Abstract
Lung and colon cancers lead to a significant portion of deaths. Their simultaneous occurrence is uncommon, however, in the absence of early diagnosis, the metastasis of cancer cells is very high between these two organs. Currently, histopathological diagnosis and appropriate treatment are the only way to improve the chances of survival and reduce cancer mortality. Using artificial intelligence in the histopathological diagnosis of colon and lung cancer can provide significant help to specialists in identifying cases of colon and lung cancers with less effort, time and cost. The objective of this study is to set up a computer-aided diagnostic system that can accurately classify five types of colon and lung tissues (two classes for colon cancer and three classes for lung cancer) by analyzing their histopathological images. Using machine learning, features engineering and image processing techniques, the six models XGBoost, SVM, RF, LDA, MLP and LightGBM were used to perform the classification of histopathological images of lung and colon cancers that were acquired from the LC25000 dataset. The main advantage of using machine learning models is that they allow a better interpretability of the classification model since they are based on feature engineering; however, deep learning models are black box networks whose working is very difficult to understand due to the complex network design. The acquired experimental results show that machine learning models give satisfactory results and are very precise in identifying classes of lung and colon cancer subtypes. The XGBoost model gave the best performance with an accuracy of 99% and a F1-score of 98.8%. The implementation and the development of this model will help healthcare specialists identify types of colon and lung cancers. The code will be available upon request.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
According to the World Health Organization, cancer is considered one of the most common causes of mortality in the world. Cancer cells acquire autonomous growth, genetic instability and significant metastatic power. Among the most frequently affected organs, colon and lung cancers account for the highest number of deaths. Lung cancer accounts for 18.4% of cancer-related deaths, while colon cancer accounts for 9.2% of all cancer-related deaths worldwide [1, 2]. The rate of simultaneous occurrence of lung and colon cancer is approximately 17%. Although this frequency is unlikely, but in the absence of an early diagnosis, cancer cells metastasis is very high between these two organs [3]. Currently, appropriate treatment and early diagnosis are the only way to reduce cancer mortality [4]. Indeed, the earlier a person is diagnosed, the better the management and the greater the chance of recovery and survival of the patient are.
Various tests such as imaging sets (x-ray, CT scan), Sputum cytology, and tissue sampling (biopsy) are done to look for cancer cells and exclude other possible conditions. While performing the biopsy, evaluation of the microscopic histopathology slides by experienced pathologists is essential to establish the diagnosis [5] and defines the types and subtypes of cancers [6]. To automatically diagnose colon and lung cancers, this study relies solely on histopathological images. Histopathological images are widely used by health specialists for diagnosis, and they are very important in predicting patients’ chances of survival. Traditionally, in order to diagnose cancer by examining histopathological images, health specialists have to go through a long process; however, it is now possible to perform this process in less time and effort with the available technological tools [3]. Recently, artificial intelligence technologies have been known for their ability to examine data faster and make decisions.
Machine learning (ML) is a subfield of artificial intelligence (AI) that allows machines to learn a specific task through experience with the datasets to which they are exposed, without explicit programming [7]. ML algorithms are used in biomedical applications for the prediction and classification of several types of signals and images. Deep learning (DL) algorithms have been developed to enable machines to handle large-dimensional data like multidimensional anatomical images, and videos. DL is a subfield of ML that structures algorithms in layers to create an “artificial neural network”, based on the structure and function of the human brain [8].
In previous research articles, most of the authors considered using DL to classify colon and lung cancer images at the same time. Some authors have focused on lung cancer classification, while others have concentrated entirely on the classification of colon cancer.
There are few works for the classification of colon cancer. For instance, Bukhari et al. [9] used three convolutional neural networks architectures: ResNet-18, ResNet-30, and ResNet50. ResNet-50 achieved the highest accuracy of 93.91%, followed by ResNet-30 and ResNet-18 with an accuracy of 93.04% each.
To classify histopathological images of lung cancer into three classes, Hatuwal et al. [10] used Convolution Neural Network (CNN). The classification result obtained was 97.2%. Nishio et al. [11] used homology-based technique and machine learning methods to classify lung tissue images into three classes. The overall classification accuracy obtained was 99.43%.
Masud et al. [12] classify colon and lung histopathological images using a deep learning-based method. They used domain transformations of two types to extract four feature sets for image classification. Then they combined the features of the two categories to arrive at the final classification results. They have achieved an accuracy of 96.33%. Mangal et al. [13] made a classification of colon and lung cancers based on histopathological images by applying a shallow neural network architecture. They achieve an accuracy of 97% and 96% in classifying lung and colon cancers, respectively. Toǧaçar [3] performed the classification of colon and lung cancers’ histopathological images by training the images with the Darknet-19 model and then obtain the feature sets, to which two optimization algorithms were applied to select the inefficient features. Then, the efficient feature sets, that have been created for each of the two optimization algorithms by distinguishing the ineffective features from the rest of the features in the set, were combined and classified using SVM classifier. He has obtained an overall accuracy of 99.69%.
The main limitation of conventional ML is that it requires pertinent features and more ongoing human intervention to get results. DL is more complex to set up but requires minimal intervention.
However, the use of ML has many advantages. The main advantage of conventional ML models over DL models is that ML models allow better interpretability of the classification model since they are based on feature engineering. The computed features have an interpretable mathematical meaning and then help us to better understand our model and to increase its predictive power. Indeed, in the medical and diagnostic field, feature engineering is crucial for doctors to make life-changing decisions because it allows them to know the importance and impact of each feature on the classification of cancer subtypes; unlike DL models which are black box networks that their working is very difficult to understand and interpret because of complex network design [14]. Indeed, DL models take automatic decisions without us being able to interpret what is going on inside the model.
Also, there are many more parameters and hyperparameters that can be learned in DL models than in ML models, and so a DL system can take a long time to train. While feature engineering-based, ML takes comparatively much less time to train, ranging from a few seconds to a few hours [14].
Additionally, ML algorithms are less complex than DL algorithms and can often run on conventional computers, while DL systems require much more powerful hardware and resources with very high performance due to the amount of data processed and the complexity of the mathematical calculations involved in the algorithms used. This need for power has led to increased use of graphics processing units (GPU) which are very expensive.
The purpose of this study is to propose a medical diagnostic support system for lung and colon imaging. In other words, it is to set up an automated system that can accurately classify the subtypes of colon and lung cancer from histopathological images using ML, and to show that with feature engineering we can find powerful accuracy results.
Our contributions are summarized as follows:
-
We proposed feature engineering based machine learning models for the classification of histopathological images of colon and lung cancers into five classes (three malignant and two benign).
-
We used the SHAP method to explain the output of our models and evaluate how the contribution of each feature affects our best model.
-
We present an experiment on the LC25000 dataset. The experiment results demonstrate that our method achieves high performance compared to state-of-the-art approaches that use DL.
Material and methods
Lung and colon cancer dataset
Lung and Colon Cancer Histopathological Image Dataset, published in 2020, is known as LC25000 dataset [15]. The LC25000 dataset images were collected at James A. Haley Veterans’ Hospital located in Tampa, Florida. The images are categorized, labeled, and augmented with rotation and flips by the authors. LC25000 dataset contains 25,000 RGB histopathology images stained with hematoxylin and eosin, of five classes of colon and lung tissues, 5,000 images of each class [16]. Images are \(768 \times 768\) pixels in size and are in JPEG file format. All images are de-identified, Health Insurance Portability and Accountability Act (HIPAA) compliant, validated and freely available for download to AI researchers. The five classes are Colon Adenocarcinoma, Benign Colonic tissue, Lung Adenocarcinoma, Benign Lung tissue, and Lung Squamous cell Carcinoma.
The most frequent type of colon cancer is Colon Adenocarcinoma, which accounts for over 95% of all cases of colon cancer. It is produced when an adenoma - a type of polyp - develops within the large intestine and eventually turning into cancer. Lung Adenocarcinoma, a type of cancer cells that represents for around 60% of all cases of lung cancers, usually grows in the glandular cells located in the outer part of the lung and then spreads to the alveoli within the lung. Lung Squamous Cell Carcinoma, which is the second most frequent type of lung cancer, develops in the airways or bronchi of the lungs and represents around 30% of all cases.
Sample of histopathological images of these five classes of colon and lung tissues that are collected from the LC25000 dataset are illustrated in Fig. 1.
Overview of the methodology
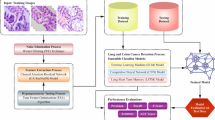
Figure 2 shows an overview of the methodology used for classifying colon and lung cancer subtypes on the basis of histopathological images. The RGB images of lung and colon cancer were fed into the system. 2500 images were used at all stages of our study, 500 images from each class. Images were resized to 200 × 200 pixels. Two preprocessing methods were tested: the Unsharp Masking and Stain Normalization. Then the images were transformed to grayscale and the features were extracted. The Recursive Feature Elimination, which is a feature selection method, is used in order to select the most efficient features. Then, a machine learning algorithm classified the image on the basis of the selected features. 20% of the dataset was used as test data and 80% was devoted to training the data (randomly chosen). The machine learning algorithm is trained using the images features of the training set. Finally, image features of the testing set are used for assessing the performance of the model. The programming language used is Python with the implementation of the following libraries: numpy, pandas, matplotlib, tensorflow, scikit-learn, scikit-image, staintools and xgboost.
Image preprocessing
After image acquisition, the images must be preprocessed. Indeed, image preprocessing is essential to improve image quality and extract important information from the images to make them more adequate for the learning algorithm. In this study, two preprocessing methods were tested: the Unsharp Masking and Stain Normalization.
Unsharp masking
The contrast of each image is enhanced using the Unsharp Masking (UM) which is an image sharpening method. Unsharp Masking enhances the contrast, and thus sharpens the original image, which can help emphasize texture and detail. The basic idea of the UM method is to subtract the original image by a blurred version of the image itself, thus resulting in only the blurred edges. The typical formula used for unsharp masking is as follows:
The outcome of the Unsharp Masking is conditioned by the radius and amount parameters. The blurring step could use any image filter method, but traditionally a Gaussian filter is used. The radius parameter in the unsharp masking filter refers to the sigma parameter of the Gaussian filter. The radius controls the degree of blurring of the original image, and therefore the dimension of the area encircling the edges that is concerned by the sharpening. The value of the enhancement effect is determined by the amount parameter, which is the value of contrast added to the edges.
In our case, in order to choose the best parameters for the unsharp masking method, we carried out a sensitivity study on the parameters: We have tested radius values from 1 to 5, and amount values from 1 to 20. We have obtained that the best values of the radius and amount parameters are 2 and 5 respectively, since the models gave the best performance using these values. Therefore, these values are used in the rest of our study. Figure 3 represents the result of enhancing a histopathological image using the Unsharp Masking method under the indicated conditions.
Stain normalization
Stain normalization is one of the preprocessing steps used by many deep learning-based algorithms to support pathology diagnoses with whole-slide images. This task reduces the color and intensity variations present in stained images, especially when they are obtained from different hospitals or laboratories or scanners, which can adversely affect the performance and accuracy of CAD systems. Stain normalization methods aim to assist CAD systems by generating images with a standardized appearance of different stains [17].
In the literature, many methods have been proposed to normalize the colors of hematoxylin and eosin stained images. The frequently used color normalization methods are Macenko [18] and Vahadane [19]. Previous state-of-the-art studies, such as Ciompi et al. [20], have shown that the classification accuracy of a machine learning-based histopathology system is improved when using stain-normalized images. When the variability between images is low, like when they belong to the same dataset from the same hospital, stain normalization can have a small impact on the ML pipeline, as also shown by Lafarge et al. [21].
In this study, we use the Vahadane method for stain normalization. The idea of Vahadane method is to first decompose the images in an unsupervised manner into sparse and non-negative stain density maps. For a given image, its stain density maps are combined with stain color basis of a target image, thus changing only its color while preserving its structure described by the maps. Figure 4 presents original and normalized images and shows how spot normalization reduces stain variability across images within the dataset.
Feature extraction
Features are measured values that can be informative for a predictive analysis to classify the attribute. Features contained in histopathological images are essential for the diagnosis of the disease, and efficient features extraction is of high importance to improve the diagnostic accuracy and assist in cancer classification [22]. In this paper, we extracted 37 features, including first order statistics, GLCM and the Hu invariant moments. The computed features for each method are shown in Table 1.
First order statistics
The features obtained from the first-order statistics provide information about the distribution of brightness in the image. The first-order statistics used are: mean, standard deviation, median, percentile 25%, percentile 50% and percentile 75%.
GLCM
In biological imaging, the Gray Level Co-occurrence Matrix (GLCM) is a widely used method for texture analysis due to its ability to capture the spatial dependence of gray level values inside an image since the pixels are considered in pairs. The co-occurrence matrix is a second-order statistical characteristics of the changes in image brightness. It gives a description of the gray level variations between each pixel in the texture of the image and its neighboring pixels. Indeed, it is a tabulation of the frequency of different combinations of pixels brightness values (gray tone) which occur within an image [23].
The co-occurrence matrix is a function of two parameters: the distance (d) that is measured in number of pixels and their orientation (\(\theta\)). The orientation \(\theta\) takes the values \(0^{\circ }\), \(45^{\circ }\), \(90^{\circ }\), and \(135^{\circ }\), which represent the four directions : the horizontal, diagonal, vertical, and anti-diagonal, respectively. The occurrence of a gray level pattern can be represented by a relative frequency matrix, \(P_{\theta ,d}(I_{1}, I_{2})\) which describes the frequency of appearance of two gray level pixels \(I_{1}, I_{2}\) in the window that are separated by a distance d in the \(\theta\) direction [24]. The computed features are: contrast, dissimilarity, homogeneity, energy, angular second moment (ASM) and correlation, of which a group of four features is calculated for each.
The contrast is a feature that measures the local level variations, and it takes high values for high contrast images. The dissimilarity provides a measure of the randomness of pixels and takes low values if we have the same pixel pairs. Homogeneity is a measure that takes high values if we have similar pairs of pixels. The ASM is used to measure the smoothness of an image and takes a low value if the region is less smooth. Correlation measures the correlation between pixels in two different directions. Since these features depend on d and \(\theta\), then their values differ if the image is returned. Thus, we will have features that are invariant to rotation.
Hu invariant moments
The moment feature generally describes the geometric characteristics in the image area. Hu invariant moments are a set of seven numbers calculated using central moments that are invariant to image transformations. Due to the invariance to translation, rotation and scaling, Hu invariant moments are largely used in the field of image pattern recognition, classification, and target recognition [25]. Therefore, in this paper, we used the Hu invariant moments to represent the characteristics of histopathological images of colon and lung cancers.
Feature selection
Recursive feature elimination (RFE) is a feature selection method that eliminates the least important features, as well as dependencies and collinearity that may exist in the model, until the desired number of features is reached. RFE is popular because it is easy to implement, and it is effective in selecting features from a training data set that are more relevant to predict the target variable.
Features are ranked using the feature_importances_ attributes of the model. RFE requires that a specified number of features be retained, but since the number of valid features is not known in advance, then to find the optimal number of features, cross validation is used with RFE to evaluate several subsets of features and select the best. RFECV performs RFE in a cross-validation loop to find the optimal number of features. The purpose of recursive feature elimination is to select features by considering recursively smaller feature sets. First, the classifier is trained on the initial feature set and the importance of each feature is obtained based on its contribution to the classification. Then, the features were sorted from high to low according to their importance, which results in a feature ranking. Lastly, the features that are least important are eliminated from the actual feature set. And then the updated features are used to re-train the model, and we obtained the classification performance using the new feature set. This process is repeated recursively on the reduced set until the desired number of features to be selected is reached. RFE needs several parameters such as estimator and scoring. A scoring function is a metric to evaluate the performance of the model such as accuracy, f1-weighted, mean squared error; in our study, accuracy is the metric used.
In this study, RFE tells us to keep only 12 of the 37 features. So, the models are trained only on these 12 features. We compared the feature non-selection and RFE method to look at the performance. The analysis performed with the two methods resulted in not using the RFE method and not reducing the feature vector, since the classification system was more efficient with the use of all features.
Classification
The features extracted from the 2500 images were used in all stages of our study. 20% of the dataset was used as test data and 80% was devoted to training the data (randomly chosen).
The features that are extracted from the images were fed into machine learning algorithms. The machine learning algorithms used are: support vector machine (SVM), Random Forest (RF), Extreme gradient boosting (XGBoost), Light gradient boosting machine (LightGBM), Linear Discriminant Analysis (LDA), and Multilayer Perceptron (MLP). These machine learning algorithms were trained using the image features of the training set.
In this study, the SVM hyperparameters were tested to select those that performed best with the experiment database; The SVM kernels:[’linear’, ’rbf’] and C: [1, 10, 100, 1000] were tested. We obtained that the best values of the SVM hyperparameters are a linear kernel and a regularization parameter C of 100, which were selected automatically by the model as they performed best, and a one-versus-one multi-class method was used.
The hyperparameters of the RF model were also tested; The n_estimators: [10, 50, 100, 300], and criterion: [’gini’, ’entropy’] were tested. The best values of the RF hyperparameters that were selected are 300 trees and a gini criterion.
The default hyperparameters for the XGBoost algorithm are provided by the implementation of xgboost. The tree-based models (gbtree) which is the type of model to run at each iteration, is the general parameter selected for the XGBoost model. The maximum depth of a tree is 6.
The default hyperparameters for the LightGBM algorithm are provided by the implementation of lightgbm. Traditional Gradient Boosting Decision Tree (gbdt) is the selected boosting type. The maximum number of leaves per tree is 31.
Also, the default hyperparameters of the LDA algorithm are provided by the implementation of sklearn.discriminant_analysis. The svd (Singular value decomposition) solver is used since it does not compute the covariance matrix, so it is recommended for data with numerous features.
The MLP classifier is composed of three hidden layers with 150, 100 and 50 neurons in each layer, using a ’relu’ activation function. The ’Softmax’ activation function is used in the last layer of the network. The solver for weight optimization used is ’adam’. The other parameters for the MLP model, such as number of epoch value of 300, and minibatch value of 200 were selected.
Support vector machine (SVM)
Support vector machine (SVM) is a parametric discriminant classifier that establishes a maximum margin separator hyperplane, known as decision boundary, between representative examples of each of the training data classes. It can carry out binary or multiple classification processes. The behavior of SVMs may differ depending on various mathematical functions which are known as the “kernel”. Popular kernel functions used in SVM classifier are linear, radial basis function (RBF), sigmoid, etc. In multi-class, the SVM method carries out a vote for the features that are in the positive or negative side of the hyperplane. Following the voting, each feature is assigned to the class in which it obtained the most votes [26]. The general equations used in the classification process are specified in Eqs. 7–10.
The separating hyperplane (H) with parameters \((w,w_{0})\) has the equation:
where w is a weight vector and \(w_{0}\) a scalar.
The objective of an SVM is to determine the parameters w and \(w_{0}\) of the hyperplane that maximize the minimum distance between the observations \(x_{k}\) and the hyperplane, i.e:
where h is the projection function on (H), \(\Vert .\Vert\) being the Euclidean norm.
After normalization, this problem is equivalent to a minimization problem of an objective function J defined by:
under the constraints:
Random forest (RF)
A random forest classifier is a supervised machine learning algorithm which is recognized as an ensemble classification technique. It uses a “parallel assembling” that constructs multiple decision tree classifiers in parallel on various subsamples of datasets and takes their majority vote for classification.
Each tree in a random forest randomly samples subsets of the training data in a process known as bootstrap aggregating (bagging). The model is fit to these smaller data sets and the predictions are aggregated. The final prediction is an average of all of the decision tree predictions. Thus, it increases the accuracy of the prediction and minimizes the overfitting problem. Therefore, a random forest learning model with several decision trees is generally more efficient than a model based on a single decision tree [26].
Extreme gradient boosting (XGBoost)
Gradient boosting is an ensemble learning algorithm that produces a final model from a series of single models, usually decision trees. The gradient allows minimizing the loss function, in the same way that neural networks optimize weights by using gradient descent. The algorithm is based on the idea of “boosting” which combined all the predictions of a set of “weak” learners for developing a “strong” learner through additive training strategies. It iteratively train an ensemble of shallow decision trees, with each iteration using the error residuals of the previous model to fit the next model. At any instant t, the model outcomes are weighed based on the outcomes of previous instant t-1. The outcomes predicted correctly are given a lower weight and the ones miss-classified are weighted higher. The final prediction is a weighted sum of all of the tree predictions.
Extreme Gradient Boosting (XGBoost) is a type of gradient boosting that minimizes the loss by computing the second order gradients of the loss function, which improves model generalization and performance, and reduces overfitting [26]. XGBoost approaches the process of sequential tree building using parallelized implementation. It is quick to integrate, and it can manage large datasets. The additive learning process in XGBoost is explained below. The first learner is fitted to the input dataset, and then a second model is fitted to these residuals to overcome the drawbacks of a weak learner. This fitting process is repeated several times. The final model prediction is obtained by the sum of the predictions of each learner. The general function for the prediction at step t is presented as follows:
where \(f_{t}(x_{i})\) is the learner at step t, \(f^{(t)}_{i}\) and \(f^{(t-1)}_{i}\) are the predictions at steps t and (t - 1) respectively, and \(x_{i}\) is the input variable.
Performance evaluation
In our study, we used several metrics to evaluate ML models like the confusion matrix and associated metric parameters, such as: Accuracy, Precision, Recall, F1-score and AUC.
-
Accuracy is a measure of the classifier’s ability to accurately predict cases into their correct category. It is the proportion of valid results obtained or correctly classified samples from total samples.
$$\begin{aligned} Accuracy = \frac{TP+TN}{TP+FP+TN+FN} \end{aligned}$$(12)Where TP, TN, FP and FN represent the True Positive, True Negative, False Positive and False negative values, respectively. True Positive (TP) indicates real disease, which means that the real value is positive, and it is classified positively i.e., that the person has the disease, and the test is positive. False negative (FN) indicates no disease while it exists, which means the actual value is positive while it is classified negatively, i.e., that the person has the disease, and the test is negative. False positive (PF) indicates a disease when it does not exist, which means that the true value is negative when it is classified positively. True Negative (TN) indicates the absence of the disease, which means that the true value is negative, and it is classified as negative, i.e., that the person is healthy, and the test is negative.
-
Precision is defined as the ratio of correctly detected samples (true positives) to samples that have been detected as positive.
$$\begin{aligned} Precision = \frac{TP}{TP+FP} \end{aligned}$$(13) -
Recall, also called sensitivity, is the percentage of positive instances of a particular class that are correctly detected. It is defined as the ratio of true positive samples to the total number of positive samples.
$$\begin{aligned} Recall = \frac{TP}{TP+FN} \end{aligned}$$(14) -
F1-score is defined as the harmonic average of the precision and the recall.
$$\begin{aligned} F1\_score = \frac{2 \times Precision \times Recall}{Precision+Recall} = \frac{2 \times TP}{2 \times TP+FP+FN} \end{aligned}$$(15) -
Area Under the Curve (AUC): AUC measures the area under the curve that is obtained by plotting the True Positive rate (TPR) compared to the False Positive rate (FPR) at numerous threshold points. In the area of machine learning, the True Positive rate is also recognized as recall or sensitivity. Similarly, the False Positive rate is the fraction of negatives that are incorrectly detected. It indicates how much the model is able to distinguish between classes.
Results
This section presents the acquired results of the six classifiers that are being investigated in this paper. Indeed, the models were evaluated on the test data to determine their performance. The 2500 RGB images, including 500 images of each class, were fed into the system. To choose the most appropriate preprocessing method, we tested the Unsharp Masking and Stain Normalization methods and compared the performance of the models using these two methods. Table 2 presents the accuracy results for the different models with the use of each preprocessing method for the same training and test dataset.
Then, we compared the performance of the models with the use of different feature extraction methods and their combinations to select the most efficient methods. Table 3 presents the accuracy results for the different models and groups of characteristics, for the same training and test database.
The performance of the classification models on the same test data using the best feature extraction methods is presented in the table is shown in Table 4.
The confusion matrix for each technique and for the same dataset is shown in Fig. 5. The confusion matrix represents the true label versus the predicted label of the images for the test data in given labeled categories. Table 5 shows the precision, recall and F1-score of the XGBoost model for the different categories of histopathological images on the test data.
To be able to compare our model with existing models in the literature, we did the same work with the same steps but using the 25,000 images of colon and lung cancer from the LC25000 database. Tables 6 with 70% for training and 30% for testing, Tables 7 and 8 with 90% for training and 10% for testing, present a comparison of our achieved results of the classification of colon and lung cancer subtypes, colon cancer classification and lung cancer classification respectively with other methods of the literature using the same dataset.
In Figs. 6 and 7, we present the confusion matrix and the Receiver Operating Characteristic (ROC) curves of the classification on the testing subset using the XGBoost model.
ROC curve of: a Colon and Lung cancer classification, b Lung cancer classification, and c Colon cancer classification, using all the images of the LC25000 dataset and with the XGBoost classifier. The classes 0, 1, 2, 3 and 4 in a represent the classes Colon_Ad, Colon_Be, Lung_Ad, Lung_Be and Lung_Sc respectively. In b the classes 0, 1 and 2 represent Lung_Ad, Lung_Be and Lung_Sc respectively
Precision, recall and F1-score of the XGBoost model for the different classes of colon and lung cancer with 70% for training and 30% for testing are shown in Table 9. Tables 10 and 11 present the precision, recall and f1-score of the XGBoost model for the different classes of colon cancer and lung cancer respectively with 10% for testing.
Discussion
After introducing the images in the system, we compared the performances of the models using the Unsharp Masking and Stain Normalization preprocessing methods to choose the most appropriate one. From Table 2, we notice that all models gave better accuracy with the use of the Unsharp Masking method than with the stain normalization method. Stain normalization method does not have much impact on the accuracy of the classification models, this might be because all histopathological images used in our study belong to the same dataset which comes from the same hospital or scanners. Unsharp masking enhances the contrast, and therefore the sharpness of the original image, which can help emphasize texture and detail. So in the rest of the study, the Unsharp Masking method is used as image preprocessing method.
After preprocessing, the enhanced images were transformed to grayscale, and three texture extraction methods including first-order statistical features, GLCM and invariant Hu moments were used for feature extraction. In order to design the most efficient feature extraction methods, we compared the performance of the models with the use of these different methods and their combinations. According to Table 3, the analysis performed with the different methods resulted in using a combination of the three feature extraction methods: statistical features, GLCM and Hu invariant moments, since the most efficient model is obtained with these three combined feature extraction methods. An accuracy of 95.6%, 95%, 94.6%, 91%, 92.2% and 93.4% is obtained respectively with the classifiers XGBoost, SVM, RF, LDA, MLP and LightGBM on the test data, using a combination of the three feature extraction methods. Indeed, we notice that by calculating the statistical characteristics with the XGBoost model, we have 87.8% of classification accuracy. By calculating the GLCMs, we have 86.8% classification accuracy. By combining these two groups of features, we obtain 94.8% of accuracy. And by adding the features of Hu invariant moments, we get 95.6% of accuracy classification. Hence, the interest of using the three combined feature extraction methods. Therefore, a concatenation of the feature vectors extracted from these three methods resulted in the combined feature set with 37 features, which are the samples of the dataset in the training and classification steps.
The features extracted from the images were fed into the machine learning algorithms. 80% of the features (randomly chosen) are used to train the machine learning algorithm and the rest 20% are used as test data to evaluate the system performance. The acquired results show that ML models perform satisfactorily and are highly accurate in identifying lung and colon cancer subtypes. As shown in Table 4, the XGBoost model has the best accuracy of 95.6% and a F1-score of 96%. As shown in the confusion matrix in Fig. 5, only 22 samples out of 500 images have been incorrectly classified with the XGBoost classifier. The Lun_Be class achieved the greatest classification result, while the Col_Ad class got the highest misclassification result.
Overall, the results of this study indicate that the ML models, especially the XGBoost model, followed by the SVM and RF models, are very accurate in identifying classes of lung and colon cancer subtypes, although there is a remaining room for improvement. Therefore, the obtained results show that ML models can be used to classify histopathological images of colon and lung cancers with high reliability and precision.
The XGBoost model showed the best performance. Indeed, the SVM is simply a linear separator. However, XGBoost is an ensemble tree-based method that uses multiple trees to take a decision. It can capture dependencies among features and build rules based on the values of these features, whereas linear models cannot.
Random Forest simply creates a large number of trees in which each tree gives a prediction, and takes their majority vote for classification. Unlike XGBoost which is based on the idea of boosting where the objective is to minimize the loss function of the model by adding weak learners using gradient descent. Boosting is an iterative learning, which means that the model will predict something initially and self analyzes its errors as a predictive tutor and gives more weight to the data points in which it made a wrong prediction in the next iteration.
Both XGBoost and LightGBM are ensemble tree methods that apply the principle of weak learner reinforcement using the gradient descent architecture. In contrast to the level-wise growth of XGBoost, LightGBM performs leaf-wise growth, which can lead to overfitting as it produces very complex trees. Therefore, XGBoost is able to build more robust models than LightGBM.
Therefore, the XGBoost model is highly recommended for classifying colon and lung cancer subtypes from histopathological images.
According to Tables 6, 7 and 8, we notice that our model XGBoost achieves superior performance over the state-of-the-art approaches that use DL. For the classification of colon and lung cancer subtypes, our model achieves an accuracy of 99%, while reference [12] which used a CNN model had an accuracy of 96.33%. Similarly, for the classification of colon cancer and lung cancer, our model achieves an accuracy of 99.3% and 99.53% respectively, which is higher than the approaches in the literature [13]-[10] that use the CNN model, which have obtained an accuracy of 96.61% and 97.2% respectively.
As seen from the confusion matrix (a) in Fig. 6, only 90 samples out of 7500 images were misclassified. The class Lun_Be had the best classification outcome; whereas, the class Lun_Ad has the highest misclassification rate. These outcomes are also apparent in the ROC curves in Fig. 7. The curves are almost touching the top-left corner, as the classifier was very successful at distinguishing their samples. Lun_Be class has the highest AUC of 99.86%. Also, we notice from the confusion matrix (b) and (c) that only 13 and 7 samples out of 1500 and 1000 images were misclassified, respectively. And the curves are almost touching the top-left corner with AUC above 99%. Overall, we can say that the Xgboost model is very accurate in identifying the different classes of lung and colon cancer. Also, from Tables 5, 9, 10 and 11, we can see that the XGBoost model works well in identifying different classes of colon and lung cancer subtypes.
In most recently published research articles, the authors have used DL to classify colon and lung cancers’ histopathological images. Indeed, these previous studies used DL, while our study used ML. Our study has proved that with feature engineering we can find results that are competitive with DL approaches. XGBoost achieved an accuracy of 99% for the classification of colon and lung cancer subtypes, 99.3% for the classification of colon cancer, and 99.53% for the classification of lung cancer subtypes. We notice that the model for each type of organs are more efficient. In fact, our objective is not to compete with existing models, but to show the interest of ML and feature engineering models and to show that it is possible to find better results using ML models.
The main advantage of using conventional machine learning models is that they allow a better interpretability of the classification model since the computed features have an interpretable mathematical meaning, which is not the case for deep learning. Indeed, in the medical and diagnostic field, feature engineering is crucial for doctors because it allows them to know the importance and impact of each feature on the classification and identification of cancer subtypes, unlike deep learning models which are black box networks.
Model explainability with SHAP
The SHAP method is used to explain the output of a machine learning model by computing the contribution of each feature to the prediction. Therefore, it allows evaluating how the contribution of each feature affects the model [27].
The importance of SHAP features is calculated as the average of the absolute Shapley values. The idea of SHAP feature importance is that important features are those with great absolute Shapley values. Figure 8 illustrates the most important features that are selected and ordered according to their importance using the SHAP method for the previously trained random forest model for colon cancer prediction. The first order statistical features are the most relevant, followed by second order features such as correlation. Percentile 50% was the most relevant feature, which modified the absolute probability of predicted cancer by an average of 8 percentage points. Thus, on the medical side, specialists and doctors can interpret the variables and know which features are more important in identifying and classifying cancer subtypes.
The SHAP Summary Plot shown in Fig. 9 combines the importance of features with their effects. Each point on the graph represents a Shapley value for a feature and an instance. The x-axis position is determined by the Shapley value. The horizontal location shows whether the effect of that value caused a higher or lower prediction. The features on the y-axis are ordered according to their importance. Each line (y-axis) on the graph points to the feature on the left and is colored according to the value of the feature - high values for that feature are red, and low values for that feature are blue. Values to the right have a “positive” impact on the output, and the values to the left have a “negative” impact on the output. Note that positive and negative refer to the direction in which the output of the model is impacted, it has no guidance on the performance.
This plot allows us to visualize the impact of the feature, as well as how the impact of the feature varies with lower or higher values. For example, a large value of mean increases the risk of predicted colon cancer and a small value reduces the risk. The features presented such as mean, correlation1 and Percentile 25% have negative impacts for low values and positive impacts for high values. So, medically, specialists can understand the variables and know how the values of each characteristic impact the identification of colon cancer subtypes.
Figure 10 presents the SHAP force_plot output for two patients from the colon cancer dataset. The prediction begins from the base value. The base value for the Shapley values is the mean of all predictions. Then the prediction is modified accordingly based on the value of each feature. Feature values that increase predictions are in red, and their visual size shows the magnitude of the feature effect. Feature values that decrease predictions are in blue.
The first patient has a high risk prediction of 0.86 of having colon cancer. Median, Mean, Percentile 25% increase his predicted risk of cancer. The greatest impact comes from the median feature. Although energy1 has a significant effect on decreasing the prediction. The second patient has a low predicted risk of 0.27. Features that increase risk are compensated by features that decrease risk, such as homogeneity1.
Thus, having a justification for the prediction of a model would give specialists confidence regarding the validity of the model’s decision. Indeed, in the medical field, decision-making processes must be transparent, and then it is important to explain the model predictions in order to support the specialists’ decision-making processes.
Conclusion
In this paper, we presented machine learning models that are based on feature engineering for the classification of histopathological images of colon and lung cancers into five classes (three malignant and two benign).
We preprocessed the dataset using an image enhancement method known as unsharp masking. Three feature sets were extracted for the classification of images. The resulting features were then concatenated to create a combined feature set that was fed into the machine learning algorithms. The XGBoost model has the best classification performance in terms of accuracy, precision, recall and F1-score for distinguishing lung and colon cancer subtypes. XGBoost achieved an accuracy of 99% and a F1-score of 98.8%. SHAP method is used to provide an explanation of the output of a ML model and to evaluate how the contribution of each feature affects the model. Using this method, specialists can then understand which features of the histopathological image contributed to its classification as cancer. Unlike previous papers where the authors used DL which is a black box network, very difficult to interpret and in the medical field specialists cannot understand what is happening inside the model.
Thus, the use of computer programs that are based on machine learning and feature engineering to analyze data and extract important information could be a very useful and crucial tool in the medical field for the immediate and accurate diagnosis of malignant tumors. Indeed, these programs will be able to provide significant help to specialists to better interpret features and know the importance and impact of each on the identification of colon and lung cancer subtypes.
In the future, it is planned to explore other feature extraction techniques that provide relevant features for the identification of colon and lung cancer subtypes from histopathological sections to improve model performance. It is also planned to evaluate the performance of our proposed approach on other histopathological images of colon and lung cancer to evaluate its efficacy.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin 68(6):394–424
Bermúdez A, Arranz-Salas I, Mercado S, López-Villodres JA, González V, Ríus F, Ortega MV, Alba C, Hierro I, Bermúdez D (2021) Her2-positive and microsatellite instability status in gastric cancer-clinicopathological implications. Diagnostics 11:944
Togaçar M (2021) Disease type detection in lung and colon cancer images using the complement approach of inefficient sets. Comput Biol Med 137:104827. https://doi.org/10.1016/j.compbiomed.2021.104827
Sánchez-Peralta LF, Bote-Curiel L, Picón A, Sánchez-Margallo FM, Pagador JB (2020) Deep learning to find colorectal polyps in colonoscopy: a systematic literature review. Artif Intell Med 108:101923. https://doi.org/10.1016/j.artmed.2020.101923
Travis WD et al (2011) International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6:244–85. https://doi.org/10.1097/JTO.0b013e318206a221
Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, Snyder M (2016) Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 7:12474. https://doi.org/10.1038/ncomms12474
Bazazeh D, Shubair R (2016) Comparative study of machine learning algorithms for breast cancer detection and diagnosis. In: 2016 5th international conference on electronic devices, systems and applications (ICEDSA), pp 1–4. https://doi.org/10.1109/ICEDSA.2016.7818560
Schmidhuber J (2015) Deep learning in neural networks: an overview. Neural Netw 61:85–117
Bukhari SUK, Asmara S, Bokhari SKA, Hussain SS, Armaghan SU, Shah SSH (2020) The histological diagnosis of colonic adenocarcinoma by applying partial self supervised learning. https://doi.org/10.1101/2020.08.15.20175760
Hatuwal BK, Thapa HC (2020) Lung cancer detection using convolutional neural network on histopathological images. Int J Comput Trends Technol 68(10):21–24. https://doi.org/10.14445/22312803/IJCTT-V68I10P104
Nishio M, Nishio M, Jimbo N, Nakane K (2021) Homology-based image processing for automatic classification of histopathological images of lung tissue. Cancers 13:1192. https://doi.org/10.3390/cancers13061192
Masud M, Sikder N, Nahid AA, Bairagi AK, AlZain MA (2021) A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors 21:748. https://doi.org/10.3390/s21030748
Mangal S, Chaurasia A, Khajanchi A (2020) Convolution neural networks for diagnosing colon and lung cancer histopathological images. arXiv:2009.03878
Dargan S, Kumar M, Ayyagari MR et al (2020) A survey of deep learning and its applications: a new paradigm to machine learning. Arch Comput Methods Eng 27:1071–1092. https://doi.org/10.1007/s11831-019-09344-w
Borkowski AA, Bui MM, Thomas LB, Wilson CP, DeLand LA, Mastorides SM (2021) Lung and colon cancer histopathological images dataset| Kaggle. https://www.kaggle.com/andrewmvd/lung-and-colon- cancer-histopathological-images
Borkowski AA, Bui MM, Thomas LB, Wilson CP, DeLand LA, Mastorides SM (2019) Lung and colon cancer histopathological image dataset (LC25000) arXiv:1912.12142v1 [eess.IV].
Janowczyk A, Basavanhally A, Madabhushi A (2017) Stain normalization using sparse autoEncoders (StaNoSA): application to digital pathology. Comput Med Imaging Graph 57:50–61. https://doi.org/10.1016/j.compmedimag.2016.05.003
Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Schmitt C, Thomas NE (2009) A method for normalizing histology slides for quantitative analysis. In: IEEE international symposium on biomedical imaging. Boston, MA 1107–1110
Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, Steiger K, Schlitter, Anna M, Esposito I, Navab N (2016) Structure-preserving color normalization and sparse stain separation for histological images. In: IEEE transactions on medical imaging, vol 35, no 8, pp 1962–1971. https://doi.org/10.1109/TMI.2016.2529665
Ciompi F, Geessink O, Bejnordi BE, Bejnordi B, de Souza GS, Baidoshvili A, Litjens G, Van Ginneken B, Nagtegaal I, Van Der Laak J (2017) The importance of stain normalization in colorectal tissue classification with convolutional networks. CoRR. arXiv:1702.05931
Lafarge MW, Pluim JPW, Eppenhof K, Moeskops P, Veta M (2017) Domain-adversarial neural networks to address the appearance variability of histopathology images. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, DLMIA, Québec City, QC pp 83–91
Alinsaif S, Lang J (2020) Texture features in the shearlet domain for histopathological image classification. BMC Med Informat Decis Making 20(S14):1–19
Madero Orozco H, Vergara Villegas OO, Cruz Sánchez VG, Ochoa Domínguez H, Nandayapa Alfaro M (2015) An automated systems for lungs nodule classifications based on wavelet feature descriptors and support-vector-machines. Biomed Eng Online 14(1):9
Aggarwal N, Agrawal RK (2012) First and second order statistics features for classification of magnetic resonance brain images. J Signal Inf Process 3(2):146–153. https://doi.org/10.4236/jsip.2012.32019
Li M, Ma X, Chen C, Yuan Y, Zhang S, Yan Z, Chen C, Chen F, Bai Y, Zhou P, et al (2021) Research on the auxiliary classification and diagnosis of lung cancer subtypes based on histopathological images. IEEE Access 9:53687–53707
Sarker IH (2021) Machine learning: algorithms, real-world applications and research directions. SN COMPUT. SCI. 2:160. https://doi.org/10.1007/s42979-021-00592-x
Molnar C (2019) Interpretable machine learning. A guide for making black box models explainable. https://christophm.github.io/interpretable-ml-book/
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study uses public databases cited in the references and therefore this section is not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hage Chehade, A., Abdallah, N., Marion, JM. et al. Lung and colon cancer classification using medical imaging: a feature engineering approach. Phys Eng Sci Med 45, 729–746 (2022). https://doi.org/10.1007/s13246-022-01139-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-022-01139-x