by qRT-PCR for twelve genes. Genes that were differentially expressed were involved in lipid metabolism and were more highly expressed in Laiwu pigs. The present study provides a comprehensive view of differences in the muscle transcriptome between two pig phenotypes. These results expand our knowledge of the genes transcribed in the skeletal muscle of two breeds and provide a basis for future research of the molecular mechanisms underlying the phenotypic differences on meat quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Comparative transcriptome analysis between different breeds plays an important role for our view of genome organization complexity, and of the transcriptional landscape of tissues and cells (Freeman et al. 2012). In the past decade, microarray hybridization technology has commonly been used for studying global gene expression (Bonnet et al. 2011; Kim et al. 2010). In recent years, sequencing technology has advanced rapidly and now offers many advantages over microarrays, largely because RNA deep sequencing (RNA-seq) focuses on the reconstruction of the entire transcriptome (Martin and Wang 2011). Therefore, RNA-seq provides a fuller comprehension of gene structures and expression patterns (Ozsolak and Milos 2011). Through RNA-seq of the transcriptome fraction it is possible to not only understand gene expression models, but to identify new transcripts, single nucleotide polymorphisms (SNPs), and alternative splicing events.
The pig (Sus scrofa) is an important domesticated animal for agricultural production, and provides large quantities of meat for human consumption. Furthermore, pigs provide an important model system for medical research (Li et al. 2012; Walters et al. 2012). To elucidate tissue- and breed-specific differentially expressed genes, researchers use whole genome profiling techniques to decipher genes related to phenotypes. In the past few years, the porcine transcriptome of different tissues has been profiled to address fundamental biological questions using a variety of different approaches. For example, transcriptome profiling of skeletal muscle and backfat tissue has been investigated using the serial analysis of gene expression method, and microarray technology (Óvilo et al. 2014; Ropka-Molik et al. 2015; SanCristobal et al. 2015; Zhao et al. 2015). This has led to many advances in our understanding of the porcine transcriptome.
Western pigs are mostly lean pig breeds. Pig breeders have emphasized fast growth during the past two decades, and there has been an associated deterioration in meat quality. Chinese indigenous pig breeds have a much higher fat content, especially intramuscular fat, than Western pig breeds (Chen et al. 2013). Furthermore, the Chinese indigenous pigs exhibit excellent meat quality (Chen et al. 2011b). The Yorkshire (YS) pig is a Western breed frequently used for commercial production. The Laiwu (LW) pig is a typical Chinese indigenous black pig breed, characterized by high intramuscular fat (IMF) content, and is mainly raised in northern China, especially in Laiwu city of Shandong Province. Because of the deterioration of meat quality in Western pigs, there has been an increasing interest in improving meat quality though use of Chinese indigenous pig breeds by producers. Unfortunately, breed-specific differences in genetics are not fully understood, and the longissimus lumborum muscle transcriptome in Laiwu pigs has not been elucidated. Therefore, elucidating the differences in the transcriptome of longissimus lumborum muscle between the LW and YS pig breeds is essential for interpreting the function of differentially expressed genes, and their relation to different phenotypes.
Here, we describe how we applied the RNA-seq approach to analyze the transcriptome of porcine longissimus lumborum muscle, and to investigate the molecular basis of differences in transcriptome profile between LW pig and YS pig. Using advanced visualization and clustering analysis techniques, we determined gene expression, identified differentially expressed genes between the two pig breeds.
Methods
Animal materials and sample collection
The animals used for RNA-seq in this study were reported in our previous research. Briefly, three LW pigs and three YS pigs, each aged 280-day-old, were reared at a LW pig breeding farm in Laiwu city, Shandong Province, China, and were fed commercial pig feed and water ad libitum. The longissimus lumborum muscles located at the last rib were sampled within 5 min of euthanasia, and immediately frozen in liquid nitrogen. The samples were stored at −80 °C until required for RNA isolation. Longissimus lumborum muscles were sampled for IMF content evaluation 24 h after slaughter. Based on the “Technical regulation for determination of pork quality (NY/T 821-2004)”, IMF content were evaluated according to the Soxhlet petroleum-ether extraction method and IMF content was expressed as the weight percentage of wet muscle tissue. Another muscle samples from 24 barrows (12 Laiwu pigs and 12 Yorkshire pigs) were used for the qRT-PCR to validate the differentially expressed genes.
RNA isolation
Total RNA was extracted from the longissimus lumborum muscle using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA). The resultant total RNA was then treated with DNase I, RNase-free (Promega, Madison, WI, USA) to eliminate genomic DNA contamination, in accordance with manufacturer’s instructions. The quantity and quality of isolated total RNAs were estimated by running on a 1% agarose gel, and using a Bio-Photometer (Eppendorf, Hamburg, Germany). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The RNA was stored at −80 °C until required.
Transcriptome sample preparation and sequencing
3 μg of RNA per sample was used as input material for the RNA sample preparations. All six RNA samples had RNA integrity number (RIN) values above eight. The libraries were sequenced using Illumina High-seq 2000 technology. Sequencing libraries were generated using an Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, USA) following manufacturer’s instructions. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads.
Fragmentation was performed using divalent cations under an elevated temperature in Illumina proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and SuperScript II. Second strand cDNA synthesis was performed using DNA polymerase I and RNase H. Overhangs were converted into blunt ends via exonuclease/polymerase activities and enzymes removed. After adenylation of 3′ ends of DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. The library fragments were purified using an AMPure XP system (Beckman Coulter, Beverly, USA). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using Illumina PCR Primer Cocktail in a ten cycle PCR reaction. Products were purified (AMPure XP system) and quantified using the Agilent high sensitivity DNA assay on an Agilent Bioanalyzer 2100 system (Santa Clara, CA, USA). Clustering of index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2000 platform, and 100 bp paired-end reads generated.
Mapping of the reads and transcripts assembly
Reference genome and gene model annotation files were downloaded from the pig genome website (http://www.ensembl.org/Sus_scrofa/Info/Index). The index of the reference genome was built using Bowtie v0.12.8 (Langmead and Salzberg 2012), and paired-end clean reads were aligned with the reference genome using TopHat v1.4.0 (Trapnell et al. 2009). The Cufflinks v1.3.0 Reference Annotation Based Transcript assembly method was used to construct and identify both known and novel transcripts from the TopHat alignment results (Trapnell et al. 2010).
Quantification and clustering of differentially expressed genes
HTSeq v0.5.3 was used to count read numbers mapped to each gene (Anders 2010). The differentially expressed genes between LW and YS pigs were qualified using the DEGseq R package (v1.28.0), using read counts per gene as a measure of expression (Wang et al. 2010). The P values were adjusted using the Benjamini and Hochberg method (Benjamini and Hochberg 1995). Corrected P value (q value) of 0.05 and log2 (Fold change) of ≥1 were set as the threshold for significantly differential expression. In addition, the hierarchical method was used for clustering analysis of differentially expressed genes.
GO and KEGG enrichment analysis of differentially expressed genes
Gene ontology enrichment analysis of differentially expressed genes was implemented by the GOseq R package (Young et al. 2010), in which gene length bias was corrected. KOBAS software was used to test statistical enrichment of differentially expressed genes in the KEGG (http://www.genome.jp/kegg/) pathways (Mao et al. 2005). The resulting P values of KEGG enrichment were adjusted, and taking corrected P value (q value) ≤0.05 as a threshold of significance.
Validation of differentially expressed genes by real-time quantitative RT-PCR
To evaluate the repeatability and reproducibility of gene expression data obtained by RNA-Seq, we performed qRT-PCR on 12 randomly selected genes. The primers were designed by Primer3web (version 4.0.0) based on the National Center for Biotechnology Information published sequences, and the primer sequences listed in Supplementary material 1. Total RNA was extracted from different pig breeds using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The total RNA was quantified by measurement of optical density at 260 nm, and the integrity of the total RNA was detected by 1% agarose gel electrophoresis. Ratios of absorption (260/280 nm) of all preparations were between 1.8 and 2.0. Then, the total RNA was reverse transcribed to cDNA according to the manufacturer’s instructions of the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). All qRT-PCRs were performed using the Mx3000p Real-Time System (Stratagene, La Jolla, CA, USA), with ACTB gene as the reference gene; the comparative Ct method was used to analyze the qRT-PCR results. The qRT-PCR was performed in triplicate with SYBR® Premix Ex Taq™ (Takara, Dalian, China) using the following program: 95 °C for 30 s; 40 cycles of 95 °C for 5 s, 58 °C for 30 s, and 72 °C for 30 s. To exclude between-run variations, all samples were amplified in triplicates and the mean was used for further analysis. The threshold cycle (CT) was determined using the default threshold settings of the Mx3000p system, and the relative expression of miRNAs was analyzed using the 2−ΔΔCT method.
Results
Loin IMF content between LW and YS pigs
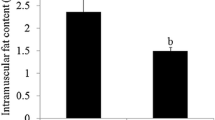
Mean live weight at slaughter was 91.08 kg (SD = 0.95 kg). There was no significant difference in live weight between LW and YS pigs. The percentage of IMF was significantly (P < 0.01) higher in LW (13.83%) pigs than YS (1.54%) pigs. The results showed the differential trend in fat deposition and meat quality.
RNA sequencing and mapping to the pig transcriptome
RNA sequencing and mapping were employed to analyze the functional complexity of the porcine muscle transcriptome. Two cDNA sequencing libraries were constructed using longissimus lumborum muscles prepared from three LW and three YS pigs. The libraries were sequenced using Illumina High-seq 2000 technology, and two sets of transcriptomics reads obtained. The total amount sequenced was 4.04 gigabases (Gb) for LW pig, and 5.90 Gb for YS pig. Sequence reads were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRP023252. All short reads were aligned onto the whole reference genome (Sscrofa 10.2). After ambiguous mapping with Tophat (Trapnell et al. 2009), more than 80% of reads were mapped to the pig reference genome (Supplementary material 2). Approximately 75.13% of reads had a unique genomic location in LW pig, and 75.35% in YS pig.
The proportion of reads mapped to annotated exons was calculated. 83.50% of the total mapped reads in LW pig were located in exons, and 85.98% in YS pig. 4.46% of the mapped reads were located in the introns of LW pig, and this was higher than the 3.34% recorded for YS pig; the remaining 10.68–12.04% of reads were assigned to intergenic regions. RNA-seq data analysis revealed the extensive expression of the whole pig genome. The density of the total mapped reads in chromosomes was also statistically analyzed. According to the distribution analysis, the longer the length of chromosome, the more the total reads within the chromosome. The mapped reads number in the chromosome analysis demonstrated that the result was in accordance with the read density in a chromosome.
Gene expression and annotation between the two pig breeds
RNA-seq analysis revealed extensive gene expression differences between the two breed’s porcine muscles. To achieve a comprehensive overview of specifically- and co-expressed genes we evaluated gene expression levels by counting the number of reads per kilobase of exon model per million mapped reads (RPKM), according to the Mortazavi et al. (2008) method. The distribution of reads in the two RNA-seq libraries had some similarities. The expression distribution revealed that approximately 97% of genes were expressed at less than 100 RPKM, and those genes were deemed as having a low abundance gene expression level; no more than 2% of genes were deemed to be expressed with medium abundance (100–1000 RPKM); and less than 1% of genes were expressed greater than 1,000 RPKM (Table 1). The maximum expression level of an annotated gene was 19,044.8 and 26,207.6 RPKM in LW and YS, respectively. There were over 16,000 genes expressed in the longissimus lumborum muscle of each pig breed. Of the genes with a mean RPKM higher than zero, 16,363 were expressed in LW pig and 17,067 in YS pig, and 15,731 genes were identical between the two pig breeds.
Differentially expressed genes analysis
To better survey the differences of muscle transcriptome, it is important to identify the differentially expressed genes between two different pig breeds. There were 178 differentially expressed genes (log2 Fold change ≥1 or ≤−1; P value < 0.05) that were detected between the two pig breeds. Of the 178 differentially expressed genes (Supplementary material 3) in muscle, there were 98 up-regulated and 80 down-regulated genes in LW pig in comparison to YS pig.
For further validation of our RNA-seq expression profile data, we performed real-time quantitative RT-PCR (qRT-PCR) assays on 12 randomly selected genes. The expression patterns of the 12 genes were generally consistent with the RNA-seq results. The qRT-PCR results were in a similar direction and magnitude compared to RNA-seq, and there was a significant correlation between the fold-change values estimated in longissimus lumborum muscle from the RNA-seq and qRT-PCR results (Supplementary material 4). Overall, these results suggesting that the results of the RNA-seq experiments were accurate and reliable.
GO and KEGG pathways analysis
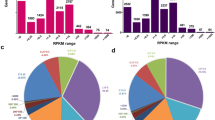
To investigate the physiological functions and biological processes of the 178 differentially expressed genes, we determined their functional category by querying associated gene ontologies. There were 222 biological process GO terms enriched between the two pig breeds in this study. The GO annotation enrichment results revealed organic acid metabolic process, protein polymerization, glucose metabolic process, and gluconeogenesis to be the most significantly enriched biological process GO terms (Fig. 1).
Gene ontology (GO) analysis of differentially expressed genes between the LW and YS pigs. The identified 178 differentially expressed genes were classified into three categories: cellular component, molecular function, and biological process. The percentage and numbers of genes in each GO term are shown above
Mining of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using the 178 differentially expressed genes retrieved 112 pathways. The number of differentially expressed genes varied from one to fifteen per pathway. The most represented pathways were the peroxisome proliferator-activated receptors (PPAR) signaling pathway, insulin signaling pathway, adipocytokine signaling pathway, propanoate metabolism, pyruvate metabolism, fatty acid metabolism, and fatty acid biosynthesis. However, among the KEGG pathways represented, only the PPAR signaling pathway involved in 8 lipid metabolism genes was significantly enriched (q < 0.05) (Table 2). These results suggested that the genes involved in PPAR signaling pathway maybe important for IMF deposition in Laiwu pigs.
Protein–protein interaction analysis
To gain a better understanding of the biological relationships between differentially expressed genes, STRING was used and the protein–protein interaction networks were identified (Fig. 2). In the core of the protein–protein interaction network, genes belonged to more than one module. ACACB was the core of the network with the highest connection degrees. In addition, ACADM and FASN were also upregulated in Laiwu pig. The qRT-PCR results show that the expression of ACACB, ACADM and FASN was significantly higher in LW than YS pigs (Supplementary material 5). Because these genes that played a role in the organic acid metabolic process and fatty acid metabolic process were up-regulated in the LW pig. Therefore, these results suggest that those genes related lipid metabolism maybe reveals the IMF deposition in LW pigs.
Protein–protein interaction network of differentially expressed genes analyzed by STRING. Lines of different color represent seven types of evidence used in predicting associations. Red line fusion evidence; green line: neighborhood evidence; blue line co-occurrence evidence; purple line experimental evidence; yellow line text mining evidence; light blue line database evidence; black line co-expression evidence. (Color figure online)
Discussion
High-throughput sequencing is widely used as a powerful tool for the identification of differentially expressed genes, alternative splicing, and other genetic traits in various organisms. We applied the method to various analyses, including gene expression assessment and AS identification; and also identified and analyzed differentially expressed genes between LW and YS pig breeds.
It is important to identify the differentially expressed genes related to meat quality, as conferred through the biological importance of muscle tissue between LW and YS pig breeds. A study identified 336 differentially expressed genes in muscle when using the 454 sequencing system (Nie et al. 2011); but another research has reported over 800 differentially expressed genes in the muscle of pigs when using RNA-seq (Chen et al. 2011a). The differentially expressed genes in our study are mainly associated with metabolic processes.
Previously, we showed that the LW pig has a higher level of intramuscular fat than the YS pig (Chen et al. 2013). In this study, our results suggest that the differentially expressed genes are likely related to the phenotypes of fat deposition, antioxidant metabolism. For example, ACADM (acyl-CoA dehydrogenase, C-4 to C-12 straight chain), and UCP3 (uncoupling protein 3) genes were up-regulated in LW when compared with YS. This is in agreement with Kim et al. (2006), who reported that ACADM gene is a functional candidate gene for fatness and body composition traits, and Nowacka-Woszuk et al. (2008), who reported that UCP3 gene is important for intramuscular fat. Moreover, these two genes were enriched for the GO term fatty acid metabolic process in the present study. Consistent with previous reports (Damon et al. 2012; Hamill et al. 2013), GO analysis indicated that the differentially expressed genes were enriched in functions related to the fatty acid metabolic process. Furthermore, KEGG functional annotation revealed up-regulation of a number of genes with functions in metabolism, including fatty acid metabolism, such as FACL2, CPT1B, PDK4, CPT1B, and FASN in LW pig. Interestingly, the PDK4 gene was also highly expressed in the skeletal muscle of Chinese Meishan pig (Lan et al. 2009); and the FASN gene is a candidate gene for lipid metabolism (Bakhtiarizadeh et al. 2013). In addition to muscle function related to intramuscular fat, oxidative phosphorylation is also a represented GO term through the UCP3 and glutathione peroxidase 1 (GPX1) genes. These two genes display an increased expression in LW pig compared with YS pig, suggesting that the ability to respond to oxidative stress increases in LW pigs, which showed a strong antioxidant capacity in our previous research (Chen et al. 2011b).
Conclusion
This study aimed at identifying biological events that underlie differences in muscle physiology and meat quality traits between LW and YS pig breeds. The RNA-seq approach proved an efficient method for gaining a comprehensive insight into the transcriptome profiles and differential gene expression of the two pig breeds. This transcriptome information will be useful for further understanding pig muscle physiology. 178 significantly differentially expressed genes were identified in the longissimus lumborum muscle of LW pig compared with YS pig.
References
Anders S (2010) HTSeq: Analysing high-throughput sequencing data with Python. http://www-huber.embl.de/users/anders/HTSeq/doc/index.html
Bakhtiarizadeh MR, Moradi-Shahrbabak M, Ebrahimie E (2013) Underlying functional genomics of fat deposition in adipose tissue. Gene 521:122–128
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Bonnet A, Bevilacqua C, Benne F, Bodin L, Cotinot C, Liaubet L, Sancristobal M, Sarry J, Terenina E, Martin P et al (2011) Transcriptome profiling of sheep granulosa cells and oocytes during early follicular development obtained by laser capture microdissection. BMC Genom 12:417
Chen C, Ai H, Ren J, Li W, Li P, Qiao R, Ouyang J, Yang M, Ma J, Huang L (2011a) A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. BMC Genom 12:448
Chen W, Zeng Y, Cui J, Chen Q, Du J, Yang L, Hu Y, Song Y, Qian Y (2011b) Effects of phospholipid hydroperoxide glutathione peroxidase mRNA expression on meat quality of M. longissimus dorsi in pigs. Eur Food Res Technol 232:433–440
Chen Q-M, Wang H, Zeng Y-Q, Chen W (2013) Developmental changes and effect on intramuscular fat content of H-FABP and A-FABP mRNA expression in pigs. J Appl Genet 54:119–123
Damon M, Wyszynska-Koko J, Vincent A, Hérault F, Lebret B (2012) Comparison of muscle transcriptome between pigs with divergent meat quality phenotypes identifies genes related to muscle metabolism and structure. PLoS ONE 7:e33763
Freeman T, Ivens A, Baillie JK, Beraldi D, Barnett M, Dorward D, Downing A, Fairbairn L, Kapetanovic R, Raza S et al (2012) A gene expression atlas of the domestic pig. BMC Biol 10:90
Hamill R, Aslan O, Mullen A, O’Doherty J, McBryan J, Morris D, Sweeney T (2013) Transcriptome analysis of porcine M. semimembranosus divergent in intramuscular fat as a consequence of dietary protein restriction. BMC Genom 14:453
Kim JH, Lim HT, Park EW, Rodríguez C, Silio L, Varona L, Mercade A, Jeon JT, Ovilo C (2006) Polymorphisms in the promoter region of the porcine acyl-coA dehydrogenase, medium-chain (ACADM) gene have no effect on fat deposition traits in a pig Iberian × Landrace cross. Anim Genet 37:430–431
Kim N-K, Park H-R, Lee H-C, Yoon D, Son E-S, Kim Y-S, Kim S-R, Kim O-H, Lee C-S (2010) Comparative studies of skeletal muscle proteome and transcriptome profilings between pig breeds. Mamm Genome 21:307–319
Lan J, Lei M-G, Zhang Y-B, Wang J-H, Feng X-T, Xu D-Q, Gui J-F, Xiong Y-Z (2009) Characterization of the porcine differentially expressed PDK4 gene and association with meat quality. Mol Biol Rep 36:2003–2010
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Meth 9:357–359
Li M, Wu H, Luo Z, Xia Y, Guan J, Wang T, Gu Y, Chen L, Zhang K, Ma J et al (2012) An atlas of DNA methylomes in porcine adipose and muscle tissues. Nat Commun 3:850
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Martin JA, Wang Z (2011) Next-generation transcriptome assembly. Nat Rev Genet 12:671–682
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Meth 5:621–628
Nie Q, Fang M, Jia X, Zhang W, Zhou X, He X, Zhang X (2011) Analysis of muscle and ovary transcriptome of Sus scrofa: assembly, annotation and marker discovery. DNA Res 18:343–351
Nowacka-Woszuk J, Szczerbal I, Fijak-Nowak H, Switonski M (2008) Chromosomal localization of 13 candidate genes for human obesity in the pig genome. J Appl Genet 49:373–377
Óvilo C, Benítez R, Fernández A, Núñez Y, Ayuso M, Fernández AI, Rodríguez C, Isabel B, Rey AI, López-Bote C et al (2014) Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics. BMC Genom 15:413
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Ropka-Molik K, Żukowski K, Eckert R, Piórkowska K, Oczkowicz M, Gurgul A, Szmatoła T (2015) Whole transcriptome analysis of the porcine muscle tissue of breeds differing in muscularity and meat quality traits. Livest Sci 182:93–100
SanCristobal M, Rohart F, Lascor C, Bouffaud M, Trouilh L, Martin PG, Lippi Y, Tribout T, Faraut T, Mercat MJ et al (2015) Exploring transcriptomic diversity in muscle revealed that cellular signaling pathways mainly differentiate five Western porcine breeds. BMC Genom 16:1055
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech 28:511–515
Walters E, Wolf E, Whyte J, Mao J, Renner S, Nagashima H, Kobayashi E, Zhao J, Wells K, Critser J et al (2012) Completion of the swine genome will simplify the production of swine as a large animal biomedical model. BMC Med Genom 5:55
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Young M, Wakefield M, Smyth G, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
Zhao Y, Li J, Liu H, Xi Y, Xue M, Liu W, Zhuang Z, Lei M (2015) Dynamic transcriptome profiles of skeletal muscle tissue across 11 developmental stages for both Tongcheng and Yorkshire pigs. BMC Genom 16:377
Acknowledgements
This study was supported by the National Animal Breed Resource Preservation Project of China (2130135), the National Project for Breeding Transgenic Pigs of China (2013ZX08006-002), Shandong Province Modern Pig Technology and Industry System Project (SDAIT-08-02) and Shandong Province Agricultural Animal Breeding Project of China (2013LZ02-015, 2014LZ03-016).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Wei Chen, Guo-Feng Fang, Shou-Dong Wang, Hui Wang andYong-Qing Zeng declares that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, W., Fang, Gf., Wang, Sd. et al. Longissimus lumborum muscle transcriptome analysis of Laiwu and Yorkshire pigs differing in intramuscular fat content. Genes Genom 39, 759–766 (2017). https://doi.org/10.1007/s13258-017-0540-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-017-0540-9