Abstract
Segmentation of organs and lesions could be employed for the express purpose of dosimetry in nuclear medicine, assisted image interpretations, and mass image processing studies. Deep leaning created liver and liver lesion segmentation on clinical 3D MRI data has not been fully addressed in previous experiments. To this end, the required data were collected from 128 patients, including their T1w and T2w MRI images, and ground truth labels of the liver and liver lesions were generated. The collection of 110 T1w-T2w MRI image sets was divided, with 94 designated for training and 16 for validation. Furthermore, 18 more datasets were separately allocated for use as hold-out test datasets. The T1w and T2w MRI images were preprocessed into a two-channel format so that they were used as inputs to the deep learning model based on the Isensee 2017 network. To calculate the final Dice coefficient of the network performance on test datasets, the binary average of T1w and T2w predicted images was used. The deep learning model could segment all 18 test cases, with an average Dice coefficient of 88% for the liver and 53% for the liver tumor. Liver segmentation was carried out with rather a high accuracy; this could be achieved for liver dosimetry during systemic or selective radiation therapies as well as for attenuation correction in PET/MRI scanners. Nevertheless, the delineation of liver lesions was not optimal; therefore, tumor detection was not practical by the proposed method on clinical data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
For organ segmentation, different methods can be adopted, ranging from manual to automated methods, each with advantages and disadvantages whereof. The artificial intelligence (AI), if used, might contribute to the integration of the advantages of both methods. Through AI, the precision of manual segmentation, which was not possible in traditional automated methods, can be achieved, along with a less erroneous iterative machine function. AI-created MRI images of the organ segmentation is a wide-ranging, controversial, and burgeoning research work. The automated segmentations could be used for dosimetry and attenuation correction purposes in nuclear medicine, assisted image interpretations, and research-oriented mass image processing [1,2,3,4,5].
The AI used previously for classification purposes [3, 4] cannot be employed for segmentation, and the denoted segmentation scripts are not sophisticated enough to delineate the boundaries of many organs, such as the liver and embedded tumors. Currently, deep learning (DL) is the optimal AI technology for segmentation. DL has been previously used for liver segmentation and the detection of liver tumors in CT scans [6, 7]; nevertheless, such an application has not been sufficiently employed for segmentation on MRI images (Table 1). There are some studies wherein 2D MRI image slices [8, 9] and machine learning [10, 11] are used. Yet three-dimensional imaging data shows more potential for correct assessment of liver tumors and also enjoys more features for improved segmentation.
One of the less studied organs is the liver. Liver lesions, although very common, are also among the most difficult tumors for segmentation as it is challenging to isolate lesions from the normal liver parenchyma, even for experienced radiologists using single imaging sequences (i.e., T1w or T2w). However, the interpreter can examine different image sequences, scroll up and down the images, and even search for previous imaging data and clinical history of the patient. It is believed that this procedure can be learned by AI using DL for segmentation of the liver and extracting liver lesions. Considering aforementioned obstacles, efficient AI segmentation of the liver and liver lesions on T1 and T2 weighted MRI images is not widely documented, in contrast to similar experiments for CT images (Table 1).
This study’s key contributions are its application of deep learning techniques for the precise segmentation of both liver and liver lesions in 3D T1w and T2w MRI images, using a specialized dataset for this dual analysis. The network used in this study, established and applied for segmentation purposes, had not previously been utilized for segmenting liver and liver lesions. Given the network’s proven efficacy in brain tumor segmentation, its successful application in training for organ and tumor segmentation would highlight its versatility for such tasks. The utilization of actual noisy clinical MRI data in the current study is unique which underscores real word DL organ segmentation strengths and drawbacks. The study results enhance segmentation accuracy in 3D clinical MRI and set a new precedent for DL in the detailed examination and differentiation of liver and liver lesions.
Materials and methods
A brief visual representation of the procedures of the current study is illustrated in Fig. 1.
Data preparation
Patients
A total of 128 patients were included in the current study from the liver transplantation and general surgery wards of Imam Khomeini Hospital Complex (IKHC), affiliated to Tehran University of Medical Sciences, Tehran, Iran. Holding to the guidelines established by the Ethics Committee of Imam Khomeini Hospital, the anonymity of the images used in the current study was guaranteed. The MRI images of the livers were collected from the picture archiving and communication system (PACS). Imaging was carried out using MAGNETOM Trio, which is a 3T total imaging matrix (Tim) eco system (Siemens, Germany), and GE Optima MR360 Advance 1.5T system (GE Healthcare, USA) with a standard imaging protocol including axial and coronal T2w, axial T1w images.
Out of 128 patients, 110 were randomly allocated to the model training data and 18 to the hold-out test group. In training data, 20% (n = 16) of the subjects were randomly allocated to the validation group, while the rest were allocated to the training cohort. Liver segmentation was carried out for 77 cases on both T1w and T2w sequences. Liver lesions were segmented in both T1w and T2w sequences across 75 cases, with 42 of these cases overlapping in both liver and lesion segmentation categories. The test group included 18 patients with 9 liver tumors and 9 patients without liver tumors.
Segmentation
The segmentation of the liver and liver lesions was semi-automatically carried out using a 3D Slicer v4.10.0 on T1w- and T2w MRI images, separately, to create the training ground truth. Masks for the images were created separately for both T1w and T2w sequences, for liver and tumor-labeled ground truths, and were saved as 3D NIfTI images. A value of 1 was assigned to the target regions of each image.
Pre-processing procedures
The variation in voxel sizes among MRI images necessitated the registration of voxel sizes for different images. To integrate different voxel sizes, the geometry-based registration of 3D Slicer software was used to resize images, by changing the voxel size into a common voxel size of 1.48 mm, 1.48 mm, 4 mm. The results ensure the input images are all of the same size, which is essential for deep learning models to function properly.
Normalization with the N4 bias field algorithm [12] was carried out in Python v3.7 to handle the heterogeneity of low-frequency data. Next, T1w and T2w images of the liver and lesion masks, with substantial misalignment of T1w and T2w images, were aligned by a Python code to match together. Liver ground truth masks were utilized separately for T1w and T2w images. To this end, it was crucial to identify the optimal shift to properly align these two sets of images. A score was established to gauge the degree of alignment between two shifted images. Various shift values were then explored to determine which one would maximize this alignment score. More precisely, the score is the result of “And” between two masks after shift. If two shifts yield the same score, the smaller of the two will be chosen for use. The final images are aligned by the best shift found. This process didn’t change the characteristics of the images.
Without normalization, the heterogeneity of MR data prevents proper training, possibly due to the training from bias. The pre-processed data were loaded. The intensity of the voxels was normalized by subtracting data from the mean divided by the standard deviation. We computed a mean and standard deviation over all the training data and applied them for normalization of both training and test datasets. The outcome values were in the range of [-1, 1].
The common field of view of medical images contains a black area around the body contour. Initial experiments indicated that it would yield better results if the black parts of the images around the body are cropped. For the segmentation of lesions, we opted not to use the segmented liver but rather the original cropped MRI images as the input for the deep learning model. This approach is consistent with the method we used for training the model in liver segmentation. The reason is that if the liver masks were used for liver lesion segmentations, large liver lesions would not be extracted efficiently due to the position of tumor at the edge of image.
In the next step, data augmentation was performed. Generally, a network with a small training sample tends to over fit. Data augmentation was applied to avoid over fitting. Data augmentation was carried out in two parts: (1) real data augmentation of flip and 90-degree rotations, augmenting the original datasets (16 times), non-90-degree random rotation, distort elastic, scale, and random noise; (2) the second part of data augmentation comprised data augmentation in an “on-the-fly” mode in the generator module to speed up the process and compensate for memory limitations.
Finally, we employed ‘white sampling’ a technique where the area of interest is specifically targeted for training, for lesion segmentation. In the process of liver segmentation, most areas of the image show a mask value of one, illustrated as a white area, because the liver comprises the major part of trans-axial images. Therefore, the segmentation process was performed without the need for additional modification. In contrast to liver segmentation, in lesion segmentation, only a small portion of the image is attributed to the lesion, and most of the image is black outside the lesion. Moreover, the position of lesions in the liver varies from one image to another. As a result, we used a method known as random white sampling for training, which focuses on the areas surrounding random values that are equal to one.
Model architecture
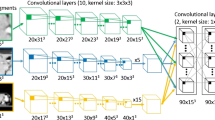
The TensorFlow library in Python v3.7 was used for this purpose. The employed DL model throughout this paper is the one developed by Isensee in 2017, which is referenced as “Isensee 2017.” This model is grounded in a U-Net-based architecture and was specifically designed and used for segmentation purposes. At its core, the network employs a convolutional neural network (CNN) framework, originally proposed for the brain tumor segmentation challenge known as BraTS [17]. The goal of BraTS is to develop a state-of-the-art method for tumor segmentation by providing a large dataset of annotated low-grade gliomas and high-grade glioblastomas. We employed parameters same as Isensee 2017, and hyper parameters are as follows: optimizer: Adam; initial learning rate: 1e− 4; loss function: weighted Dice coefficient loss; activation function: sigmoid; dropout rate: 0.2; Batch size: 8; and number of epochs: 400. Patch size of 96 × 96 × 32 for liver and 64 × 64 × 32 for liver tumors were employed; moreover, 16 filters were implemented, the size of which varied across each layer. The network depth was set to 5; the “weighted Dice coefficient loss” was used as loss function.
In the final training effort, the network was trained with 400 epochs for both liver and lesion datasets. T1w and T2w data of each patient were fed into the network via two channels. Training continued until the Dice coefficient calculated for the validation data remained constant with no increment. The Dice coefficient [18] was calculated based on the following equation:
where Ytrue is the image annotation or ground truth, Ypred is the resulting mask (i.e., liver or lesion mask), ϵ is a small value added to prevent the denominator from being equal to zero, ∩ is an intersection, and| ⋅| determines the cardinality of its argument (i.e., number of non-zero elements in the mask). The T1 and T2 images were used via two different channels of CNN input to predict two different outputs: segmentation for T1 images and T2 images. To quantify the final performance of the network, Dice coefficient for the hold-out test dataset was calculated using the binary average of T1w and T2w sequences.
Due to the limited size of our MRI dataset and the preference to make the most of our labels, we adopted the strategy of using the weights from liver segmentation training as the initial weights for tumor segmentation. This approach enables the model to continue learning from the point where it previously stopped.
The network was trained on a desktop PC (NVIDIA 2080 Ti GPU, 32 GB RAM).
Results
The code and weight of the trained network were uploaded to GitHub. Finally, the liver and lesion segmentations were tested on 18 cases. The average Dice coefficient for the binary average of T1w and T2w sequences was 88% for the liver and 53% for liver tumors. Diagnostic accuracy and statistics for the performance of the trained network for the test data is presented in Table 2. Nine patients without liver tumor were involved. However, since normal liver was not considered in the training, the results of segmentation were not optimal for them. Figures 2 and 3 illustrate loss-validation diagrams for liver and lesion segmentation training. Figures 4 and 5 present the segmentation results for the liver and liver tumors from T1w and T2w sequences, and Fig. 6 presents the binary averages. Training with the mentioned specifications using the method described for either liver or lesion segmentation (except data preparation) continued for about three days. Segmentation of the liver or liver lesion for a never-seen-before T1w or T2w image took almost five minutes.
A typical slice of the original MRI (a & e), grand truth labels (b & f), and network-generated liver segmentation (c & g); the upper row denotes T1w, and the lower row denotes T2w of same patient; d and h present the difference between ground truth labels and network predictions. The gray color (float number) in f is related to the registration process
A typical slice of original MRI (a & e), ground truth labels (b & f), and network-generated lesion segmentation (c & g); the upper row denotes T1w, and the lower row denotes T2w of same patient as in Fig. 3; d and h present the difference between the ground truth label and network prediction. The gray color (float number) in f is related to the registration process
Discussion
The performance of the Isensee 2017 network for segmentation of the liver was more effective than lesion segmentation. Visually, the majority of lesions were detected, despite failure to depict their shapes and boundaries. The roughly typical shape of the liver in different individuals, besides the large white portion of the liver on images due to the larger liver size compared to variable sized and shaped lesions, plausibly facilitates the detection of liver boundaries compared to the liver lesions by the network.
Comparing other studies, the Dice score of the current U-net deep CNN is not optimal (88% vs. generally 94–97%; Table 1). The reasons for the difference can be listed as below:
(1) Segmentation with CT data which provides higher resolution is more accurate compared to segmentation using MRI data. Reasonably, the Dice similarity coefficients were higher in the studies cited in Table 1 which segmented CT images compared to the current study; (2) MRI data from low-noise public databases differ essentially from clinical MRI images with higher noise and uncontrolled clinical conditions including different image voxel sizes; and (3) the use of MRI and CT data together facilitates detection of the boundaries but reduces the clinical applicability because MRI and CT images are not usually available simultaneously. Plausibly, the study of Wang K et al. [11] employing both CT and MRI data had better result. The high-resolution CT data prevail the noisy MRI data to learn segmentation. (4) Few pieces of research reporting liver segmentation worked on a 3D MRI dataset using DL [8,9,10,11]. It should be noted that in real clinical scenarios, the whole (i.e. 3D) liver should be segmented and 2d segmentation would cause boundary indentations and final inaccuracies. If artificial intelligence is to segment the liver in real clinical application, 3D segmentation should be targeted and the obstacles become solved. The reason why 2D datasets are more commonly employed is that the prepared 3D datasets are scant in online libraries, particularly for MRI images. The data of the current study could be used for this purpose in future studies. Furthermore, manual annotation of the liver and liver lesion slices is very cumbersome which was performed in the current work; and additionally, training, based on 3D datasets, requires advanced systems with more data processing specifications. Essentially, comparison of the results of the training based on 3D MRI data with 2D datasets cannot be performed robustly since the corresponding networks have a different number of parameters and a different number of convolutional layers.
A novel approach provided by the current study is that the original liver and lesion segmentation tasks on 3D MRI, applied simultaneously for T1w and T2w data, have not been reported for DL yet. The only available study using DL on MRI data for 3D liver and lesion segmentation was carried out by Christ et al. [19]. They reported Dice coefficients of 86% and 69% for the liver and liver lesions, respectively, which are comparable with results of current study. While their cascaded fully convolutional neural networks (CFCNs) surpassed our current model in lesion detection, our method showed a slight advantage in liver segmentation performance. Since their CNN was trained with diffusion-weighted MR (DW-MR), considering liver tumors better visualization in DW-MR compared to T1w and T2w images, their superior results is reasonable. It should be considered that Crist et al. trained their MRI model using a smaller dataset, consisting of only 31 cases.
In the current study, the Isensee 2017 network was opted because it is a pure 3D U-Net-based CNN, developed for segmentation. Comparing to the results obtained from previous studies employing machine learning (ML) based approach on MRI images, the network performance in the current study was superior to those reported in studies by Pratondo et al. and Häme et al. [20, 21]. The reason behind the current finding is that the use of DL is superior to ML due to the generalizability of the network for a wide spectrum of data; moreover, there is no need for feature extraction in DL [7, 20,23,24].
The preparation of MRI data for DL was challenging and time-consuming since real hospital-based MRI images of the patients were utilized (uploaded to https://www.cancerimagingarchive.net; accepted and available for the reviewer by correspondence). The use of MRI was important because the up-to-date most accurate imaging for hepatic lesions is MRI. The method could be used for express research and dosimetry purposes. Furthermore, the corresponding coefficient of liver attenuation could be employed for liver masking during the attenuation correction process in PET/MR instruments. Although poor lesion delineation hinders lesion dosimetry, such a failure would negligibly change attenuation correction-based application for the proposed network.
A drawback to the current study is that the performance of the trained network for lesion detection on images without liver lesion was ignored. In future assessments, the confusion of application of lesion detection algorithm to the images without lesion should be sought further. Furthermore, both T1w and T2w images were used; however, odds are that one may use one of these images, for example only T1w images, for the purpose of AI training. The comparison between the suggested applications of T1w and T2w images may provide further insights in future studies. Lastly, adding classifiers to find detection rate of tumor segmentation will lead to more insight on model performance.
Conclusion
In the current study, T1w and T2w 3D images were prepared and learned, via two channels, employing the Isensee 2017 network. The results indicated the capability of DL to utilize T1w and T2w data simultaneously for each specific patient in order to segment the liver. Such a method can be applied to researches on MRI images, dosimetry, and attenuation correction in PET/MRI scanners in future studies.
References
Tang X et al (2020) Whole liver segmentation based on deep learning and manual adjustment for clinical use in SIRT. Eur J Nucl Med Mol Imaging 47:2742–2752
Rahmani R et al (2019) Improved diagnostic accuracy for myocardial perfusion imaging using artificial neural networks on different input variables including clinical and quantification data. Revista Española De Medicina Nuclear E Imagen Molecular (English Edition) 38(5):275–279
Eftekhari M et al (2018) Automated interpretation of myocardial perfusion images with multilayer perceptron network as a decision support system. J Med Imaging Health Inf 8(9):1844–1849
Gholipour C et al (2009) Prediction of conversion of laparoscopic cholecystectomy to open surgery with artificial neural networks. BMC Surg 9(1):1–6
Zhen S-h et al (2020) Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front Oncol 10:680
Hu P et al (2016) Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol 61(24):8676
Dou Q et al (2016) 3D deeply supervised network for automatic liver segmentation from CT volumes. In: Ourselin S et al (eds) Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, October 17–21, 2016, Proceedings, Part II 19. Springer
Jansen MJ et al (2019) Liver segmentation and metastases detection in MR images using convolutional neural networks. J Med Imaging 6(4):044003–044003
Jansen MJ, Kuijf HJ, Pluim JP (2019) Optimal input configuration of dynamic contrast enhanced MRI in convolutional neural networks for liver segmentation. In: Medical imaging 2019: image Processing. International Society for Optics and Photonics
Masoumi H et al (2012) Automatic liver segmentation in MRI images using an iterative watershed algorithm and artificial neural network. Biomed Signal Process Control 7(5):429–437
Wang K et al (2019) Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiology: Artif Intell 1(2):180022
Tustison NJ et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Ahmad M et al (2019) Convolutional-neural-network-based feature extraction for liver segmentation from CT images. In: Eleventh International Conference on Digital Image Processing (ICDIP 2019). SPIE
Ahmad M et al (2018) Deep-stacked auto encoder for liver segmentation. In: Wang Y et al (eds) Advances in Image and Graphics Technologies: 12th Chinese conference, IGTA 2017, Beijing, China, June 30–July 1, 2017, Revised Selected Papers 12. Springer
Ahmad M et al (2019) Deep belief network modeling for automatic liver segmentation. IEEE Access 7:20585–20595
Ahmad M et al (2022) A lightweight convolutional neural network model for liver segmentation in medical diagnosis. Computational Intelligence and Neuroscience, 2022
Isensee F et al (2018) Brain tumor segmentation and radiomics survival prediction: Contribution to the BRATS 2017 challenge. In: Crimi A et al (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Third International Workshop, BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 14, 2017, Revised Selected Papers 3. Springer
Drozdzal M et al (2016) The importance of skip connections in biomedical image segmentation. In: Carneiro G et al (eds) Deep Learning and Data Labeling for Medical Applications. DLMIA LABELS 2016. Springer
Christ PF et al (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv Preprint arXiv:1702.05970,
Pratondo A, Chui C-K, Ong S-H (2017) Integrating machine learning with region-based active contour models in medical image segmentation. J Vis Commun Image Represent 43:1–9
Häme Y (2008) Liver tumor segmentation using implicit surface evolution. Midas J: p. 1–10
Fallahpoor M et al (2022) Generalizability assessment of COVID-19 3D CT data for deep learning-based disease detection. Comput Biol Med 145:105464
Han X (2017) Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv preprint arXiv:1704.07239,
Vorontsov E et al (2018) Liver lesion segmentation informed by joint liver segmentation. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE
Acknowledgements
This study was supported by the Elite Research Grant Committee, with an award number (958298) from the National Institute for Medical Research Development (NIMAD), Tehran, Iran.
The authors keep the memory of their colleague, Dr. Habibollah Dashti, who passed away in his fight against cancer before reading the final version of the manuscript. We as many of his patients remember his expertise and essence of humanity in liver surgery and practice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with Ethical Standards
This article does not contain any studies on human participants or animals performed by any of the authors. The study was performed on the previously acquired images after the anonymization of the data.
Conflict of interest
The authors declare no conflicting interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fallahpoor, M., Nguyen, D., Montahaei, E. et al. Segmentation of liver and liver lesions using deep learning. Phys Eng Sci Med 47, 611–619 (2024). https://doi.org/10.1007/s13246-024-01390-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-024-01390-4