Abstract
DNA binding with one finger (Dof) transcription factors are unique in plants, characteristically composed of a Dof domain in structure, functionally involved in mediating many processes of plant growth and development. In this study, we report a genome-wide analysis for Dof gene family in castor bean (Ricinus communis L., Euphorbiaceae) genome. In total, 21 RcDof genes were identified and their constructional characterization, classes and phylogenetic relationships were documented. The expression profiles of the 21 RcDof genes among different tissues were inspected using the high throughput RNA-Seq data. Finally, SqRT-PCR analysis was performed to test the transcriptional responses of the 21 RcDof genes to abscisic acid (ABA) and/or gibberellic acid (GA) signals, resulting in the identification of 18 RcDof genes responsive to ABA and/or GA signals. Thus, this study would provide basic information in understanding the molecular basis of the RcDof family and their potential function in regulating the growth and development of castor bean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA binding with one finger (Dof) transcription factors, characterized by the conserved DNA-binding domain called Dof domain, exist uniquely in plants. The Dof domain contains a single C2/C2-type (CX2CX21CX2C-type) zinc-finger-like motif composed of 52 amino acid residues, binding specifically to the core sequence AAAG (Yanagisawa and Schmidt 1999; Umemura et al. 2004), with an exception of recognizing an AGTA element (the pumpkin AOBP) (Kisu et al. 1998). Four absolute conserved cysteine residues can covalently bind to the zinc ion, essential for the activity of Dof proteins (Yanagisawa 1995, 2004). The Dof proteins are structurally characterized by an N-terminal conserved DNA-binding domain and a C-terminal transcription-regulating domain (Yanagisawa 2002). The Dof domain is known to be a bi-functional domain that mediates not only DNA-binding but also protein–protein interactions (Vicente-Carbajosa et al. 1997). The amino acids of the C-terminal transcription-regulating domain are variable, leading to the functional diversities of Dof members in plants, such as carbon–nitrogen metabolism (Yanagisawa 2000; Kumar et al. 2009), light responses and photoperiodic flowering (Yang et al. 2010; Song et al. 2012), nutrition storage during seed development (Dong et al. 2007; Wang et al. 2007; Marzábal et al. 2008), pollen maturation (Chen et al. 2012), stomata guard cell specific gene regulation (Plesch et al. 2001), and cell cycle regulation (Skirycz et al. 2008).
Since the first event of Dof protein isolation (ZmDof1) from maize (Yanagisawa and Izui 1993), many Dof genes have been functionally identified or predicted in a genome-wide scale from green unicellular algae to higher plants. Only one Dof gene was identified in the green alga Chlamydomonas reinhardtii (Shigyo et al. 2007), and there were 19, 8 and 8 Dof genes respectively in the moss Physcomitrella patens (Shigyo et al. 2007), the fern Selaginella moellendorffii and the gymnosperm Pinus taeda (Moreno-Risueno et al. 2007b). In angiosperms, the number of Dof genes seems widely variable among species. For instance, there were 36 (in Arabidopsis, Lijavetzky et al. 2003), 30 (in rice, Lijavetzky et al. 2003), 41 (in poplar, Yang et al. 2006), 26 (in barley, Moreno-Risueno et al. 2007b), 31 (in wheat, Shaw et al. 2009), 28 (in sorghum, Kushwaha et al. 2011), 78 (in soybean, Guo and Qiu 2013) and 34 (in tomato, Cai et al. 2013) Dof genes identified from genome-based surveys. Based on the sequence similarity of amino acids in the Dof domain region, Dof family was categorized into seven subfamilies A-G (Moreno-Risueno et al. 2007b). These subfamilies might represent the functionally different groups in regulating the growth and development of plants.
Castor bean (Ricinus communis L.), a model plant in the family Euphorbiaceae, is an important non-edible oil seed crop. Due to the high economic value, castor bean is widely cultivated in many countries, particularly in India, China and Brazil (Qiu et al. 2010). The biosynthesis of lipids in oilseeds is highly regulated by physiological and genetic factors, like light, oxygen availability and transcriptional factors (Baud and Lepiniec 2010). Previous study indicated that the Dof genes (GmDof4 and GmDof11) might be involved in regulating lipid biosynthesis in soybean seeds (Wang et al. 2007). Several PBFs (prolamin-box binding factors) were implicated in regulation of storage proteins at the beginning of seed filling (Yamamoto et al. 2006; Dong et al. 2007; Marzábal et al. 2008). Though the genome sequencing has been completed (Chan et al. 2010), functions of most genes are unclear. In particular, the mechanism of oil accumulation in oilseeds is far from understanding. Based on the genomic and transcriptional data, this study focuses on identification and characterization of Dof transcription factors in castor bean to dissect the molecular basis of the Dof genes and their potential function in regulating the growth and development in plants.
Materials and methods
Database search for castor bean Dof genes
The Dof domain sequences were used as query to perform a blastp search in the castor bean genome with default blast settings and an expect value set 1.0 at TIGR database (http://castorbean.jcvi.org/index.php). The SMART (http://smart.embl-heidelberg.de/) and pfam (http://pfam.sanger.ac.uk/) were further applied to confirm the presence of Dof domain in these putative Dof proteins. The previously identified Arabidopsis 36 Dof protein sequences were obtained from the DATF database (http://datf.cbi.pku.edu.cn/). To predict cis-acting regulatory elements in the promoter regions of all castor bean Dof genes, 1,500 bp genomic DNA sequences upstream of the initiation codon (ATG) were surveyed against the PlantCARE database (Lescot et al. 2002).
Phylogenetic analysis and sequence structure
Multiple sequence alignments were performed in Clustal W (version 2.0, Larkin et al. 2007) with default parameters. Phylogenetic trees were constructed with Neighbor-Joining (NJ) criteria using MEGA (version 5.2, Tamura et al. 2011), with 1,000 bootstrap replicates. The conserved motifs among those Dof proteins (with E value <0.1) were identified using the online MEME analysis tool (http://meme.ebi.edu.au/meme/intro.html) with following parameters: optimum motif width set to ≥6 and ≤200; maximum number of motifs set to 50. To predict the exon/intron arrangements of the Dof genes, alignments of coding sequences against the genomic sequences were performed using the online GSDS analysis tool (http://gsds.cbi.pku.edu.cn/).
Global gene expression analysis
To gain insights into the expression patterns of Dof genes among different tissues during development, the global gene expression analysis was performed using the high throughput RNA-Seq data from five tissues/organs including developing endosperms at the early stage of oil accumulation and the oil fast accumulation stage, germinating seeds, leaves and developing male flowers, available online at the European Nucleotide Archive (http://www.ebi.ac.uk/ena) with accession number ERA047687 (Brown et al. 2012). The expression profiles of Dof genes among different tissues were visualized by a heatmap according to the [Fragments Per Kilobase of exon per Million (FPKM) fragments mapped] values.
Plant materials, treatments and SqRT-PCR
Castor bean seeds were germinated and grown in a greenhouse in pots. For treatments, 100 µM ABA (abscisic acid) and 100 µM gibberellic acid (GA) were sprayed on leaves of seedlings at the three-leaf stage. The leaves were harvested after 40 h and immediately frozen in liquid nitrogen and stored at −80 °C for further use. Total RNAs were extracted using RNAiso Plus (Takara, Dalian) according to the manufacturer’s protocol. To obtain the first-strand cDNA, 1 µg of RNA was used as the template for cDNA synthesis with the cDNA Synthesis SuperMix (Transgen, Beijing). For the SqRT-PCR, gene specific primers were given in Supplementary Table S1, and a castor bean Actin gene was used as an internal control. The PCR reaction conditions were set: denaturing at 94 °C for 3 min, followed by 25 cycles of denaturing at 94 °C for 30 s, annealing temperature of each pair of primers for 30 s and extension at 72 °C for 1 min, and a final elongation step at 72 °C for 5 min. Each reaction was performed in triplicates. The amplified products were tested on a 1.0 % agarose gel and then the signal intensity was quantified using Bio-Rad Quantity One software (Bio-Rad Laboratories, CA, USA).
Results
Identification of Dof genes in castor bean
By searching putative Dof genes against castor bean genome database using Dof domain sequence, 23 Dof amino acid sequences were identified. Each of identified 23 Dof amino acid sequences exhibited a Dof domain with significant E value (<1.0). However, two proteins encoded by genes 29813.m001503 and 30025.m000581 were found to have incomplete Dof domain, both lacking of the two cysteine residues near the N terminal. Consequently, the two genes were excluded for further analyzed, and the other 21 were consecutively named as RcDof-1, RcDof-2 to RcDof-21 based on their gene locus (Table 1). The 21 RcDof genes encoding proteins ranged from 148 (RcDof-11) to 506 (RcDof-14) amino acids (aa) in length with an average of 323. Domain analysis revealed that all the RcDof proteins had a typical zinc-finger Dof-type profile with four conserved cysteine residues, essential for the zinc finger configuration and required for loop stability (Fig. 1). The Dof domain sequences showed highly conserved sequences with 28 out of 52 amino acids being 100 % identical in the 21 RcDof proteins, and all presented before 200 aa of N terminal. Dof genes identified from Arabidopsis were used to assign orthologous for castor bean (detailed information is listed in Table 1).
Phylogenetic relationships and gene structure analysis
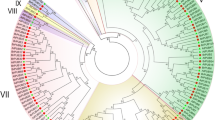
Considering the diverse structures and functions of RcDof genes, the RcDof domain sequences were used for phylogenetic analysis of RcDof genes. Based on the NJ tree generated, RcDof genes were clustered into four major classes (I–IV; Fig. 2a). Correspondingly, major groups (MOCGs A, B, C and D) were labeled (see Fig. 2a) according to the nomenclature and classification system of Dof genes in Arabidopsis. Class I contained the group C, class II consisted of the group B1, and group A and D2 were clustered into Class III; while Class IV included the group D1. The classes I and III respectively containing 8 and 6 members were dominant in RcDof gene family. While as illustrated in Supplementary Fig. S1, the phylogenetic relationships of Dof genes between castor bean and Arabidopsis generated was nested by five distinct major clades. Following the nomenclatures for each clade in Arabidopsis, clades A, B, C and D contained both Dof genes from Arabidopsis and castor bean, while group C3 was composed of four AtDof genes (AT4G21030, AT4G21040, AT4G21050 and AT4G21080). In addition, group C and D were further divided into three (C1, C2.1 and C2.2) and two (D1 and D2) subgroups respectively. These results showed that Dof genes were highly conserved between castor bean and Arabidopsis.
Phylogenetic tree and exon–intron structure of RcDof genes. a The phylogenetic tree was constructed by the alignments of Dof domain amino acid sequences of the 21 RcDof proteins with the neighbor-joining method. The classes were named as I–IV, and subgroups were named coincide to the nomenclature in Arabidopsis. Bootstrapping values are indicated as percentages (>50 %) along the branches. b Intron–exon structure of RcDof genes. The green bars indicate the exons and the lines represent the introns. The red bars represent the Dof domain, and the size can be estimated using the horizontal scale. Numbers are splicing phases
Since gene structural diversity might be a key factor for the evolution of multi-gene families, we compared the exon–intron organization for each Dof genes identified. As shown in Fig. 2b, 12 genes were intronless and 9 had only one intron, particularly, most of the members in the same classes or subgroups shared the similar exon–intron organization. For instance, all members in subgroups A, C2.2 and D2 were intronless, and genes in subgroup C2.1 contained only one intron. Previous analysis for Dof genes revealed that most of Dof genes had none or one intron, and two at most in rice and Arabidopsis (Lijavetzky et al. 2003). However, the Dof gene CrDof1 identified in green algae (Chlamydomonas reinhardtii) had four introns, implying that the introns of Dof genes might be lost during the evolution of plants.
Conserved motifs in RcDof proteins
The identified 21 RcDof proteins and 36 AtDof proteins were analyzed using MEME software to predict the presence of conserved motifs in Dof transcription factors. In total, 33 motifs were identified (see Fig. 3 and Supplementary Table S2), of which motif 1 was the conserved Dof domain present in each Dof member. Most of motifs were shared between castor bean and Arabidopsis, except six motifs (6, 8, 13, 25, 28 and 33) which were uniquely present in Arabidopsis. Motifs 6, 8, 13, 25 and 28 were distributed in Arabidopsis exclusive of subgroup C3 members (Fig. 3). These specific motifs strongly implied that the subgroup C3 in Arabidopsis might be unique or species-specific. Besides, some motifs were group-specific, for example, motifs 2, 3, 4, 5, 11, 12 and 20 were unique to subgroup D1, and motifs 14 and 24 were specific to group C. As illustrated in previous study (Kishimoto et al. 1985), the motifs 2, 23 and 26 represented the protein kinase C phosphorylation sites ([ST]-x-[RK]); the amino acid residue S or T was a specific phosphorylation site; and motifs 7, 11, 15, 26, 29 and 33 also contained phosphorylation sites for casein kinase II ([ST]-x(2)-[DE]). The similar motifs present in subgroups might imply the functional conservation, although the function of most motifs was still unknown.
Analysis of cis-regulatory elements
To explore the potential molecular mechanism underlain the functional differentiation of RcDof genes, cis-regulatory elements within 1,500 bp upstream sequences before the initiation codon (ATG) of 21 RcDof genes identified were comprehensively predicted. In addition to the typical eukaryotic cis-regulatory elements like TATA-motif and CAAT-motif, another 61 distinct elements were identified, which could be divided into four major physiological function classes, including light responses, hormone responses, stress responses, and involvement of tissue specific gene expression (see Table 2). The light responsive elements were found in all RcDof genes (Supplementary Table S3). Studies have shown that some hormones, such as GA, salicylic acid (SA), are strongly related to Dof genes (Washio 2001; Kang et al. 2003). Correspondingly, responsive elements to these hormones (mainly GA, methyl jasmonate, salicylic acid and ABA) were widely identified in the promoter regions of RcDof genes. Also, the stress responsive elements were identified in most of RcDof genes, including responses to cold, drought, heat, and fungus infection. Additionally, cis-regulatory elements like O2-site, RY-element, Skn-1_motif and GCN4_motif, which were involved in regulating endosperm specific gene expression, were identified also in most of RcDof genes. In addition, functions of eight genes (RcDof-4, 5, 6, 8, 12, 15, 17 and 18) were possibly involved in the development of meristem-specific because they had gene expression (see Supplementary Table S3 for details). Since cis-regulatory elements usually are critical factors in regulating gene expression, the diverse cis-regulatory elements identified from the promoter regions of RcDof genes might be a basis of functional differentiation in RcDof genes.
Expression pattern of RcDof genes in tissues
Dissecting the expression profiles of RcDof genes among tissues could offer critical evidence to understand their potential function. According to the normalised RNA-Seq data (Supplementary Table S4), a hierarchy cluster was performed to globally visualize the expression profiles of RcDof genes, resulting in the expression of 19 RcDof genes were detected in different tissues (Fig. 4). The expression profiles showed that 14 out of 19 RcDof genes were constitutively expressed in every organ tested. Four genes (RcDof-6, 12, 18 and 20) were differentially expressed and preferred in leafs. The RcDof-2 was expressed only in male flower tissues. Three genes (RcDof-3, 16 and 17) were co-expressed in leaf and/or male flower tissues.
Heatmap of expression profiles for 19 RcDof genes in five castor bean tissue samples. The heatmap was generated by R using the expression data of the castor bean gene models by Illumina RNA-Seq from European nucleotide archive database (accession number ERA047687, Table S4), and the normalized log2 transformed values were used with hierarchical clustering represented by the color scale (1.0–5.0). Green indicates low expression, and red indicates high expression. Tissue samples were: developing endosperm stage II/III, E II/III; developing endosperm stage V/VI, E V/VI; developing MF male flowers, GS germinating seed, L leaf
Comparing the expressional difference of RcDof genes at the two different developing stages of endosperm, we found that the expression levels of RcDof genes were apparently higher in the early stage than in the mid-late stage. In particular, the RcDof-9 was highly and specifically expressed at the early stage of seed development. Also, RcDof-10, 13, 8, 15 and 20 were up-regulated at the early stage of seed development. However, none Dof gene was up-regulated in the mid-late stage of seed development.
Expression of RcDof genes under ABA or GA treatment
Studies have shown that Dof genes are widely participated in ABA and GA signal pathways (Gualberti et al. 2002; Moreno-Risueno et al. 2007a; Gabriele et al. 2010), implying that the functions of Dof genes might be mediated by ABA and GA signals. To dissect the possible involvement of RcDof genes in the regulation of hormone responses, the expression levels of RcDof genes were investigated in response to ABA and GA treatments respectively. SqRT-PCR analysis showed a number of genes not detected in normal leaves. In response to ABA treatment, SqRT-PCR analysis showed 13 genes were up-regulated in contrast to the controls, in particular, the RcDof-5, 6, 13 and 20 were highly induced (Fig. 5), indicating that these genes might be high sensitive to ABA signaling. Also, two genes (RcDof-15 and 18) were down-regulated compared with the controls. In response to GA treatment, seven genes were inducible and five genes were inhibited in comparison with the controls. It is noteworthy that the seven inducible genes under GA treatment were up-regulated in response to ABA treatment. Notably, the RcDof-20 was up-regulated in response to ABA treatment, whereas down-regulated in response to GA treatment. These results indicated that some RcDof genes were involved in cross-talking among pathways of different hormone signal transduction.
Discussion
The present study reported 21 RcDof genes identified in castor bean genome, and classified them into four classes and seven subgroups based on the similarities of conserved Dof domain region and gene structures (Fig. 2). Compared with the number of Dof genes in rice (30 genes) and Arabidopsis (36 genes), the size of the castor bean Dof family is slightly smaller. For a given transcription factor, rich introns on genome could delay regulatory responses and be selected against in genes whose transcripts require rapid adjustment for survival of environmental challenges (Jeffares et al. 2008). The Dof genes having no intron, or having a few introns in higher plants suggested that they may be highly sensitive to stresses responses. The less intron within RcDof genes in castor bean might be related to its strong ability in adapting diverse environments.
The global expression profile analysis showed that most of RcDof genes were widely expressed in different tissues, indicating that they might be involved in diverse physiological functions. Cis-elements analysis also suggested the functional diversities of RcDof genes, mainly involving in light responses, hormone responses and stress responses. The light responsive elements were extensively presented in all RcDof genes, in accordance with RcDof genes highly expressed in leaves (Fig. 5), indicating that Dof genes might be involved to light responses and seedling morphogenesis (Park et al. 2003; Ward et al. 2005). Four genes (RcDof-6, 12, 18 and 20) were identified to be specifically expressed in leaves. On inspecting the homologues of RcDof-6, 12, 18 and 20 in Arabidopsis, we found that these genes were closely homologous with the group B members in AtDof, which were functionally well characterized as responsive to hormone signals. In particular, the OBP2 (At1g07640, a homologue of RcDof-6) and OBP3 (At3g55370, a homologue of RcDof-12) were highly expressed in leaves and roots, and sensitively responsive to auxin and salicylic acid in Arabidopsis seedlings (Kang and Singh 2000; Kang et al. 2003). Notably, there was also evidence for the involvement of OBP2 gene in regulating glucosinolate biosynthesis (Skirycz et al. 2006). However, RcDof-2 was highly expressed in male flowers, and RcDof-3, 16 and 17 showed co-expressed differential expression in leaves and/or male flowers. Previous studies had showed that the homologues of RcDof-3 and 17 in Arabidopsis could interact with F-box protein in regulating photoperiodic flowering (Imaizumi et al. 2005; Fornara et al. 2009; Yang et al. 2011). In addition, the expression levels of two genes (RcDof-5 and RcDof-11) were not detected in any of the five tissues tested. The homologous gene COG1 (AT1G29160) of RcDof-11 might possibly function as a negative regulator in phyA- and phyB-signaling pathways (Park et al. 2003). The function of the Arabidopsis homologous of RcDof-5 seems to be uncertain.
Considering the importance of endosperm in storage reserve accumulation during seed development, comparing the expression of different endosperm stages may reveal some of the related genes associated with storage material metabolism. The storage materials rapidly accumulated at the mid-late stage, but none of RcDof gene was up-regulated during the mid-late stage of endosperm development suggesting that the functional regulation of Dof genes likely are not involved in storage material metabolism in castor endosperm. However, several RcDof genes such as RcDof-10, RcDof-13, RcDof-8, and RcDof-15 were highly expressed in the early stage of endosperm development with their functions remain to be tested.
When testing the responses of RcDof genes to ABA and GA signals in castor bean seedlings, 18 genes in total, were identified to be responsive to ABA and/or GA signals. In particular, the RcDof-20 gene was significantly up-regulated under ABA treatment, while down-regulated by GA. The rice homology of RcDof-20 played a regulatory role in the expression of the CPD3 (type3 carboxypeptidase) gene under the control of GA (Washio 2001, 2003). It was notable that the expression of both RcDof-5 and RcDof-11 (which were not detectable in the any of the five tissues tested) were detected in seedlings. Furthermore, RcDof-5 and RcDof-11 were both up-regulated with ABA treatment, meaning their potential functions involving in responding to ABA signal.
Conclusions
As key roles in regulating the expression of target genes at transcription levels, identification and functional characterization of transcription factors is essential for understanding the transcriptional regulatory networks. We identified and characterized 21 RcDof genes by a genome-wide survey in castor bean, classified these Dof genes into four groups and seven subgroups based on their structural features. Further, the phylogenetic relationships of Dof genes between castor bean and Arabidopsis, gene structure, conserved motifs, cis-regulatory elements were analyzed and characterized. In particular, the expression profiles of RcDof genes among different tissues were assessed based on high throughput RNA-Seq data. The expressional responses of RcDof genes to ABA and GA signals in castor bean seedlings were tested by SqRT-PCR technique. On the whole, the current study provides valuable information to understand the RcDof functions in regulating the growth and development of castor bean.
References
Baud S, Lepiniec L (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49:235–249
Brown AP, Kroon JTM, Swarbreck D, Febrer M, Larson TR, Graham IA, Caccamo M, Slabas AR (2012) Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One 7:e30100
Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z (2013) Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 55:552–566
Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G et al (2010) Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol 28:951–956
Chen X, Wang D, Liu C, Wang M, Wang T, Zhao Q, Yu J (2012) Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul 66:271–284
Dong G, Ni Z, Yao Y, Nie X, Sun Q (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63:73–84
Fornara F, Panigrahi K, Gissot L, Sauerbrunn N, Rühl M, Jarillo J, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17:75–86
Gabriele S, Rizzal A, Martone J, Circelli P, Costantino P, Vittorioso P (2010) The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J 61:312–323
Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14:1253–1263
Guo Y, Qiu LJ (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS One 8:e76809
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297
Jeffares DC, Penkett CJ, Bähler J (2008) Rapidly regulated genes are intron poor. Trends Genet 24:375–378
Kang HG, Singh KB (2000) Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J 21:329–339
Kang HG, Foley RC, Oñate-Sánchez L, Lin C, Singh KB (2003) Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J 35:362–372
Kishimoto A, Nishiyama K, Nakanishi H, Uratsuji Y, Nomura H, Takeyama Y, Nishizuka Y (1985) Studies on the phosphorylation of myelin basic protein by protein kinase C and adenosine 3′:5′-monophosphate-dependent protein kinase. J Bio Chem 260:12492–12499
Kisu Y, Ono T, Shimofurutani N, Suzuki M, Esaka M (1998) Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 39:1054–1064
Kumar R, Taware R, Gaur VS, Guru SK, Kumar A (2009) Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol Biol Rep 36:2209–2220
Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D (2011) Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep 38:5037–5053
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3:17
Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD (2008) The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol Biol 67:441–454
Moreno-Risueno MÁ, Díaz I, Carrillo L, Fuentes R, Carbonero P (2007a) The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J 51:352–365
Moreno-Risueno MÁ, Martínez M, Vicente-Carbajosa J, Carbonero P (2007b) The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 277:379–390
Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG (2003) The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34:161–171
Plesch G, Ehrhardt T, Mueller-Roeber B (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 28:455–464
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10:278
Shaw LM, McIntyre CL, Gresshoff PM, Xue GP (2009) Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomic 9:485–498
Shigyo M, Tabei N, Yoneyama T, Yanagisawa S (2007) Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol 48:179–185
Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47:10–24
Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, de Veylder L, Witt I et al (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56:779–792
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336:1045–1049
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Umemura Y, Ishiduka T, Yamamoto R, Esaka M (2004) The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J 37:741–749
Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94:7685–7690
Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52:716–729
Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17:475–485
Washio K (2001) Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. BBA Gene Struct Expr 1520:54–62
Washio K (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein–protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133:850–863
Yamamoto MP, Onodera Y, Touno SM, Takaiwa F (2006) Synergism between RPBF Dof and RISBZ1 Bzip activators in the regulation of rice seed expression genes. Plant Physiol 141:1694–1707
Yanagisawa S (1995) A novel DNA-binding domain that may form a single zinc-finger motif. Nucl Acids Res 23:3403–3410
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21:281–288
Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7:555–560
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45:386–391
Yanagisawa S, Izui K (1993) Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem 268:16028–16036
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17:209–214
Yang X, Tuskan GA, Cheng ZM (2006) Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142:820–830
Yang J, Yang MF, Wang D, Chen F, Shen SH (2010) JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol Plant 139:324–334
Yang J, Yang MF, Zhang WP, Chen F, Shen SH (2011) A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci 181:667–674
Acknowledgments
We gave many thanks to Zibo Academy of Agricultural Sciences, Shandong, China for providing the seeds of castor bean var. ZB306 elite inbred line. This work was supported by the “Hundreds of Talents” program of the Chinese Academy of Sciences (AL).
Conflict of interest
All authors declared that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, Z., Chandrasekaran, U. & Liu, A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genom 36, 527–537 (2014). https://doi.org/10.1007/s13258-014-0189-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0189-6