Abstract
Electroencephalographic (EEG) activity recorded during the entire sleep cycle reflects various complex processes associated with brain and exhibits a high degree of irregularity through various stages of sleep. The identification of transition from wakefulness to stage1 sleep is a challenging area of research for the biomedical community. In this paper, spectral entropy (SE) is used as a complexity measure to quantify irregularities in awake and stage1 sleep of 8-channel sleep EEG data from the polysomnographic recordings of ten healthy subjects. The SE measures of awake and stage1 sleep EEG data are estimated for each second and applied to a multilayer perceptron feed forward neural network (MLP-FF). The network is trained using back propagation algorithm for recognizing these two patterns. Initially, the MLP network is trained and tested for randomly chosen subject-wise combined datasets I and II and then for the combined large dataset III. In all cases, 60 % of the entire dataset is used for training while 20 % is used for testing and 20 % for validation. Results indicate that the MLP neural network learns with maximum testing accuracy of 95.9 % for dataset II. In the case of combined large dataset, the network performs with a maximum accuracy of 99.2 % with 100 hidden neurons. Results show that in channels O1, O2, F3 and F4 (A1, A2 as reference), the mean of the spectral entropy value is higher in awake state than in stage1 sleep indicating that the EEG becomes more regular and rhythmic as the subject attains stage1 sleep from wakefulness. However, in C3 and C4 the mean values of SE values are not very much discriminative of both groups. This may prove to be a very effective indicator for scoring the first two stages of sleep EEG and may be used to detect the transition from wakefulness to stage1 sleep.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The sleep architecture is a complex process that varies from person to person depending on age, sex, and due to sleep disorders [1]. Sleep disorder manifests itself as a condition with abnormal sleep patterns which are seen in humans as well as in animals. Obstructive sleep apnea disorder is not only that which affects sleep but may reflect the source of other major pathologies [2] such as hypertension, cardiac dysfunction, cognitive deficits and memory loss. In all these cases, sleep examination helps in identifying the disease associated with a specific sleep disorder. Polysomnography (PSG) is a test commonly used to detect some of these sleeps disorders. PSG test requires multiple electrode placements on the human body to record electroencephalogram (EEG), electrooculogram (EOG) and electrocardiogram (ECG) signals, along with respiratory movements and oxygen saturation for the entire duration of night. In PSG recordings, sleep time is divided into short epochs (fixed at 30 s) and classified as a specific stage of sleep according to the guidelines proposed by Allan Rechtschaffen and Anthony Kales [3]. The sleep scoring and staging must be done by a highly trained expert and manual scoring consumes a lot of time. Apart from the time factor, it is also prone to inter and intra scorer variability [4]. In addition, the fixed 30 s epoch may cover the onset of a succeeding stage, which makes it difficult to score such epochs with a specific stage. In order to overcome this problem many researchers have proposed automated sleep scoring system. There are two different approaches to automate a sleep scoring and staging system. In the first approach, manual scoring and staging may be mimicked and translated into an automatic process whereas in the second approach, relevant information is extracted as features from PSG signals using signal processing techniques, followed by classifiers to determine sleep stages [1, 2, 4–11].
Numerous studies reported on sleep onset detection use various time and/or frequency, parametric and statistical processing methods on sleep EEG datasets [12–17]. Linear methods such as spectral power estimation using Fourier transform have been used for sleep stage classification, to study changes in event related oscillations of EEG in sleep apnea subjects [18–21]. In these studies multiple frequency bands and their normalized band powers [19] and ratios of the frequency bands [20] are computed. Various methods have been adopted for an automated scoring of sleep based on single EEG channel data. Studies [15–17] on wake-sleep transition using spike rhythmicity reported that the rhythmicity, a time domain attribute proved to be statistically significant for recognizing both patterns in the occipital region using a single channel. In a study using parametric methods [22] the mean frequencies of a single EEG are used to train autoregressive hidden markov model (HMM) to detect arousal states of human, claiming a wake-drowsiness detection rate of 70 %. In another study [2] using a single-channel EEG, kalman filter model of wake-sleep transition followed by HMM detection resulted in an accuracy of 60.14 %. Another study on sleep stage detection used a Gaussian observation HMM [23] with a reported accuracy of 86 % for wake but only 22 % for stage N1. The decrease in power of all frequency bands except delta band has been reported during sleep onset [24] especially in the frontal region. Parametric modeling [25], has been used to model PSG recordings as time-varying autoregressive moving average model with recursive particle filtering for modeling sleep onset intervals. Results showed performance metrics claiming 93.18 % accuracy in best cases. Attempts have been made to use multiple feature sets to score sleep stages using time-based, stochastic, spectral and chaotic features [26]. The achieved mean error rates were reported to be 50 % for detection of stage 1 (N1), to 10 % for slow wave sleep. Another study with multiple features from time, frequency domain along with nonlinear features reported average accuracy rates of 95:88 % upon a single EEG channel [27] using SVM classifier. Recently, a method [28] reported 93.4 % accuracy for automatic analysis of sleep macrostructure using full set of PSG signals in a fuzzy reasoning classifier. A comprehensive study of multimodal correlates of sleep onset and the experimental details of their characterization has been reported by [29]. Another study [30] reports the use of a fuzzy logic inference engine for early detection of sleep onset in people driving a car or a public transportation vehicle using power spectrum density of Heart Rate Variability (HRV) signal and autonomous nervous system frequency activity reflected by the HRV signal. Sleep onset detection showed the same detection rate as clinically collected data with 90 % true detection on a set of ten analyzed ECGs.
The EEG activity recorded during the entire sleep duration exhibits significant irregularity and complexity. It becomes more regular and rhythmic as the subject slips into stage1 sleep from wakefulness. Non-linear methods like approximate entropy (ApEn), correlation dimension, largest lyapunov exponent, hurst exponent (H), and fractal dimension [31] have also been used successfully for quantifying EEG complexity variation for sleep stage scoring. Many studies have used ApEn for EEG analysis [31–33] to characterize different types of epileptic seizures and analyze EEG regularity in Alzheimer’s disease subjects. ApEn is found to be an effective measure for quantifying complexity in short and noisy data sets [34] and has been applied in the analysis of sleep stages [31] and it is also shown that the sleep stages are characterized by not only different mean ApEn values in each sleep stage but also differ within the sleep stages of normal subjects. Our previous study on the same database used in this study with only three subjects [17] uses hurst exponent as feature on a single channel EEG to obtain 99.96, 71.8 % classification accuracies with k-NN and LDA classifiers respectively.
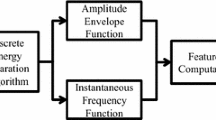
Many of these studies above have concentrated on single EEG channel for sleep scoring. As the transition of wake to stage1 sleep correlates with the cortical neuronal activities of different regions of the brain (mainly occipital and frontal region), the current study uses a multichannel EEG study with spectral entropy (SE) feature for wake-stage1 sleep characterization instead of a single channel. Our motivation to study the effect of spectral entropy in wake-stage1 sleep transition emerges from anesthetic studies effectively using it as a distinguishing feature [35–40]. Recently a number of different entropy estimators such as the sample entropy, tsallis entropy and Bispectral index [35, 36] and more recently the SE index [37–39] monitors have been released commercially to quantify the complexity and depth of anesthesia. Studies have shown that the anesthetic drug administration decreases the SE along with a decrease in cortical and global cerebral blood flow indicating that the change in SE can depict global changes in neuronal activity induced by the drugs. Hence an attempt is made here to study the complexity of the human brain during the first two stages of sleep using SE as a complexity measure. The block diagram of the proposed method is shown in Fig. 1.
Stages of sleep
During different stages of sleep, thalamus, cortex and pons interact with each other at the neuronal level that manifests itself as EEG activity with distinct characteristics that lead to differentiate whole night sleep into different stages viz., awake, NREM stage consisting of stage 1, 2, 3 and 4 followed by REM stage. The reticular formation possesses neurons with both sensory and motor functions. During active wakefulness, the reticular activation system inhibits reticular nucleus and excites neurons of the sensory thalamus, allowing an uninterrupted flow of signal from brainstem to cerebral cortex. Both beta waves (13–28 Hz) and alpha waves (8–13 Hz) are observed with beta waves being more desynchronized and possessing lesser amplitude when compared to alpha waves. During relaxed wakefulness, alpha waves become more synchronous in their pattern, indicating gradual decrease in the frequency of brain waves.
Awake/stage1 sleep stages
In stage1 sleep the EEG signal consists of theta waves (4–8 Hz) with greater amplitude than the alpha waves. Stage 2 sleep is characterized by two phenomena: sleep spindles and K complexes with theta waves in background. During deep sleep, delta waves (0.5–4 Hz) appear with higher amplitude than other waves. The next stage is the REM sleep during which the acetyl cholinergic RAS becomes active again. The EEG signals during REM sleep appear to be similar to that of wakefulness with rapid eye movements, reduced muscle activity with the predominance of alpha and beta waves once again.
Data acquisition
In the PSG data used in this study, multiple physiological parameters related to sleep such as EEG, EOG, chin EMG, respiratory movements (thorax and abdomen), nasal airflow, ECG and measurements of oxygen saturation are recorded for the entire duration of sleep. Usually EEG, EOG, and EMG signals are used for sleep stage classification.
Database
This study used data from ten normal subjects acquired from overnight Polysomnographic recordings done in clinical sleep labs of M S Ramaiah Hospitals, Bangalore. The artefact-free PSG signals were acquired by using Sandman’s software. The recordings included six EEG channels (C3, C4, F3, F4, O1 and O2 locations with A1 and A2 as reference electrode positions according to the 10–20 International standard for electrode placement), two EOG channels, two EMG channels, two ECG channels, oxygen saturation value (SaO2) and pulse rate. For our study only awake and stage1 EEG recordings of sleep data are considered. The sampling rate of each channel is 256 Hz. Sleep staging was manually done by expert scorers of the neurology department, at every 30 s interval based on the Rechtschaffen and Kales criteria.
In the present study, dataset I (combined datasets of subjects 1, 3, 6) and dataset II (combined datasets of subjects 2, 5, 8) consist of three randomly selected subjects 8-channel EEG sleep data whereas dataset III consists of all ten subjects (combination of datasets I, II and subjects 4, 7, 9, 10) EEG data epochs to form a large dataset. The number of data epochs used for training and testing in both the cases is shown in Table 1. Cerebral montages O1, O2, F3, F4, C3 and C4 were used for EEG analysis from the ten PSG records. Artifact free EEG records with minimal EMG activity were selected. Figure 2a, b display segments of awake and stage1 sleep EEG and EOG data of a normal subject used for analysis. For each subject, one or more 30 s segments of simultaneous EEG and EOG epochs were chosen for both awake and stage1 sleep. The EEG and EOG signals were sampled at 256 Hz. A total of 15 min of EEG data were analyzed for each subject containing 10 min of wake and 5 min of stage1 sleep. Thus each of the datasets I and II consists of a total of 30 min wake and 15 min stage1 sleep from three randomly selected PSGs whereas the large dataset contains a total of 100 min of wake and 50 min of stage1 sleep from ten PSGs. The raw EEG epochs are passed through a 6th order elliptic band pass filter with pass band ranging from 0.7 to 47 Hz to ensure the rejection of any stray frequency components outside the frequencies of interest.
SE estimation and pattern identification
Entropy was first introduced in information theory by Shannon [41] and further applied to compute the SE by Johnson and Shore [42]. Entropy refers to the degree of disorderliness in a thermodynamic system. However for a temporal signal like EEG, entropy value reflects the predictability or regularity of a signal. Higher entropy indicates higher complexity (irregularity) of the signal while smaller entropy values show that the signal under consideration is more predictable and hence less complex. The approximate entropy and Shannon entropy are time domain measures of complexity whereas SE is used in frequency domain. According to Shannon, the entropy (H) is expressed as:
where p k are probabilities of individual frequencies in bin k. It is reported that the entropy decreases at the neuronal level as the human cortex becomes unconscious [43]. It implies that the real time information transfer within the cortex associated with a change in information entropy may be precisely reflected in EEG. The information entropy, applied in frequency domain defines the microstates in terms of rates-of-change for the entropy [40].
SE feature extraction
SE is the normalized version of Shannon entropy formula, applied to the power spectral density of EEG signal. Following are the steps involved in computing the SE features from one-second segments of 6-active channel EEG sleep data as follows [38, 44]:
The spectrum of 1-second sequence of each channel, {x(t i ) = x(t 1), x(t 2)…x(t n )}, Fs = 256 Hz is estimated using 256-point FFT with a non-overlapping Hamming window, using Eq. (1),
The power spectrum P(f i ) is obtained as the square of the amplitudes of each component in X(f i ).
\(X^{'} \left( {f_{i} } \right)\) represents the complex conjugate of \(X\left( {f_{i} } \right)\) Since entropy calculation requires computation of probabilities (p k) within the power spectrum such that \(\mathop \sum \nolimits p_{\text{k}} = 1,\) it becomes necessary to normalize the power spectrum with a constant C n, such that the sum of the normalized power spectrum over the interval of frequencies of interest [f1, f2] is equal to one:
Next, the normalized SE within the frequency interval [f1, f2] is computed as follows:
where N[f 1, f 2] represents the number of frequency components within \(\left[ {f_{1} ,f_{2} } \right].\)
The normalized entropy value equals one for maximum irregularity) and zero for minimum irregularity. Each one-second, 6-channel EEG data epoch is represented by a 6-component SE vector. Figures 3 and 4 show sample plots of the power spectral densities for O1 and O2 channels and SE values for O1 channel in awake/stage1 sleep stages respectively. Prior to recognizing the two patterns using the neural network, the mean value of SE coefficients of all 6-channel EEG data is computed for the large dataset to evaluate the statistical significance associated with first two stages of sleep.
One minute sample plots of SE values in O1-O2 channels for awake and Stage1 sleep EEG Fig. 5. Training and validation performance of MLP network
MLP-FF neural network with back propagation algorithm for pattern identification
MLP-FF neural networks use back propagation algorithm for pattern matching and recognition tasks. Initially the network is trained by using labelled example patterns and as training progresses, the network learns by adapting its weights. The training session ends based on the mean squared error between the labelled outputs and actual outputs of the network. During training, initially small values of random weight vectors are normalized and used for the first iteration.
Next, the input pattern is applied and layer outputs are calculated during forward pass. The mean squared error (MSE) is computed between the labelled output and the actual output. During backward pass, MSE is propagated back to the preceding layers and their corresponding weights change till the cumulative error becomes less than the user defined error. In other words with each training cycle, the difference between the actual output of each neuron and its target output must decrease to achieve good training accuracy. The network keeps training all the patterns repeatedly until the total error falls to a user defined value and then it stops. The trained network exhibits good generalization if it successfully recognises not only those patterns used in training but also corrupted or noisy versions. Usually better training accuracies can be obtained if the patterns belonging to different classes are submitted to the network in a random order. After the training session ends, the network is tested and validated with set of patterns (testing set and validation set) not used in training. This helps in avoiding network overfitting [45]. The MSE reaches a minimum upon the successful training of the network with validation set. However, if the network is over trained the MSE starts increasing (Fig. 5). If the network is exposed to large size datasets with similar features, the network suffers from overfitting due to which it won’t handle noisy data well.
From all night sleep EEG (6-channel) recording of three subjects, awake and stage1 sleep artifact free data are manually extracted based on the scoring done by trained experts. The SE values are computed from 6-channel EEG data for each second of awake and stage1 sleep data. The entropy values thus computed from 6-channels constitute the feature vector for each data epoch of one second. Next, the feature vectors of both stages are grouped subject-wise to form 2 feature datasets of dimension shown in Table 1. Also the SE vectors of both awake and stage1 data of all ten subjects are combined to form a feature matrix of 6 × 38400 dimension to form the combined large dataset.
Network training
Initially, datasets I and II are individually used to train the network for awake and stage1 SE pattern identification. The network is further trained with the combined large dataset for identifying potential classes of patterns that discriminate awake and stage1 sleep. For each of the cases above, the entire dataset is divided randomly into 60 % training vectors, 20 % testing vectors and 20 % validation vectors. For each training cycle, 60 % of the SE vectors are randomly chosen from the dataset. This method automatically performs the validation as each training cycle has different sets of training and testing vectors and minimizes the problem of overfitting.
Levenberg–Marquardt back propagation algorithm is used to train the network. The network performance is evaluated for different number of hidden neurons. The MLP network with back propagation algorithm has the ability to generate complex decision boundaries in the feature space and therefore can be used as a classifier. The MLP-FF network architecture used in this study is as shown in Fig. 6. Six input nodes are used for the 6-channel SE coefficients computed for each epoch of 1 s. The performance of the network is evaluated by selecting different number of hidden neurons for all the cases. The hidden layer neurons use tansig transfer (activation) function. One linear neuron used in the output layer responds with continuous output values varying between 0 and 1 upon training. The output values are scaled to 0 or 1 after applying a threshold of 0.5. The training parameters and stopping criterion for the network are shown in Table 2.
Results
Preliminary studies are very impressive as far as using SE feature for discriminating alcoholic/control and wake/stage1 sleep patterns. The higher SE values in O2 (right occipital), F3 (left frontal) and F4 (right frontal) locations during wake state represent the active state of human brain during wakefulness. It indicates complexities in the neuronal activities of the region correlated to unsynchronized beta and gamma activity during wakefulness.
The statistical significance of awake and stage1 sleep states in terms of the mean are evaluated using an error bar chart for all the channels independently and a box plot for O2 channel alone (Figs. 7, 8). The difference between groups is statistically significant (p < 0.05) if the error bars do not overlap within a confidence interval (CI) of 95 %. It is seen from Fig. 7 that the error bars corresponding to the awake/stage1 of O2, F3 and F4 channels do not overlap and hence exhibit a statistical significance value of p < 0.05. The box plot for the mean value of SE in O2 channel (Fig. 8) indicates that the median in awake and stage1 slightly differ but the variance upon the mean is quite different in both cases. The maximum and minimum values that are not outliers are significantly different in stage1 than in awake. The significant difference in the mean of both groups in occipital and frontal regions also indicate the changes in event related oscillations reflecting neuronal activities associated with them. This directly correlates with the reduced oscillations from beta to gamma to alpha range and subsequent transition into low-voltage mixed frequency signals that characterize the onset of sleep [46].
In order to recognize awake and stage1 sleep patterns, MLP network with 20 hidden neurons is trained first by using 60 % of subject-wise database. During training, the network performs with classification accuracies 95.2, 95.9 % respectively for the combined datasets I and II under consideration. For large dataset, the performance improves with an increase in the number of hidden neurons from 20 to 100 in steps of 20. The results are tabulated in Table 3. It is seen that the testing accuracy of the MLP network improves from 92.9 to 99.2 % with the increase in hidden neurons. In terms of the computation time, the classification of wake/sleep patterns consumes more time with increase in the number of hidden neurons. The statistical significance of the mean value in O2, F3 and F4 correlates well with the accuracies obtained in the MLP neural network.
Upon training the network successfully, network performance is evaluated for identifying the two groups of patterns and plotted in Fig. 9. It can be seen that the output layer linear neuron responds with continuous outputs between 0 and 1. The outputs are finally scaled using a threshold of 0.5 to determine the accuracy. In some cases with misclassification, it is seen that the network performs with greater than 0.5 outputs, while its labeled output is zero. Similarly for labeled outputs of +1, there are some misclassifications responding with less than 0.5. All the testing classification accuracies shown are with respect to 50 % holdout cross validation. The holdout validation method is simple to perform that ensures faster computation. While determining the computation time, an average of ten runs was taken to account for the slight variation in elapsed time for each run of the code due to the inherent instability of the clock inside the processor. The entire computation was performed on Matlab platform with Intel core i3 350 MHz CPU at 2.27 GHz clock speed.
Discussion
In literature, previous studies have concentrated on single EEG channel for sleep scoring of the same database using only three healthy subject’s data. Studies [15, 16] on wake-sleep transition using spike rhythmicity and amplitude respectively reported that the rhythmicity, a time domain attribute proved to be statistically significant for recognizing wake-stage1 patterns. The drawback of the above studies is that the database is small and only single channel O1 is used for the study. Similarly the results obtained using kalman filter and a single channel [2], around 60 % classification accuracy is obtained. In our previous study [17] using Hurst exponents as features on the same three subject’s data, classification accuracies of 99.96, 71.8 % with k-NN and LDA classifiers respectively are reported using only O1 and O2 channels. A study [48] evaluated the accuracy of sleep staging for multiple features based on EEG alpha activity during sleep onset in healthy, insomniac and schizophrenic patients using artificial neural networks classifier. Results show an agreement of only 81.3 % with the human expert scoring, where studies are concentrated on alpha bands only. Another study [49] used relative concentration change of oxy- and deoxy-hemoglobin transform (using functional near infrared spectroscopy) to extract information on heart rate and EEG spectral bands to monitor the alertness of driver under normal and sleep deprived conditions. Results indicate that the beta and alpha bands change in normal and sleep deprived conditions. Also the hemodynamic change is more stable in normal condition and heart rate decreases in sleep deprivation condition. The alpha band and heart rate are different in sleepless and sleepy states. Here it is observed that the sleep deprived condition presents a different sample paradigm than that of a normal sleep scoring process. In a study on safe driving performance estimation and alertness [50], eight EEG-band power-related features, viz., beta, alpha, theta, delta, (alpha plus theta)/beta, alpha/beta, (alpha plus theta)/(alpha plus beta) and theta/beta are extracted from the preprocessed EEG signals by employing FFT. Fisher score technique chooses the most descriptive features for further classification using support vector machine (SVM) to quantify drowsiness level. Experimental results show that the quantitative driving performance can be correctly estimated through analyzing driver’s EEG signals. However the results are prone to artifacts as claimed by authors. In the proposed study, estimation of SE has a unique property that it does not depend on the absolute values of amplitude or frequency of the signal. This property helps to study inter-subject variations in the absolute frequencies of the EEG. In the proposed study, the size of the database is increased from 3 to 10 subjects and a multi-channel approach is used to study the wake-sleep transition. With the increased data size, better generalization and hence better classification accuracy is achieved. Also the multichannel detection of wake sleep states helps in better localization of differences between groups.
Decrease in SE values is observed as the subject slips into stage 1 sleep from wakefulness. This is in confirmation with the theory that as stage1 sleep sets in from wakefulness, EEG becomes more rhythmic and regular resulting in a decreased value of entropy. A decrease in SE denoting entry into stage1 sleep from wakefulness is also associated with the transition of beta and gamma activity to a more synchronized alpha activity of low frequency. It also reflects a decrease in the complexity of brain functions with the onset of sleep. The F3 and F4 channels along with O2 exhibit differing means for wake/stage1 sleep states. In a study using spectral analysis of EEG signals for the detection of wake-sleep transition [47], a decrease in sleep onset power of all frequency bands except delta range has been reported especially in the frontal region confirms with the result of our study. It also correlates to reduced activity in stage1as reported in medical literature.
Conclusion and future work
The spectral entropies of awake/stage1 sleep are computed for each second in all the six channel EEG data of ten normal subjects. These SE patterns for both stages are used to train a MLP-FF neural network with back propagation algorithm. Results of pattern identification of both stages are very promising and indicate that the SE may be used as a discriminative feature for the identification of awake/stage1 sleep. The spectral entropies decrease with subjects going from wakefulness to stage1 sleep. This is in confirmation with the theory that as stage1 sleep sets in from wakefulness, the EEG becomes more rhythmic and regular resulting in a decreased value of entropy. In future, the number of subjects can be increased to improve the generalization capability of the neural network. Also, the results strongly indicate that it may be beneficial to use SE for detecting the transition from awake to stage1 sleep. It may also help to study the underlying complexities of the human sleep process by computing spectral entropies for all stages of sleep.
References
Redline HL, Kirchner SF, Quan SE, Gottlieb DJ, Kapur V, Newman A (2004) The effects of age, sex, ethnicity, and sleep-disordered breathing on sleep architecture. JAMA Intern Med 164:406–418
Rossow AB, Salles EOT, Coco KF (2011) Automatic sleep staging using a single-channel eeg modeling by kalman filter and HMM. In: International conference IEEE Engineering in Medicine and Biology Society, vol 1 pp 1-6
Rechtschaffen A, Kales A (1968) A manual of standardized terminology techniques and scoring sleep stage of human subjects. Public Health Service US Govt Printing Office, Washington DC
Le QK, Truong QDK, Van Toi V (2011) A tool for analysis and classification of sleep stages. In: International conference on advanced technologies for communications, pp 307–310
See AR, Liang CK (2011) A study on sleep EEG using sample entropy and power spectrum analysis. In: International conference on defence science research, pp 1–4
Khalighi S, Sousa T, Oliveira D, Pires G, Nunes U (2011). Efficient feature selection for sleep staging based on maximal overlap discrete wavelet transform and SVM. 33rd Annual international conference IEEE Engineering Medicine Biology Society, pp 3306–3309
Liang SF, Kuo CE, Hu YH, Cheng YS (2011) A rule-based automatic sleep staging method. In: International conference of the IEEE Engineering in Medicine & Biology Society, pp 6067–6070
Herrera LJ, Mora AM, Fernandes C, Migotina, D (2011) Symbolic representation of the EEG for sleep stage classification. In: International conference on intelligent system design and applications, pp 253–258
Sloboda J, Das M (2011). A simple sleep stage identification technique for incorporation in inexpensive electronic sleep screening devices. In: Proceedings IEEE national aerospace and electronics conference, pp 21–24
Langkvist M, Karlsson L, Loutfi A (2012) Sleep stage classification using unsupervised feature learning. Adv Artif Neural Syst. doi:10.1155/2012/107046
Phan H, Do Q, Do TL, Vu DL (2013). Metric learning for automatic sleep stage classification. In: International conference IEEE Engineering in Medicine Biology Society, pp 3–7
Christopher Letellier, Claudia Lainscek (2012) Method and system for automatic scoring of sleep stages, the Patent Cooperation Treaty (PCT). World Intellectual Property Organization International Bureau, Geneva
Modarres-Zadeh M (2005) A neuro-behavioral test and algorithms for quantification of sleepiness and characterization of wake-sleep transitions. In: Proceedings of IEEE EMBS 27th annual conference, Shanghai, pp 5746–5749
De Gennaro L, Ferrara M, Ferlazzo F, Bertini M (2000) Slow eye movements and EEG power spectra during wake-sleep transition. J Clin Neurophysiol 111:2107–2115
Purnima BR, Sriraam N, Krishnaswamy U, Radhika K (2014) A measure to detect sleep onset using statistical analysis of spike rhythmicity. Int J Biomed Clin Eng 3:27–41
Sriraam N, Purnima BR (2014) Sleep wake transition using relative spike amplitude. In: International conference on medical imaging, health & emerging communication systems, pp 437–441
Sriraam N, Purnima BR, Uma K, PadmaShri TK (2014) Hurst exponents Based detection of wake-sleep—A pilot study IEEE. In: Proceedings international conference on circuits, communication, control & computing, pp 1–4
Svanborg E, Guilleminault C (1996) EEG frequency changes during sleep apneas. J Sleep 19(3):248–254
Dingli K, Assimakopoulos T, Fietze I, Witt C, Wraith PK, Douglas NJ (2002) Electroencephalographic spectral analysis: detection of cortical activity changes in sleep apnea patients. J Eur Respir 20:1246–1253
Morisson F, Lavigne G, Petit D, Nielson T, Malo J, Montplaisir J (1998) Spectral analysis of wakefulness and REM sleep EEG in patients with sleep apnea syndrome. J Eur Respir 11:1135–1140
Fell J, Roschke J, Mann K, Schaffner C (1996) Discrimination of sleep stages: a comparison between spectral and nonlinear EEG measures. J Clin Neurophysiol 98:401–410
Huang RS, Kuo CJ, Ling-Ling T, Chen OTC (1996) EEG pattern recognition-arousal states detection and classification. In: IEEE international conference on neural networks
Flexer A, Sykacek P Rezek I, Dorffner G (2000) Using hidden markov models to build an automatic, continuous and probabilistic sleep stager. In: International joint conference on neural networks, vol 3, pp 627–631
Germain A, Nielson TA (2001) EEG Power associated with early sleep onset images differing in sensory content. J Sleep Res Online 4:83–90
Chaparro-Vargas R, Dissayanaka PC, Penzel T, Ahmed B (2014) Sleep onset detection based on time-varying autoregressive models with particle filter estimation. In: IEEE conference on biomedical engineering. & sciences, pp 436–441
Krakovska A, Mezeiova K (2011) Automatic sleep scoring: a search for an optimal combination of measures. J Artif Intell Med 53(1):25–33
Koley B, Dey D (2012) An ensemble system for automatic sleep stage classification using single channel EEG signal. J Comput Biol Med 10:1–10
lvarez-Estevez DA, Fernandez-Pastoriza JM, Hernandez-Pereira E, Moret-Bonillo V (2013) A method for the automatic analysis of the sleep macrostructure in continuum. J Expert Syst Appl 40(5):1796–1803
Ogilvie RD (2001) The process of falling asleep. J Sleep Med Rev 5:247–270
Malcangi M, Smirne S (2012) Fuzzy-logic inference for early detection of sleep onset in car driver. J Eng Appl Neural Netw 311:41–50
Acharya R, Faust O, Kannathal N, Chua T, Laxminarayan S (2005) Non-linear analysis of EEG signals at various sleep stages. J Comput Methods Progr Biomed 80:37–45
Diambra L, Bastos de Figueiredo JC, Malta CP (1999) Epileptic activity recognition in EEG recording. J Phys A 273:495–505
Abasolo D, Hornero R, Espino P, Poza J, Sanchez CI, de la Rosa R (2005) Analysis of regularity in the EEG background activity of Alzheimer’s disease patients with approximate entropy. J Clin Neurophysiol 116:1826–1834
Pincus SM, Goldberger A (1994) Physiological time series analysis: what does regularity quantify? Am J Physiol 266:1643–1656
Mathew BA (2006) Entropy of electroencephalogram (EEG) signal changes with sleep state. University of Kentucky Master’s Theses Graduate School
Rampil Ira J (1998) A primer for EEG signals processing in anesthesia. J Am Soc Anesthesiol 89:980–1002
Vakkuri A, Yli-Hankala A, Talja P (2004) Time-frequency balanced spectral entropy as a measure of anesthetic drug effect in central nervous system during sevoflurane, propofol, and thiopental anesthesia. J Acta Anaesthesiol Scand 48:145–153
Oja-Viertio H, Maja V, Sarkela M, Talja P, Tenkanen N, Tolvanen-Laakso H, Paloheimo M, Vakkuri A, Yli-Hankala A, Merilainen P (2004) Description of the entropytm algorithm as applied in the Datex-Ohmeda S/5TM entropy module. J Acta Anesthesiol Scand 48:154–161
Maksimow K, Kaisti S, Aalto S, Maenpaa M, Jaaskelainen S, Hinkka S, Martens M, Sarkela H, Oja V, Scheinin H (2005) Correlation of EEG spectral entropy with regional cerebral blood flow during sevoflurane and propofol anesthesia. J Anesth 60:862–869
Sleigh J, Olofsen, WE, Dahan EA, de Goede J, Steyn-Ross A (2001) Entropies of the EEG: the effects of general anesthesia. In: Fifth conference on memory, anesthesia and consciousness, New York
Shannon CE (1948) A mathematical theory of communication. J Bell Syst Technol 27:623–656
Johnson RW, Shore JE (1984) Which is the better entropy expression for speech processing: S log S or log S? IEEE Trans Acoust 32:129–137
Ross Steyn ML (1999) Theoretical electroencephalogram stationary spectrum for a white-noise-driven cortex: evidence for a general anesthetic-induced phase transition. J Phys Rev 60:7299–7311
Anier A, Lipping T, Ferenets R, Puumala P, Sonkajarvi E, Ratsep I, Jantti V (2012) Relationship between approximate entropy and visual inspection of irregularity in the EEG signal, a comparison with spectral entropy. Brit J Anesth 109:928–934
Haykin S (1999) Neural networks, a comprehensive foundation, 2nd edn. Prentice Hall, New Jersey
Carskadon MA, Dement WC (2011) Monitoring and staging human sleep. J Princ Pract Sleep Med 5:16–26
Tinguely G, Finelli LA, Landolt HP, Borbély A, Achermann P (2006) Functional EEG topography in sleep and waking: state-dependent and state independent features. J Neuro Image 32:283–292
Dissayanaka C (2015) Sleep onset detection with multiple EEG alpha-band features: comparison between healthy, insomniac and schizophrenic patients. In: Biomedical circuits and systems conference (BioCAS ) 2015 IEEE, Atlanta, pp 1–4
Nguyen T, Ahn S, Jang H, Jun SC, Kim JG (2015) Sleep onset detection based on simultaneous EEG-NIRS measurement, In: International congress of ophthalmology and optometry, China
Sun H, Bi L, Chen B, Guo Y (2015) EEG-based safety driving performance estimation and alertness using support vector machine. Int J Secur Appl 9:125–134
Acknowledgments
We thank Dr. Uma Maheshwari and Sleep Lab staff of MS Ramaiah Medical College and Hospitals, Bangalore, India, for providing us with the PSG recordings of sleep data.
Authors contribution
N. Sriram and T.K. Padma Shri contributed equally in terms of the data collection, analysis and performing the classification. Uma Maheshwari Krishnaswamy was involved in clinical validation of the data and ensuring the statistical analysis of the data. All the three authors contributed equally in terms of manuscript preparation and finalization.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We hereby declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Sriraam, N., Padma Shri, T.K. & Maheshwari, U. Recognition of wake-sleep stage 1 multichannel eeg patterns using spectral entropy features for drowsiness detection. Australas Phys Eng Sci Med 39, 797–806 (2016). https://doi.org/10.1007/s13246-016-0472-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-016-0472-8