Abstract
In this paper, a joint use of the discrete cosine transform (DCT), and differential pulse code modulation (DPCM) based quantization is presented for predefined quality controlled electrocardiogram (ECG) data compression. The formulated approach exploits the energy compaction property in transformed domain. The DPCM quantization has been applied to zero-sequence grouped DCT coefficients that were optimally thresholded via Regula-Falsi method. The generated sequence is encoded using Huffman coding. This encoded series is further converted to a valid ASCII code using the standard codebook for transmission purpose. Such a coded series possesses inherent encryption capability. The proposed technique is validated on all 48 records of standard MIT-BIH database using different measures for compression and encryption. The acquisition time has been taken in accordance to that existed in literature for the fair comparison with contemporary state-of-art approaches. The chosen measures are (1) compression ratio (CR), (2) percent root mean square difference (PRD), (3) percent root mean square difference without base (PRD1), (4) percent root mean square difference normalized (PRDN), (5) root mean square (RMS) error, (6) signal to noise ratio (SNR), (7) quality score (QS), (8) entropy, (9) Entropy score (ES) and (10) correlation coefficient (r x,y ). Prominently the average values of CR, PRD and QS were equal to 18.03, 1.06, and 17.57 respectively. Similarly, the mean encryption metrics i.e. entropy, ES and r x,y were 7.9692, 0.9962 and 0.0113 respectively. The novelty in combining the approaches is well justified by the values of these metrics that are significantly higher than the comparison counterparts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The incorporation of wireless communication technology in the field of the tele-cardiological platform has played a vital role in the timely monitoring of electrocardiogram (ECG) signal, especially for remote areas [1, 2]. The ECG signal monitors the electrical activity of the heart. It is usually characterized by its various set points (P, QRS, T) and intervals (PR interval, QT interval, and RR interval) that reflect the rhythmic electrical depolarisation and repolarisation of atria and ventricles [3]. The clinical diagnosis of cardiac abnormalities is based upon the morphological information obtained from these fiducial points [4, 5]. By exploiting the inter, and intra-beat correlation the redundant information in ECG could be removed thereby resulting in the compression of ECG data [6].
In general, ECG data compression can be classified into lossy and lossless techniques [7]. The lossless compression ensures the integrity of reconstructed data. The obtained compression ratio (CR) is nearly 2–4 with 0 % reconstruction error measured in terms of percentage root mean square difference (PRD) [8]. On the other hand, lossy compression has high CR with varying level of reconstruction error [7]. ECG signal compression techniques widely fall into the three categories of the direct method [9–12], transformed method [10, 13–18] and parameter extraction method [19, 20]. The lossy data compression results in reduction of bit rate per sample by utilizing the property of energy compaction and quantization. The significant ECG diagnostic features are well preserved at the minimum possible bit rate using this approach. The other methods for compression include the grouping of the ECG data samples followed by encoding them using standard ASCII characters [21, 22].
All the prior work reported on ECG compression demonstrate that there is a trade-off between CR and the reconstruction error. Considering, in [15] Lee et al. have used the discrete cosine transform (DCT) between the current peak and the previous peak of the ECG by changing window size from 100 to 15 %. The CR ranged from 5.19 to 27.9 while reconstruction error varied from 0.23 to 2.93. Batista et al. [14], used Lagrange multipliers for the minimization of entropy in ECG signal for a given distortion rate and then employing arithmetic coder for the lossless encoding of the quantized coefficients. They achieved an average CR of 9.3 at PRD equal to 2.5 % of all records of MIT-BIH arrhythmia database. In [23] optimized DCT coefficient quantization was introduced along with Golomb-Rice coding, that leads to PRD equal to 2.5 with CR of 10.8 for particular record 100 from the same database as mentioned earlier. ASCII character encoding in the combination with the set of signal processing techniques like downsampling, differencing, normalization and encoding have been employed in the works by Mukhopadhyay et al. [21] and Gupta et al. [24] that is further used for efficient transmission and storage.
There are challenges of accomplishing higher CR while keeping low reconstruction error. An efficient and simplistic approach that utilizes the energy compaction property along with zero sequence encoding and optimum quantization is needed to mitigate this drawback. A joint application of transform domain method for ECG data compression such as DCT and direct method such as efficient zero sequence grouping followed by optimum differential pulse code modulation (DPCM) encoding and ASCII coding provides a breakeven point between CR and PRD.
After efficient lossy compression mechanism, there is a need for suitable encryption method for concealing of the essential diagnostic ECG features, as biometric identification can be obtained using those features [25–27]. Statistical investigation validates the stated inherent encryption mechanism. Sufi et al. [28] proposed an efficient ECG obfuscation method that involved detection of P, QRS complex, and T-wave feature from ECG and then replacing them with their noisy versions. The advantage of the proposed approach over existing encryption method was that the corrupted ECG appears as regular ECG though it is an encrypted ECG. Chen et al. [29] used quantization-based digital watermark encryption technology on the ECG to protect patient rights and information. The confidential patient identity information was treated as a watermark for medical data. They concluded that the difference between the watermarked ECG and the original one was subtle and negligible for physiological diagnostics.
Existing ECG encryption possesses the disadvantage of distorting the diagnostic features along with the generation of enormous side information. The present work alleviates this drawback via exploration of inherent ECG features. The proposed combined technique employs a block-wise DCT for energy compaction of significant coefficients.
Depending upon user defined reconstruction error the significance of DCT coefficients are decided through the optimal threshold parameter, calculated via the Regula-Falsi method as reported in the works by [30]. For higher UPRD, higher is the value of optimal threshold and lesser would be the retained DCT coefficients for reconstruction. After that by zero sequence grouping, DPCM based quantization, lossless Huffman coding followed by ASCII Encoding scheme results in efficient encryption of the ECG segments.
The rest of the paper is organized as follows: the “ECG database and performance measures” consists database along with the performance measure description, “The proposed ECG compression scheme” introduces proposed ECG compression scheme in detail. Results discussions and comparisons with previous work are reported in “Results and discussion”. Finally, “Conclusion” concludes the paper.
ECG database and performance measures
In the emerging area of medical engineering, research issues like cardiac arrhythmia detection [31], heart rate variability [32], cardiovascular diagnosis dynamics in telecardiology [33], artificial intelligence based medical decision support and ECG compression, etc. are of major interest [15]. The Massachusetts Institute of Technology (MIT) provides some valuable resources for such research projects. These resources include databases and software for obtaining, viewing, and analysing various physiological signals. In the present study, ECG samples from arrhythmia database [34] (http://ecg.mit.edu) have been taken for investigating the efficiency of the proposed method. The database contains 48 half-hour excerpts of two-channel ambulatory ECG recordings, obtained from 47 subjects studied by the BIH arrhythmia laboratory. The 11-bit resolutions over 10 mV range at sampling frequency 360 Hz per channel has been recorded. To access the efficiency of proposed compression technique, first 65,0000 samples (30 min duration) has been chosen and evaluated using seven ECG data compression metric [8, 10, 14–16]. A baseline of 1024 is added, and gain of 200 is multiplied to each sample of ECG data for normalization. The purpose of normalization is to scale the ECG sample value in the smallest possible positive integer for proficient storage.
Compressor performance measures
As stated in literature the proficiency of any ECG data compression algorithm is estimated using both the visually and the quantitatively. Seven performance measures exist in the literature [15, 21, 35, 36].
Compression ratio (CR)
The CR provides information about the extent by which the compression algorithm eliminates the redundant data. Higher the CR lesser is the number of bits required to store or transmit the data and is defined as follows
where B 0 is the total number of bits required to represent original ECG data and B c total number of bits required to represent compressed ECG data stream along with the side information needed for retrieval of original data. Moreover, CR is alternatively defined as
where f s is the sampling frequency, N is the total number of samples in ECG data, \(B_{c}\) is the total number of bits in compressed ECG data and B s is the bits required to store the side information.
Percentage root mean square difference (PRD)
It quantifies the acceptable fidelity of the reconstructed signal. It measures the degree of distortion in the reconstructed signal; that was introduced during the compression and decompression procedures.
Percentage root mean square difference without base (PRD1)
As MIT-BIH arrhythmia database incorporates an offset value of 1024 in each ECG record. The PRD1 provides a more reliable parameter than PRD. PRD1 measures the compressor efficiency by offsetting this effect by subtracting this value from the original signal in the denominator.
Percentage root mean square difference, normalized (PRDN)
It is a normalized version of PRD. The difference from PRD is that PRDN does not depend upon the signal mean value \(\overline{X}\).
Root mean square error (RMS)
This measure provides the error in the reconstructed signal with respect to the original signal.
Signal to noise ratio (SNR)
SNR, measure of degree of noise energy introduced by compression mechanism in decibel (dB) scale.
Quality score (QS)
It is the ratio of the CR and PRD. The QS is a very rational performance indicator when it is difficult to estimate the best compression method while taking into account the reconstruction errors as well. Higher the QS, superior is the performance of compression method.
Encryption performance measures
The randomness in the compressed ECG can be evaluated using the existing encryption performance measures like histogram, entropy, and correlation analysis [37–39]. Out of these three, histogram analysis was used for visual inspection and the remaining measures were used for statistical analysis.
Histogram analysis
Histogram analysis provides the probability distribution of given data samples. The non-uniformity in histogram accounts due to the higher correlation in the time series. Further, the encrypted time series represents true randomness as depicted via uniform histogram.
Entropy analysis
This measure accounts for the randomness present in the signal. Higher the entropy greater will be the randomness. The information entropy of the data is defined as
where M is the total number of samples \(n_{i}\) ∈ \(n_{{}}\); \(p(n_{i} )\) denotes the probability of occurrence of the sample \(n_{i}\) and log with base 2 signifies that the entropy expressed in terms of bits. Considering an infinite population, with the uniform distribution of random data having 1024 amplitude levels (10-bit resolution or 10-bit quantization level), then ideally the value of \(E(n)\) = 10-bits. For the fair comparison of entropy values with two different information sources, which were having different quantization levels (resolution), a new parameter called entropy score (ES) has been proposed. It is defined as
where resolution is represented as the number of bits per sample for the given information source. The ES accounts for the normalization of entropy value, against different quantization levels. The maximum value of ES is unity, indicating a uniform distribution of samples. The minimum value of ES i.e. 0, denotes that the same sample value is repeated over the entire sample space. Higher the ES, more is the randomness and lesser is the correlation in the given time series.
Correlation analysis
Correlation analysis has been reported in the present work so as to estimate the proficiency of the encryption mechanism. Correlation values close to zero indicates that the signal has a strong ciphering capability. The correlation coefficient \(r_{x,y}\) of the pair of data points is given as:
where x and y are two adjacent data points in the sample space. \(E(x)\) and \(D(x)\) are expectation and variance of x. M is the total number of considered samples. The encrypted time series has a lesser value of the correlation coefficient indicating that the data is highly uncorrelated.
The proposed ECG compression scheme
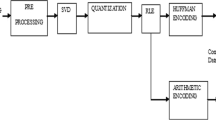
The application of optimal threshold based DCT and ASCII Encoding in ECG data compression address the issue of achieving the desired PRD corresponding to defined CR. As shown in Fig. 1a, the proposed ECG compression scheme is primarily composed of six stages. The original ECG signal, \(X_{s} \left( n \right)\) obtained through predefined lead configuration can be represented in terms of samples as:
where \(n = 1, \, 2, \ldots N\). Here N is the product of sampling frequency f s and time duration (in seconds) of ECG data.
Down sampling
The obtained ECG is down-sampled by factor 2 resulting in 50 % reduction in ECG data. To analyse the effect of this procedure on compression parameters, the signal is reconstructed using cubic spline interpolation mechanism. On an average CR equal to 2 with minimal PRD value equal to 0.12 was achieved when estimated for all the 48 records.
Block wise DCT and optimal selection of threshold
After the downsampling stage, the selection of optimal threshold was done using the iterative Regula-Falsi [40] numerical method. DCT coefficients having a value higher than the calculated threshold are retained for further processing. The detailed algorithm for optimal threshold selection for given PRD is similar to that reported in [30]. The analogous procedure is adopted for the present work that can be diagrammatically outlined in the form of a flowchart in Fig. 1a, where TH 0 corresponds to the optimal threshold value. The initial two temporary thresholds (TH_Temp0, TH_Temp1) are chosen as any random number that finally converges to TH 0 via this approach.
The PRD at different values of the threshold is calculated as needed, irrespective of the initialization of threshold. The algorithm is designed to reiterate until the reconstruction error falls within the tolerable limits (\(\varepsilon\)) of the PRD as reported in works by [30]. As experimented on all the 48 records of the MIT-BIH database the condition gets satisfied in less than 100 iterations thereby giving the optimal threshold value. The predefined \(\varepsilon\) is kept to 10 % of the user defined PRD (UPRD) [30]. Next, the signal is formulated and re-assembled according to the UPRD, and the appropriate block size is computed using inverse DCT. The iterative approach for obtaining TH
0 is illustrated in following steps 
Zero sequence grouping
The aim of this stage is to encode three or more consecutive zeros and to formulate a new series that encodes the location of three or more consecutive zeros. The side information generated by this step works as an inherent encryption key that provides the location, as well as the number of zero sequences in between the non-consecutive zeros DCT coefficients. For illustration, consider.
DCT coefficient before zero sequence grouping with block size and UPRD equal to 64 and 0.4 respectively: {−144.20 −34.47 40.79 25.11 25.81 0 17.99 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 17.17 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0}.
DCT coefficients after zero sequence grouping: {−144.20 −34.47 40.79 25.11 25.811 0 17.99 17.17}.
Location of three or more consecutive zeros: {8 16}.
Number of consecutive zeros: {15 42}.
The generated output sequence results in three different time series; First is the significant DCT coefficients with non-consecutive zeros, second indicates the location of three or more consecutive zeros, and the last denotes the number of consecutive zeros at the location of three or more successive zeros. The second and third output acts as an inherent encryption key that is unique to each and every ECG data.
Quantization of un-grouped DCT coefficients
After the zero sequence grouping stage, the ECG data can be represented in two portions, (1) firstly the thresholded DCT coefficients, (2) secondly the location and length of consecutive zeros. Furthermore, the DCT thresholded coefficients are quantized using DPCM [10, 41].
The quantization resolution is controlled by the number of bits per sample (m). Because of this step reconstruction error \(\varepsilon_{q}\) due to quantization can be expected. DPCM quantization works by using a variable width of bits, which are suitable for the nature of each data. The entire sample space which is quantized using m bits is converted into the binary equivalent. Furthermore, chunks of 8 bit each is extracted for ASCII encoding. The eight bits were formulated by concatenating the bits from the next samples where ever needed. Additionally, zero padding was done at the end of the DCT sample space to make it suitable for ASCII code conversion .
.
Huffman coding and ASCII encoding
For the final process of compression, Huffman coding along with the ASCII encoding has been utilized. The procedure assigns a high-bit format (i.e. a long string of bits) to fewer occurring samples while allocating a low bit format (i.e. a short string of bits) to more frequently occurring data samples. The sample size reduction was 19.98 % on an average when encoded using this algorithm. Afterward, the algorithm is designed such that these encoded symbols are converted to the ASCII characters that can be transmitted through the existing transmission models.
Reconstruction of the compressed ECG data
The reconstruction of raw ECG data from the compressed file is accomplished by following the inverse operation of compression stages. The first stage of restoration procedure was to separate and estimate the side information and compressed ECG signal from the received file. The second stage involves ASCII decoding and inverse Huffman coding, which defined in the corresponding data table, as the frequency of occurrence of the data samples. The third stage converts the Huffman decoded samples back to m-bit quantization levels from 8-bit data type, followed by DPCM based decoding. This decoding is done using the code book and predictor, which was referred to the encoding procedure. The fourth stage deals with appending zeros, which are grouped on the encoding side at a particular location. The fifth step proceeds by taking the inverse DCT using the same encoded DCT block size followed by reverting the down-sampling process of time series using spline interpolation. Finally, the estimated ECG signal is available for cardiac analysis.
Proposed ECG data compression algorithm with typical example
A typical example of record 117 is illustrated in Fig. 2 with 128 original ECG samples at UPRD = 0.40. Step by step data processing is provided using 64 block size followed by zero sequence grouping and DPCM based quantization. Finally, lossless encoding using Huffman and ASCII encoding is done. The overall CR and PRD 16.66 and 0.47 were achieved during the due course.
Inherent ECG data encryption
The proposed method serves as an ECG encryption mechanism also, with the following merits.
-
Due to randomness in the nature of the side information, it’s hard to predict the encryption key.
-
There is no need for extra encryption mechanism (advanced encryption standard or data encryption standard) due to the inherent data-hiding mechanism.
-
The average size of the encryption key is only 1.6 % of the original data length.
The length of original data is calculated as \(f_{s} \times T\) where \(f_{s}\) is the sampling frequency and \(T\) is the time in seconds. The side information is calculated as number of location of three or more consecutive zeros (\(L_{t}\)) and number of consecutive zeros (\(C_{z}\)) along with Huffman coding dictionary (\(H_{cd}\)). All the three components are added to provide the side information as represented by the R in the equation below, which is 1.6 % of the original data length when averaged over all the 48 records of the MIT-BIH database.
\(K\) in the above equation refers to the encryption key.
Zero sequence detection algorithm serves the purpose of lossless grouping thereby facilitating inherent ECG data encryption; zero sequences encoded as the location of three or more consecutive zero and number of consecutive zero as sequence encoding table. A correct encoding table is needed for accurate ECG signal reconstruction otherwise, it is impossible to decipher the original ECG signal.
To reconstruct the ECG signal authorized users should be facilitated with sequence encoding table and block size to decode compressed encrypted ECG data. Furthermore, the encryption key was generated according to the data to be fetched, which varies from one ECG data to other.
Results and discussion
This section describes the results of several sets of experiments, in order to assess the effectiveness of the proposed method over all the 48 record of the standard arrhythmia MIT-BIH database. Both the compression and encryption performance are presented.
Compression analysis
The considered time duration of ECG segments was 30 min and was varied in accordance with the framework used in literature. The comparison of proposed work with existing ECG coder was also evaluated. For the visualization of the algorithm efficiency, the results are presented for the typical record117.
Figure 3 shows the results of the proposed algorithm for different chosen values of UPRD (0.2, 0.4 and 1) for record 117. Reconstruction error is clearly visualized at an optimum threshold value of 46.66 with UPRD equal to 1.0. Whereas for UPRD 0.20, the significant difference exists between reconstructed and the original ECG signal, which cannot be easily visualized when the threshold values were lesser than 15.21 were used.
Original signal and reconstructed signal segment of record no. 117 at different PRD for quantization resolution of 8 bit for 30 min duration. a Original data. b CR = 10.92, PRD = 0.30, UPRD = 0.2 and TH = 5.14. c CR = 16.64, PRD = 0.45, UPRD = 0.4 and TH = 15.21. d CR = 23.57, PRD = 1.01, UPRD = 1 and TH = 46.66
The difference between PRD from UPRD was mainly due to the quantization resolution. If the higher number of quantization bits (m) are assigned per sample, then the degree of closeness of PRD to UPRD increases. Figure 4, illustrates the variation in CR with respect to PRD at different levels of m. The various values of m are chosen to explore the effect over CR and PRD. From the analysis, it can be concluded that the decrease in m results in an increase in the performance of the compressor (CR/PRD ratio) up to a certain level. To achieve low reconstruction error, there is a need to increase the quantization resolution. For achieving the maximum compression efficiency of the algorithm with PRD greater than 1.4, lower values of m must be chosen as justified in Fig. 4. For 6-bit quantization resolution, linear relationship between CR and PRD curve is achieved after PRD equals to 0.6.
At lower PRD, linear relationship between CR and PRD curve is achieved after PRD equals to 0.6 in the case of the 6-bit quantization resolution. The linear relationship for 8-bit quantization resolution begins from PRD = 0.38 for record 117. For lesser PRD (<0.6), 8-bit quantization is preferred in contrast to 6-bit quantization for the same PRD as shown in Fig. 4. This is in well accordance with the concept that higher resolution is needed for lower PRD. At lower PRD, linear relationship between CR and PRD curve is achieved after PRD >0.6 in case of the 6-bit quantization resolution, and the same linear relationship is retained for 8-bit quantization resolution that begins with PRD = 0.38. It is observed that morphological features are preserved better for the higher number of quantization bits at the cost of lower CR.
Three sets of experiments were done to explore the different combinations of UPRD and m namely (1) UPRD = 0.40 and m = 6, (2) UPRD = 1.0 and m = 6 and (3) UPRD = 1 and m = 8. For these chosen experimental values, the reconstructed ECG segments are shown in Figs. 5, 6 and 7. As seen in Fig. 5, typically a CR value of 16.66, PRD = 0.47 was achieved at m = 6 for an optimal threshold value of 15.46. Furthermore, with an increase in the value of m to 8, reduction in the value of CR was noticed with improvement in PRD. The values of CR, PRD and optimal threshold are equal to 12.95, 0.41 and 15.46 respectively as shown in Fig. 6. Therefore, it can be concluded that by varying the value of m from 6 to 8, at a fixed value of UPRD equal to 0.4 results in an improvement in the quality of reconstructed signal at the cost of the CR as shown in Figs. 5 and 6.
First 1280 ECG samples of record 117 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 16.66, UPRD = 0.40, PRD = 0.47 and threshold = 15.46. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
First 1280 ECG samples of record 117 and its DCT coefficients for block size = 64, with quantization resolution (m) of 8 bit for 10 min duration, CR = 12.95, UPRD = 0.4, PRD = 0.41 and threshold = 15.46. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
First 1280 ECG samples of record 117 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 23.97, UPRD = 1.0, PRD = 1.01 and threshold = 47.59. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
Also, at the fixed value of m and the higher value of UPRD equal to 1.0 the quality of the reconstruction degrades in comparison to that obtained using UPRD equal to 0.4. Typical findings are shown in Fig. 7 for which a CR equal to 23.97 and PRD = 1.01 has been achieved with an optimal threshold value equal to 47.59 at m = 6.
The reconstructed signal quality was also estimated, both visually and quantitatively for records 100 and 119 in addition to the typical record 117. From the previous observations for record 117, the quantization resolution ‘m’ equal to 6 yielded the best results. For all other records, the analyzes were quoted only for m = 6 as it represented the best trade-off between CR and PRD. Effect of the variations in the UPRD, on the signal reconstruction quality can be visualized in Figs. 8 and 9 for record 100 and in Figs. 10 and 11 for record 119.
First 1280 ECG samples of record 100 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 18.15, UPRD = 1.0, PRD = 1.0 and threshold = 41.32. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
First 1280 ECG samples of record 100 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 11.86, UPRD = 0.40, PRD = 0.42 and threshold = 15.11. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
First 1280 ECG samples of record 119 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 17.61, UPRD = 1.0, PRD = 1.15 and threshold = 41.47. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
First 1280 ECG samples of record 119 and its DCT coefficients for block size = 64, with quantization resolution (m) of 6 bit for 10 min duration, CR = 11.61, UPRD = 0.40, PRD = 0.72 and threshold = 15.11. a Original ECG segment. b DCT coefficients without thresholding. c DCT coefficients estimation after optimal thresholding by Regula-Falsi method. d Reconstructed ECG segment. e Reconstruction error in ECG segment
From the graphical scrutinization of the reconstructed signal at different values of UPRD equal to 0.40 and 1.0, it was seen that the signal restoration quality degrades. Typically a value of CR = 18.15, PRD = 1.01 was obtained for an optimal threshold value equal to 41.32 at UPRD equal to 1.0 as shown in Fig. 8.
Similarly, the decrease in value of CR and PRD equal to 11.86 and 0.42 was noticed when UPRD declined to 0.40. All these results were obtained at an optimal threshold value of 15.11. The obtained trend can be visually inspected from Fig. 9.
To generalize the proposed method for different ECG records possessing different morphological features; the experiments were also performed on record 119. Figure 10, 11 shows that same trend followed as verified in the previous records, i.e. increase in UPRD results in the increase in CR and PRD. This increase in CR is at the cost of degradation of the reconstruction signal quality. The typical values obtained are clearly indicated in figure captions.
Table 1 shows the comparative analysis of CR, PRD, PRDN, RMS, SNR and QS for all records of MIT-BIH arrhythmia database for quantization bit resolution of 6. The mean values of CR, PRD1, UPRD, PRD, PRDN, RMS, SNR and QS of 48 subjects were 18.59, 12.01, 1, 1.06, 17.09, 10.20, 15.95, 17.57, and 44.96 respectively. The efficacy of the proposed method was duly justified by its comparison with the existing state of art compression algorithms as given in Table 2.
In Table 2, fifth column, a brief description of ECG test sets and duration is presented, and the last column represents the quantization resolution used (#b). Same database, sampling frequency, lead configuration and time duration were used for ECG facilitating the fair comparison with existing techniques in the literature. As these factors heavily affect the CR and PRD, accordingly anticipation is required in the compressor.
The advantage of the proposed method is its superior CR, PRD and QS performance; which is in agreement with the wide consensus that “QS is a better performance indicator while taking into consideration the both aspect reconstruction distortions (PRD) as well as a quantitative description of the compression (CR).” Furthermore, the proposed method performs excellent by obtaining better QS as compared to other methods. From the formulated results in Table 2, it can be concluded that values of CR, PRD and QS is significantly better for proposed method in comparison to various lossy, lossless and tends to lossless methods.
Encryption analysis
The statistical and visual security analysis for developed inherent ECG data encryption is analysed in this section. For detection of ECG samples, there is need to have advance knowledge of block size, m, the location of zeros and number of consecutive zeros. As block size and m remains same for entire ECG compression algorithm, the main inherent security is achieved by location and number of consecutive zeros. For example, the detection of 4 min ECG data with 360 Hz sampling frequency, having 64 DCT block size and 6-bit quantization resolution (for record 117 with UPRD = 1.0), the needed random combinations were 846 × 846 for correct detection of encoded ECG. Which implies that the possibility of correct ECG data reconstruction is 1/(846 × 6 × 846 × 6) ≈ 3.88 × 10−08, which tends to zero.
Key sensitivity
Figure 12 gives the visual representation of key sensitivity. Only a small change in the particular sample either in location or number of consecutive zeros can affect the reconstructed ECG data. For a typical example, the location of zeros were changed in one of the location, the results were revealed for records (1) 100, (2) 117 and (3) 119 shown in Fig. 12. It indicates the original signal (a), the reconstructed signal with incorrect key (g) and the error signal (h). However a tiny change in the inherent key space sample will lead to a drastic change in reconstructed ECG data, so there is a need for accurate handling the inherent key space. Thus, it can be concluded that inherent key space has high key sensitivity.
Visual ECG data security representation for typical records (1) 100, (2) 117 and (3) 119 with block size 64, UPRD = 0.40. a Original ECG data, b histogram representation of original ECG, c compressed ECG data after Huffman coding, d histogram of compressed ECG, e reconstructed ECG data with correct key space, f error signal, g reconstructed ECG with incorrect key, h error signal between original ECG signal (a) and reconstructed ECG data with incorrect key (g)
Histogram analysis
The generated histogram for original and inherently encrypted ECG data depicted in Fig. 12b, d respectively.
The histogram of the encrypted data should be uniform which means equiprobable data points are present. Encryption relates to in deciphering capability of any algorithm. The histogram analysis is one of the subjective measures for estimation of such capability. The more is the spread in the histogram lesser is the correlation and higher is the encryption.
In Fig. 12d all the three records shows uniform histogram as compared to original ECG data. That shows the novelty of the proposed ECG data compression scheme.
Entropy analysis
Entropy is being used to check the presence of randomness in the time series. An ES close to one means better randomness property. Simulation results for records 100, 117 and 119 are listed in Table 3. It shows that, the entropy as well as ES for original and inherently encrypted ECG data that were having the resolution of 11 bits per sample and 8 bits per sample respectively. From the results, the ES for all the inherently encrypted ECG records are close to one. The average entropy for encrypted ECG obtained by proposed method is 7.9692, which very close to ideal value 8. Which validates the proposed scheme is having the potential for inherent encryption. This feature prevents information leakage from the ECG data.
Correlation coefficient analysis
To test the correlation in ECG time series between two adjacent randomly selected data points, the following procedure is carried out. Firstly randomly select 2000 pairs of samples of adjacent data points from original ECG samples and inherently encrypted ECG samples. Then the correlation coefficient \(r_{x,y}\) for each pair is calculated using Eq. 11, and the results are listed in Table 4.
Figure 13 represents the correlation coefficient of two randomly selected adjacent ECG data points. It is obvious that the neighbouring data points in inherently encrypted ECG have very less correlation.
Discussion of essential features for proposed technique
Performance comparison
After scrutinizing the findings reported in Table 2, it can be concluded that the proposed approach outperforms the existing works reported by Batista et al. [14], Lee et al. [15], Mukhopadhyay et al. [22], Gupta et al. [24], Hilton [42], Ku et al. [43], Sadhukhan et al. [44]. and Aggarwal et al. [45] in terms of CR, PRD and QS. The work shows an extensive comparison at varying levels of PRD and resolution. Experimentation have been done for selected records as quoted in the literature for fair comparison. Significantly for record 121 lead-1 the obtained PRD was 0.69 in comparison to 3.023 [24] at CR equal to 19.80.
Merits of the proposed work
The designed approach applies zero sequence grouping mechanism by identifying the location and number of three or more consecutive zeros which makes it computationally simple and efficient. Moreover, the formulated approach does not involve sign encoding for any element as used previously in the literature [24, 33, 46]. Due to the addition of offset value that is also termed as the dc value, the total magnitude remains in defined dynamic range. The dc value is selected as the minimum value of the corresponding grouped DCT coefficient sequence. The elimination of sign encoding stage provides two inherent advantages, firstly, in the encryption part, as the minimum value offset acts as the key to decrypt or carry out the reverse operation. Secondly reduction in the de-compression time is achieved as there as there is no need to find the sign byte and to reconvert the location associated with positive and negative numbers. Lastly, the devised approach eliminates the need for QRS detection and achieves customizable distortion rate. QRS detection stage is avoided by selection of fixed block size DCT, which remain constant for the entire range of the considered ECG data. Also, the distortion rate in the reconstructed ECG signal can be controlled by formatting the data with UPRD via an iterative Regula-Falsi numerical method.
Short coming of proposed work
Approximately 2–3 % of segment size (ECG time duration) is needed for key formation. Efforts can be made to reduce the key size, thereby ensuring the speedy decryption and decompression of ECG data for real-time applications.
Conclusion
In this paper, an algorithm based on joint use DCT domain, zero sequence encoding, DPCM quantization and ASCII encoding is presented to improve the ECG data compression efficiency along with the efficient inherent encryption. The experimental results show that the proposed algorithm performs better than existing DCT domain ECG compression techniques and ASCII encoding.
Some performance degradation can be tolerated for higher CR for given PRD due to downsampling, thresholding and quantization. Moreover, following points are the key contribution:
-
Elimination of the QRS detection process from ECG signal
-
The guaranteed-retrieved quality is ensured by matching the desired, fixed in advance, PRD within a tolerance limit of 1 %. The convergence is done after few iterations by using numerical methods such as bisection, false position.
-
The simple and efficient DPCM quantization approach is applied to the ungrouped DCT coefficients.
-
The Huffman encoding and ASCII coding was applied to the quantized DCT coefficients, which improve the compression efficiency.
-
Overall, obtained inherent encryption for compressed ECG was statistically validated.
Doctor’s Comment on the original and reconstructed ECG signal.
ECG record detail.
2 min of record 100, 117, and 119 ECG signal from MIT-BIH Arrhythmia database.
References
Baig MM, Gholamhosseini H, Connolly MJ et al (2013) A comprehensive survey of wearable and wireless ECG monitoring systems for older adults. Med Biol Eng Comput 51:485–495
Karthikeyan P, Murugappan M, Yaacob S (2013) Analysis of stroop color word test-based human stress detection using electrocardiography and heart rate variability signals. Arab J Sci Eng 39:1835–1847
Li Y, Yan H, Hong F, Song J (2012) A new approach of QRS complex detection based on matched filtering and triangle character analysis. Australas Phys Eng Sci Med 35:341–356
Martis RJ, Acharya UR, Adeli H (2014) Current methods in electrocardiogram characterization. Comput Biol Med 48:133–149
Matsuyama A, Jonkman M (2006) The application of wavelet and feature vectors to ECG signals. Australas Phys Eng Sci Med 29:13–17
Alesanco A (2008) Automatic real-time ECG coding methodology guaranteeing signal interpretation quality. IEEE Trans Biomed Eng 55:2519–2527
Sayood K (2010) Introduction to data compression, 3rd edn. Elsevier, Amsterdam
Koski A (1997) Lossless ECG encoding. Comput Methods Programs Biomed 52:23–33
Kumar V, Saxena SC, Giri VK (2006) Direct data compression of ECG signal for telemedicine. Int J Syst Sci 37:45–63
Jalaleddine SM, Hutchens CG, Strattan RD, Coberly WA (1990) ECG data compression techniques-a unified approach. IEEE Trans Biomed Eng 37:329–343
Kumar V, Saxena SC, Giri VK, Singh D (2005) Improved modified AZTEC technique for ECG data compression: effect of length of parabolic filter on reconstructed signal. Comput Electr Eng 31:334–344
Barr RC, Blanchard SM, Dipersio DA (1985) SAPA-2 is the Fan. IEEE Trans Biomed Eng 32:337
Reddy BRS, Murthy ISN (1986) ECG data compression using Fourier descriptors. IEEE Trans Biomed Eng 33:428–434
Batista LV, Melcher EUK, Carvalho LC (2001) Compression of ECG signals by optimized quantization of discrete cosine transform coefficients. Med Eng Phys 23:127–134
Lee S, Kim J, Lee M (2011) A real-time ECG data compression and transmission algorithm for an e-health device. IEEE Trans Biomed Eng 58:2448–2455
Duarte RCM, Matos FM, Batista LV (2007) Near-lossless compression of ECG signals using perceptual masks in the DCT domain. In: IV Latin American Congress on Biomedical Engineering. IFMBE Proc. 18. IFMBE Proceedings, pp 229–231
Lu Z, Kim DY, Pearlman WAP (2000) Wavelet compression of ECG signals by set partitioning in hierarchical trees algorithm. IEEE Trans Biomed Eng 47:849–856
Mamaghanian H, Khaled N, Atienza D, Vandergheynst P (2011) Compressed sensing for real-time energy-efficient ECG compression on wireless body sensor nodes. IEEE Trans Biomed Eng 58:2456–2466
Iwata A, Nagasaka Y, Suzumura N (1990) Data compression of the ECG using neural network for digital Holter monitor. IEEE Eng Med Biol Mag 9:53–57
Al-Shrouf A, Abo-Zahhad M, Ahmed SM (2003) A novel compression algorithm for electrocardiogram signals based on the linear prediction of the wavelet coefficients. Digit Signal Process 13:604–622
Mukhopadhyay SK, Mitra S, Mitra M (2011) A lossless ECG data compression technique using ASCII character encoding. Comput Electr Eng 37:486–497
Mukhopadhyay SK, Mitra S, Mitra M (2013) ECG signal compression using ASCII character encoding and transmission via SMS. Biomed Signal Process Control 8:354–363
Batista LV, Carvalho LC, Melcher EUK (2003) Compression of ECG signals based on optimum quantization of discrete cosine transform coefficients and Golomb-Rice coding. In: Proceedings of the 25th Annual International Conference of the IEEE. Eng Med Biol Soc (IEEE Cat No03CH37439) 3:2647–2650
Gupta R, Mitra M (2014) Wireless electrocardiogram transmission in ISM Band: an approach towards telecardiology. J Med Syst 38:90
Sufi F, Khalil I (2008) Enforcing secured ECG transmission for realtime telemonitoring: a joint encoding, compression, encryption mechanism. Secur Comm Networks 1:389–405
Chen C, Lin C, Chiang C, Lin S (2012) Personalized information encryption using ECG signals with chaotic functions. Inf Sci (Ny) 193:125–140
Ibaida A, Khalil I (2013) Wavelet-based ECG steganography for protecting patient confidential information in point-of-care systems. IEEE Trans Biomed Eng 60:3322–3330
Sufi F, Khalil I (2008) A new feature detection mechanism and its application in secured ECG transmission with noise masking. J Med Syst 33:121–132
Chen S-T, Guo Y-J, Huang H-N et al (2014) Hiding patients confidential data in the ECG signal viaa transform-domain quantization scheme. J Med Syst 38:38–54
Benzid R, Messaoudi A, Boussaad A (2008) Constrained ECG compression algorithm using the block-based discrete cosine transform. Digit Signal Process 18:56–64
Fensli R, Gundersen T, Snaprud T, Hejlesen O (2013) Clinical evaluation of a wireless ECG sensor system for arrhythmia diagnostic purposes. Med Eng Phys 35:697–703
Singh B, Singh D (2012) Effect of threshold value r on multiscale entropy based heart rate variability. Cardiovasc Eng Technol 3:211–216
Sufi F, Fang Q, Khalil I, Mahmoud SS (2009) Novel methods of faster cardiovascular diagnosis in wireless telecardiology. IEEE J Sel Areas Commun 27:537–552
Goldberger AL, Amaral LA, Glass L et al (2000) PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101:e215–e220
Filho EBL, Rodrigues NMM, da Silva EAB et al (2008) ECG signal compression based on Dc equalization and complexity sorting. IEEE Trans Biomed Eng 55:1923–1926
Blanco-Velasco M, Cruz-Roldán F, Godino-Llorente JI et al (2005) On the use of PRD and CR parameters for ECG compression. Med Eng Phys 27:798–802
Zhou Y, Bao L, Chen CLP (2014) A new 1D chaotic system for image encryption. Sig Process 97:172–182
Liao X, Lai S, Zhou Q (2010) A novel image encryption algorithm based on self-adaptive wave transmission. Sig Process 90:2714–2722
Wang Y, Wong K, Liao X, Chen G (2011) A new chaos-based fast image encryption algorithm. Appl Soft Comput 11:514–522
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (1992) Numerical Recipes in C—The Art of the Scientific Computing, 2nd edn. Cambridge University Press, Cambridge
Peric Z, Denic D, Nikolic J et al (2013) DPCM quantizer adaptation method for efficient ECG signal compression. J Commun Technol Electron 58:1241–1250
Hilton ML (1997) Wavelet and wavelet packet compression of electrocardiograms. IEEE Trans Biomed Eng 44:394–402
Ku C, Hung K, Wu T, Wang H (2010) Wavelet-based ECG data compression system with linear quality control scheme. IEEE Trans Biomed Eng 57:1399–1409
Sadhukhan D, Pal S, Mitra M (2015) Electrocardiogram data compression using adaptive bit encoding of the discrete Fourier transforms coefficients. IET Sci Meas Technol 9:866–874
Aggarwal V, Patterh MS (2012) Quality controlled ECG compression using essentially non-oscillatory point-value decomposition (ENOPV) technique. Digit Signal Process 22:878–884
Mitra M, Bera JN, Gupta R (2012) Electrocardiogram compression technique for global system of mobile-based offline telecardiology application for rural clinics in India. IET Sci Meas Technol 6:412
Mukhopadhyay SK, Mitra S, Mitra M (2012) An ECG signal compression technique using ASCII character encoding. Measurement 45:1651–1660
Acknowlegments
The authors deeply acknowledge the efforts by Dr. Shashi Prakash in providing the interpretations and evaluations on the final reconstructed ECG records.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pandey, A., Singh, B., Saini, B.S. et al. A joint application of optimal threshold based discrete cosine transform and ASCII encoding for ECG data compression with its inherent encryption. Australas Phys Eng Sci Med 39, 833–855 (2016). https://doi.org/10.1007/s13246-016-0476-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-016-0476-4