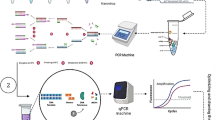
Abstract
Lactobacillus paracasei is a mesophilic lactic acid bacterium technologically active in food fermentation. Culture-independent methods have rapidly been recognized as a valuable alternative to culture-dependent methods for lactic acid bacteria enumeration. In the present work, the efficacy of different protocols to extract DNA from yoghurt were compared, real-time PCR (qPCR) targeting tuf gene for L. paracasei enumeration was evaluated, and qPCR and plate counts of L. paracasei in yoghurt samples were compared. Total DNA concentrations from commercial yoghurts were higher using DNAzol method 2 than using the other tested methods. Standard curves presented suitable mean efficiency values of 91 % (pure L. paracasei strain CTT 7501), 95 % (pure L. paracasei strain FNU), and 103 % (yoghurt with L. paracasei strain FNU). Limit of detection is 3 log DNA copy number, corresponding to 2.78 log CFU, a suitable range of CFU enumeration for probiotic bacteria in yoghurt samples, considering that they should be present in large amounts. The L. paracasei (CFU) enumerated by qPCR were compared to culturable L. paracasei enumerated by plate counts at 7, 14, 21, and 28 days of yoghurt manufacture. Differences between qPCR and plate counts were observed only 28 days after yoghurt preparation, counts were similar at 7, 14, and 21 days. In conclusion, this qPCR assay is a useful and rapid tool to enumerate L. paracasei in yoghurt, although it does not distinguish dead and viable cells.
Similar content being viewed by others
Introduction
Lactobacilli species are involved in both spontaneous fermentation and large-scale fermentation processes for the preservation and transformation of many raw materials such as milk (Furet et al. 2004; Rushdy and Gomaa 2013). Lactobacilli strains have been used commercially over the last years as they are believed to possess probiotic features (Francesca et al. 2013; Herbel et al. 2013). Probiotics are defined as “live micro-organisms that when administered in adequate amounts, confer a health benefit on the host”. However, to exert health benefits, the concentration of live probiotic bacteria needs to be of approximately 6 log CFU g−1 of the product at the time of consumption (Roy 2005).
Among the various types of food products, yoghurt or similar products have been used as the most popular vehicles for the incorporation of probiotic microorganisms (Kristo et al. 2003). Yoghurt has long history of recognition as a dietary product with many desirable effects. It is made from the symbiotic growth of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. These yoghurt starter bacteria might not survive the gastric passage or colonize the gut and consequently may not play a role in the human gut. Hence, the recent trend is to add probiotic bacteria to yoghurt (Ashraf and Shah 2011).
Lactobacillus paracasei is a mesophilic lactic acid bacterium technologically active in food fermentation. L. paracasei subsp. paracasei NTU 101 and its fermented products proved to be effective for the management of blood cholesterol and pressure, prevention of gastric mucosal lesion development, immunomodulation and alleviation of allergies, prevention of osteoporosis, and inhibition of fat tissue accumulation (Chiang and Pan 2012). L. paracasei M7 was shown to inhibit the adhesion of Salmonella to epithelial cells (Xue et al. 2015). L. paracasei FNU was shown to possess desirable in vitro resistance to low pH and bile salts (Ilha et al. 2014).
Currently, despite their economic impact, most of the assays that are used to identify and quantify lactobacilli are classical microbiological methods. Lactobacilli are often hard to be distinguished by classical microbiological techniques since most of them have similar nutritional and growth requirements (Kao et al. 2007; Poltronieri et al. 2008). Generally, dairy products, such as yoghurt and cheese, contain a population of several species of intimately related lactic acid bacteria. Furthermore, phenotypic characteristic analysis is always time-consuming and labor-intensive.
Culture-independent methods have rapidly been recognized as a valuable alternative to culture-dependent methods (Agrimonti et al. 2013; Ceccherini et al. 2013; De Medici et al. 2015; Ilabaca et al. 2014; Ke et al. 2014; Reitschuler et al. 2014; Rodriguez-Lazaro et al. 2015; Ruiz et al. 2014). These methods can be based on the direct analysis of DNA extracted from the food matrix (Achilleos and Berthier 2013; Rodríguez et al. 2012). DNA extraction is a crucial step for reliable DNA quantification by real-time PCR, called qPCR (Cankar et al. 2006; Garcia et al. 2013; Oliveira et al. 2013; Tian et al. 2013). Bacterial DNA amplification by PCR from milk samples can be affected by the presence of inhibitory substances such as Ca2+, fat, and proteins (Machado et al. 2013), so it is a challenge to obtain good quality DNA from dairy products for PCR purposes (Pirondini et al. 2010; Quigley et al. 2012).
In order to monitor lactic acid bacteria on dairy products, molecular methods based on bacterial DNA amplification by real-time PCR have been successfully developed for Lactobacillus acidophilus, L. brevis, L. delbrueckii subsp. bulgaricus, L. helveticus, and L. reuteri in yoghurt (Herbel et al. 2013), and for S. thermophilus, L. delbrueckii, Lactobacillus casei, L. paracasei, L. rhamnosus, L. acidophilus, and L. johnsonii in fermented milk products (Furet et al. 2004). Several species-specific primer pairs were designed based on the variability of 16S rRNA sequences for differentiating five strains of lactobacilli that were added into probiotic products in Taiwan (Kao et al. 2007). It was simple to identify L. acidophilus and L. delbrueckii by species-specific primers, but it could not be used to distinguish L. casei, L. paracasei, and L. rhamnosus (Kao et al. 2007). The yycH gene was proposed as an additional molecular marker for L. casei group species discrimination that provides higher resolution than 16S rRNA (Huang et al. 2014). Recently, a real-time PCR assay to quantify L. paracasei in cheese was developed targeting an elongation factor gene (Achilleos and Berthier 2013). The elongation factor (tuf) gene was shown to be highly variable among lactic acid bacteria, especially among closely related species such as those of the L. casei group (Chavagnat et al. 2002; Yu et al. 2012).
In the present study, L. paracasei FNU was incorporated as a probiotic strain in yoghurt. This strain was previously characterized as being resistant to low pH and bile salts (Ilha et al. 2014). The objectives of the present study were to verify L. paracasei FNU growth and viability in yoghurt by plate count, to compare the efficacy of protocols to extract DNA from yoghurt, to evaluate qPCR targeting the tuf gene for L. paracasei enumeration, and to compare qPCR and plate counts of L. paracasei in yoghurt samples. The novel aspects of this work are the comparison of bacterial DNA extraction methods giving different extraction yields; the bacterial count to check survival of the added culture during 28 days; and the use of Tuf primers for L. paracasei DNA quantification in a new matrix, yoghurt containing high population of L. delbrueckii subsp. bulgaricus and S. thermophilus, instead of cheese as proposed previously (Achilleos and Berthier 2013).
Material and methods
Bacterial strains and culture conditions
For the primer specificity test, bacterial strains Lactobacillus paracasei FNU, isolated from grape sourdough (Ilha et al. 2014); Lactobacillus paracasei CCT 7501 acquired from the collection of cultures of André Tosello Foundation (Campinas, São Paulo, Brazil); Lactobacillus paracasei LY0 750 provided by Danisco (Cotia, São Paulo, Brazil); and Lactobacillus plantarum (ATCC 8014) purchased from ATCC (Manassas, VA, USA), were grown in De Man, Rogosa, and Sharpe (MRS) broth (Merck, Germany) at 30 °C for 24 h. Furthermore, Pseudomonas spp. was grown in Luria-Bertani broth (USB, Cleveland, OH, USA) at 28 °C, Escherichia coli (ATCC 25922, Manassas, VA, USA) was grown in Brain-Heart Infusion broth (Himedia, PA, USA), while Bacillus cereus (ATCC 14579, Manassas, VA, USA) was grown in nutrient broth (Himedia, PA, USA) at 37 °C for 24 h. Optical density (OD) of bacterial cell culture was measured at 600 nm using Hitachi U2910 Spectrophotometer (IL, USA).
Lactobacillus acidophilus (LA-5, Chr. Hansen, Hónsholm, Denmark) and Bifidobacterium animalis subsp. lactis (BB-12, Chr. Hansen, Hónsholm, Denmark) were maintained in UHT whole milk at − 20 °C and activated at 37 °C for 2 h. The thermophilic starter culture was prepared using Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilus (Yo-Flex® L812, Chr. Hansen, Horsholm, Denmark) according to the manufacturer’s protocol.
Preparation of L. paracasei FNU culture for addition to the yoghurt
Cell suspension was prepared in MRS broth by growing the strain 24 h at 30 °C three times to reach 1 L of culture. Afterwards, cells were harvested by centrifugation (17,920 g, 20 min, 4 °C), washed twice with sterile peptone water (0.01 g mL−1), suspended in 500 mL whole UHT milk, and kept under − 20 °C in sterile glass bottles. The viable cell count in culture suspended in UHT whole milk after freezing was carried out in MRS agar.
Yoghurt preparation
Yoghurt was prepared with UHT milk with added starter culture containing L. delbrueckii subsp. bulgaricus and S. thermophilus, according to manufacturer’s instructions. The mixture was incubated at 45 °C, until reaching a pH of 4.6, followed by cooling to 4 °C and storage at 4 °C for 24 h. The prepared yoghurt was then divided into two lots: yoghurt without addition of L. paracasei FNU (control yoghurt) and yoghurt with addition of milk suspension containing L. paracasei FNU (8 log CFU mL-1 final concentration). Yoghurts were distributed in 200 mL sterile glass bottles, kept at 4 °C, and sampling was performed at days 1, 7, 14, 21, and 28 after yoghurt preparation. Samples were used immediately for plate counting or stored at − 20 °C for DNA extraction.
Commercial probiotic yoghurts, called commercial yoghurt A and commercial yoghurt B, were acquired in local supermarkets and used only for evaluation of DNA extraction protocols.
Evaluation of DNA extraction protocols
DNA extraction protocols were evaluated: DNeasy Mericon Food Kit (Qiagen, Venlo, Netherlands), DNeasy Mericon Food Kit (Qiagen, Venlo, Netherlands) with some modifications (20 μL of proteinase K instead of 2.5 μL and incubation at 65 °C instead of 60 °C), DNAzol® method 1 (Villegas-Rivera et al. 2013), DNAzol® method 2 (Achilleos and Berthier 2013), and CTAB method (Lipp et al. 1999). Samples of milk, commercial yoghurt, and bacterial culture medium were used to evaluate DNA extraction protocols in duplicate.
Bacterial culture DNA extraction protocol
For genomic DNA extraction in duplicate from bacterial culture medium, 2 mL medium aliquots (OD 0.8) were centrifuged (6000 g, 3 min, 4 °C) and pellets were stored at − 80 °C until DNA extraction. Pellets were suspended in 100 μL of ultrapure water, frozen at − 80 °C for 20 min, and immediately heated in boiling water (100 °C) for 10 min (Pereira et al. 2014). Cell suspension was cooled to room temperature, centrifuged (13,000 g, 10 s) and supernatant was used for DNA extraction using Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. DNA concentration and purity were determined on a Thermo Scientific NanoDrop 2000 spectrophotometer (Wilmington, DE, USA) with measurements at 260 and 280 nm.
DNA extraction protocol for yoghurt: DNAzol method 2
For genomic DNA extraction from yoghurt, starter culture and probiotic bacteria activated in milk, DNA was extracted in duplicate according to a method adapted from (Achilleos and Berthier 2013), named DNAzol® method 2. Ten milliliters of yoghurt or milk samples, 25 mL of 0.9 % NaCl, 8 mL of 25 % trisodium citrate, 2 g of polyethylene glycol 8000, and water, to a final volume of 50 mL, were homogenized for 5 min, separated into two aliquots of 25 mL, and centrifuged (9700 g, 15 min, 4 °C). Pellets were stored at − 20 °C until DNA extraction. Frozen pellets were thawed for 15 min at room temperature, suspended in 1 mL of DNAzol® reagent (Life Technologies, Carlsbad, CA, USA), and protocol was followed as described by (Achilleos and Berthier 2013).
Real-time PCR quantification
In order to detect and quantify by qPCR the presence of L. paracasei in yoghurt, Tuf primer pair was used, (TCCGGGAACTGCTCAGC and TGTTTCACGAACAGGTG) (Achilleos and Berthier 2013), which amplifies a fragment of 161 bp of the elongation factor Tu (tuf) gene. Quantitative real-time PCR was performed in ABI PRISM 7500 Detection System (Applied Biosystems, Foster City, CA, USA). Amplification reactions were carried out in duplicate in a final volume of 25 μL containing 12.5 μL of 2X SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), 300 nM of TufF and 150 nM of TufR, water, and template DNA (10 ng). The amplification program was: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The fluorescence signal was measured at the end of each 60 °C step. Melting curve analysis was performed automatically by continuous heating from 65 °C to 95 °C. All real-time PCR runs were analyzed using automatic software settings.
Construction of standard curves
Standard curves were prepared with serial dilutions of genomic DNA isolated from L. paracasei FNU, L. paracasei CCT 7501 and yoghurt. The number of bacterial DNA copies were calculated on the basis of the size of the L. paracasei strain 8700:2 (GenBank accession number NC_022112.1) genome (3.025 Mbp) using Avogadro’s constant (6.023 × 1023) and the molecular weight of DNA (660 Da/bp). Genomic DNA was tenfold serially diluted in ultra-pure water to final concentrations ranging from 107 to 100 genome copies per 2 μL, equivalent to concentrations of 33.2 to 3.32 × 10−6 ng.
The Cq versus log CFU of L. paracasei was estimated using genomic DNA extracted from the bacteria culture grown until stationary growth phase (OD 0.8) or from yoghurt with L. paracasei day 1. Ten times serial dilutions of DNA extracted from L. paracasei was performed and the corresponding CFU was calculated based on plate counting of the same sample, bacterial culture or yoghurt with L. paracasei day 1.
Standard curves were generated by the plot cycle threshold (Cq) values versus logarithm of bacterial DNA copy number (pure culture) or CFU (pure culture and yoghurt). Amplification efficiencies were determined using the following equation: E = 10(−1/S) − 1\( , \) where E is the efficiency and s is the slope obtained from the standard curve.
L. paracasei enumeration by qPCR
Genomic DNA was extracted from the yoghurt with L. paracasei from day 1 and serial dilutions were prepared, equivalent to concentrations of 33.2 to 3.32 × 10−6 ng of total DNA. Taking into account CFU obtained by plate counting from the same sample, CFU per reaction was calculated using the plot Cq versus CFU.
Bacterial count (CFU mL−1) in control yoghurt and yoghurt with L. paracasei collected in different days were determined using the following equation: \( bacterial\kern0.5em count=\frac{A\ast B\ast C}{D\ast E} \), where A is CFU per reaction well obtained from Cq of the DNA sample using standard curve (Cq versus log CFU), B is the extracted DNA concentration (ng μL-1), C is total volume of extracted DNA (μL), D is template DNA mass in reaction well (ng), and E is yoghurt volume (mL) used for DNA extraction.
L. paracasei enumeration by plate counting
The enumeration of L. paracasei FNU in yoghurt was carried out as previously described (Van de Casteele et al. 2006) with modifications. Control yoghurt was prepared with the addition of starter culture and yoghurt with L. paracasei was prepared with the addition of starter culture and L. paracasei FNU in a final concentration of 8 log CFU mL−1. Lactobacillus casei (LC) medium (Ravula and Shah 1998), adjusted to pH 5.0 using 5 M HCL, and supplemented with a membrane-filtered sterile solution of 10 % (w/v) D (−) ribose (Sigma Aldrich; 1 % final concentration) and 0.2 % (v/v) bromocresol green solution (Sigma Aldrich; 0.04 % (v/v) final concentration) was used. The samples were tenfold diluted and aliquots were pour plated on LC medium with an overlay of LC medium and incubated at 30 °C for 3–4 days.
Enumeration of L. paracasei CCT 7501 and L. paracasei FNU in pure cultures were made by pour plate technique on MRS agar (Merck, Darmstadt, Germany) with an overlay of MRS agar and incubation for 48 h at 30 °C. Enumerations were performed in triplicate.
Results
Comparison of yield and purity of DNA extracted using five protocols
Protocols were tested to compare their relative efficiency with respect to extracting DNA on the basis of yield and purity from milk, commercial yoghurt, and bacterial culture medium. The first tested protocol was DNeasy Mericon Food Kit, but it provided a very low DNA yield from commercial yoghurt (Table 1), despite the modifications realized to this method. Using DNAzol method 1 (Villegas-Rivera et al. 2013) and CTAB method (Lipp et al. 1999), it was not possible to extract DNA from L. paracasei bacterial culture medium or L. paracasei activated in milk. Using DNAzol method 2 (Achilleos and Berthier 2013), total DNA concentrations from commercial yoghurts A and B were higher than DNA concentrations obtained using modified DNeasy Mericon kit from the same samples, although DNA purity was better using modified DNeasy Mericon kit. With respect to DNA yield from milk containing bacteria (L. paracasei FNU, L. acidophilus LA-5, Bifidobacterium BB-12, or starter culture), DNA concentrations varied from 27 to 371 ng μL−1 (Table 1).
Using DNAzol method 2, total DNA concentrations from yoghurt prepared with thermophilic starter culture, with or without L. paracasei, varied from 37 to 154 ng μL−1 (Table S1).
Genomic DNA was successfully extracted from pure bacterial culture medium using Wizard kit from L. paracasei, L. plantarum, E. coli, B. cereus, and Pseudomonas spp. (Table S2), and they were used to test primer specificity.
Primer specificity for L. paracasei
Specificity test was conducted using DNA extracted from L. paracasei strains and other bacterial genera to verify if the Tuf primer pair was able to detect exclusively L. paracasei. Specificity test was done using bacterial DNA extracted from medium cultures or milk (10 ng of template DNA). L. paracasei strains FNU, CCT 7501, and LYO750 presented mean Cq values ± standard deviation equal to 16.7 ± 0.7, 18.2 ± 0.4, and 15.7 ± 0.2, respectively, and amplicon presented Tm of 78.3 ± 0.2, 78.2 ± 0.4, and 78.2 ± 0.2, respectively (Table 2). Bifidobacterium BB-12 and L. plantarum did not present amplification as expected for all qPCR assays. Other negative samples presented some unspecific amplification, with late Cq and different Tm values compared to L. paracasei Tm value of 78.2. Bacillus cereus presented 2 positives out of 6 total repetitions, with Cq higher than 31.7. Starter culture (L. delbrueckii subsp. bulgaricus and S. salivarius subsp. thermophilus) presented three positives out of six total repetitions, with late Cq (Cq > 38.4) and different Tm values. E. coli, Pseudomonas spp., and L. acidophilus also presented unspecific amplification at late Cq values (Table 2). L. acidophilus showed a late Cq (Cq > 34.3) in three reactions out of six repetitions. E. coli and Pseudomonas presented late Cq (Cq > 33.9 and Cq > 32.7, respectively) for all six repetitions with different Tm values.
qPCR parameters for L. paracasei enumeration
The reaction parameters (efficiency and correlation coefficient) of the qPCR assay using Tuf primer pair were determined based on standard curves obtained from tenfold serial dilution of bacterial DNA isolated from pure culture of L. paracasei CCT 7501 (Fig. 1) and L. paracasei FNU (Fig. 2a), performed in three and five qPCR runs on different days, respectively. The standard curves presented suitable linear correlation coefficient (R2) and mean efficiency (E), R2 > 0.93 and E of 91 % for L. paracasei CCT 7501 and R2 > 0.95 and E of 95 % for L. paracasei FNU (Table 3). The limit of detection (LOD) for both L. paracasei strains was 3 log DNA copy number, corresponding to 3.32 pg of DNA. For L. paracasei FNU, LOD corresponded to a mean Cq of 29.1. The Cq versus log CFU of L. paracasei FNU (Fig. 2b) was estimated using genomic DNA extracted from the L. paracasei FNU bacterial culture plate counted in parallel, so tenfold serial dilutions of bacterial DNA were performed and the corresponding CFU values were calculated based on plate counting. LOD corresponded to 2.78 log CFU of L. paracasei (Fig. 2b).
In order to evaluate qPCR enumeration of L. paracasei in yoghurt samples, amplification efficiency value was determined by the construction of standard curve of serial dilution of DNA extracted from yoghurt prepared with L. paracasei (day 1). In this case, the efficiency value was 103 % and R2 was 0.98 (Fig. 3), while for yoghurt sample (day 1) the log CFU was calculated based on plate counting.
Enumeration of L. paracasei in yoghurt samples
The average count of L. paracasei FNU in frozen UHT culture used for yoghurt preparation was 9.13 log CFU mL−1, obtained by plate counting. Yoghurt samples were collected 1, 7, 14, 21, and 28 days after preparation, and L. paracasei enumeration in these yoghurt samples was performed by plate counting and qPCR. DNA was extracted in duplicate for each sample using DNazol method 2 (Table S1) and submitted to qPCR assay (Table 4). Control yoghurt samples collected at 1, 7, 14, 21, and 28 days did not present amplification or presented unspecific amplification with late Cq (Cq > 31.9). Tm values of control yoghurt samples were different for L. paracasei FNU mean Tm value of 78.3 ± 0.2 (Table 2) and also different for yoghurt with L. paracasei mean Tm value of 78.6 ± 0.2 (Table 4). Regarding yoghurt samples with L. paracasei, Cq values ranged from 18.9 to 23.4, and all replicates presented amplification with similar Tm value, mean Tm of 78.6 ± 0.2 (Table 4).
The standard curve equation Cq versus log CFU of samples of yoghurt prepared with L. paracasei day 1 (Fig. 3) was used to calculate CFU per reaction well of all yoghurt samples from obtained Cq values. It was possible to obtain L. paracasei count (CFU mL−1of yoghurt) of yoghurt samples by qPCR (Table 5) using the equation described in material and methods. Bacterial count was obtained from the same yoghurt samples by plate counting (Table 5) based on a plate count method previously described (Van de Casteele et al. 2006). L. paracasei counts were similar by qPCR and plate count for samples collected from 7, 14, and 21 days after yoghurt preparation (Table 5); however, L. paracasei enumeration was lower by plate count (7.75 log CFU mL−1) than qPCR count (9.73 log CFU mL−1) for sample collected 28 days after yoghurt preparation. L. paracasei FNU viable cells were presented in high quantity (7.75 log CFU mL−1) even 28 days after yoghurt preparation as determined by plate count.
Discussion
The first requirement of a DNA-based method for bacterial quantification in food is an efficient DNA extraction method from that food (Garcia et al. 2013; Oliveira et al. 2013; Quigley et al. 2012). Bacterial DNA extraction from a dairy product is a special challenge to obtain DNA without PCR inhibitors such as calcium and fat (Pirondini et al. 2010). In the present study, different protocols were assessed to compare their relative success with respect to DNA extraction from commercial yoghurt. Using DNAzol method 2, total DNA concentrations from commercial yoghurts were higher than DNA concentrations obtained using the other tested methods (Table 1). DNAzol method 2 was chosen to extract DNA from yoghurt (Table S1) because it was used for qPCR enumeration assay of Lactobacillus paracasei and Lactococcus lactis in cheese (Achilleos and Berthier 2013). A qPCR assay targeting the tuf gene was successfully employed for L. paracasei enumeration in cheese (Achilleos and Berthier 2013). Several other lactic acid bacteria were tested as negative controls by other authors (Achilleos and Berthier 2013), and this qPCR assay using Tuf primers were specific enough for the identification of L. paracasei. We tested the specificity of these primers using other bacterial species (Table 2), and some bacterial species presented unspecific amplification; however, Cq values were always above the Cq value corresponding to LOD (Cq = 29), and they presented amplicons with different Tm values compared to L. paracasei samples. The ΔCq observed between DNA samples (10 ng) of L. paracasei strains (Cq < 24) and the other bacteria (Cq > 31) is sufficient to reinforce the use of this Tuf primer pair because these amplifications of other bacterial DNA are unspecific and they are easily distinguishable by their Tm. Unspecific amplification of other bacterial DNA with late Cq were also observed for qPCR assays targeting tuf gene developed for L. helveticus and L. rhamnosus even using hydrolysis probe (Desfosses-Foucault et al. 2012).
The choice of DNA extraction method can influence the quantification by real-time PCR and it is essential that the procedure results in an optimal yield of DNA and in removal of substances that could influence PCR efficiency (Cankar et al. 2006; Rodriguez-Lazaro et al. 2007). Comparing qPCR efficiency values obtained for this qPCR assay when target DNA is DNA extracted from L. paracasei pure cultures (Table 3 and Fig. 2) or DNA extracted from yoghurt prepared with L. paracasei FNU (Fig. 3), standard curves presented mean efficiency values of 91 % (pure strain CTT 7501), 95 % (pure strain FNU), and 103 % (yoghurt with strain FNU); suitable efficiency values shall be between 90 % and 110 % (Rodríguez et al. 2012). The qPCR efficiency values obtained by Achilleos and Berthier (2013) ranged from 81.1 % to 99.5 % for L. paracasei Tuf qPCR assay. The qPCR assay applied to one matrix may not be suitable for other matrices. In order to verify applicability of this qPCR assay for bacterial DNA quantification in yoghurt samples, standard curves using total DNA extracted from yoghurt were performed and an efficiency value of 103 % was obtained, indicating the absence of inhibitors from yoghurt. This can be evidence that the matrix did not significantly affect the PCR (Agrimonti et al. 2013).
Limit of detection (LOD) is the lowest amount of sample that can be reliably detected; in this qPCR assay the LOD is 3 log DNA copy number. This L. paracasei qPCR assay ensured the reliably detection of L. paracasei DNA ranging between 7 log genome copies (33.2 ng) to 3 log genome copies (3.32 pg) in the reaction well. It is a suitable range of genome copy number enumeration for probiotic bacteria in yoghurt samples, once they should be presented in large amounts. Similar to our results, the qPCR assays targeting the Tuf gene developed for L. helveticus and L. rhamnosus presented a range of quantification from 8 log to 3 log copy number (Desfosses-Foucault et al. 2012).
The L. paracasei FNU were presented in the range 8.30–9.73 log CFU mL−1 of yoghurt as determined by plate count at 7, 14, 21, and 28 days after yoghurt preparation. This concentration of live probiotic bacteria is considered enough to exert health benefits (Roy 2005). The L. paracasei FNU quantity (CFU mL−1) enumerated by qPCR was compared to L. paracasei enumerated by plate counts at different days of yoghurt manufacture (Table 5). Difference between qPCR and plate count was observed only 28 days after yoghurt preparation; counts were similar at 7, 14, and 21 days. A possible explanation on the statistical significant difference between qPCR and plate count at 28 days is the presence of dead cells that could not be distinguished from viable cells by qPCR, since it amplifies DNA from both dead and viable bacteria as DNA remains stable after the death of bacteria (Li et al. 2013). An alternative approach to detect only viable bacteria is viability qPCR using dyes that intercalate DNA of dead cells, such as ethidium monoazide and propidium monoazide (Barbau-Piednoir et al. 2014; Elizaquivel et al. 2014). Another strategy to detect viable bacteria is the analysis of 16S transcripts by RNAseq (Gosalbes et al. 2011), although it is an expensive strategy. In conclusion, this qPCR assay is a useful and rapid tool to enumerate L. paracasei in yoghurt, although it does not distinguish dead and viable cells.
References
Achilleos C, Berthier F (2013) Quantitative PCR for the specific quantification of Lactococcus lactis and Lactobacillus paracasei and its interest for Lactococcus lactis in cheese samples. Food Microbiol 36:286–295. doi:10.1016/j.fm.2013.06.024
Agrimonti C, Bortolazzi L, Maestri E, Sanangelantoni A, Marmiroli N (2013) A Real-Time PCR/SYBR Green I Method for the rapid quantification of Salmonella enterica in poultry meat. Food Anal Methods 6:1004–1015. doi:10.1007/s12161-013-9583-y
Ashraf R, Shah NP (2011) Selective and differential enumerations of Lactobacillus delbrueckii subsp bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt - A review. Int J Food Microbiol 149:194–208. doi:10.1016/j.ijfoodmicro.2011.07.008
Barbau-Piednoir E, Mahillon J, Pillyser J, Coucke W, Roosens NH, Botteldoorn N (2014) Evaluation of viability-qPCR detection system on viable and dead Salmonella serovar Enteritidis. J Microbiol Methods 103:131–137. doi:10.1016/j.mimet.2014.06.003
Cankar K, Stebih D, Dreo T, Zel J, Gruden K (2006) Critical points of DNA quantification by real-time PCR - effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. Bmc Biotechnology 6 doi:3710.1186/1472-6750-6-37
Ceccherini MT, Luchi N, Pantani O-L, Ascher J, Capretti P, Pietramellara G (2013) Upward movement of Verticillium dahliae from soil to olive plants detected by qPCR. World J Microbiol Biotechnol 29:1961–1967. doi:10.1007/s11274-013-1342-0
Chavagnat F, Haueter M, Jimeno J, Casey MG (2002) Comparison of partial tuf gene sequences for the identification of lactobacilli. Fems Microbiol Lett 217:177–183. doi:10.1111/j.1574-6968.2002.tb11472.x
Chiang S-S, Pan T-M (2012) Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. Appl Microbiol Biotechnol 93:903–916. doi:10.1007/s00253-011-3753-x
De Medici D, Kuchta T, Knutsson R, Angelov A, Auricchio B, Barbanera M, Diaz-Amigo C, Fiore A, Kudirkiene E, Hohl A, Horvatek Tomic D, Gotcheva V, Popping B, Prukner-Radovcic E, Scaramaglia S, Siekel P, To KA, Wagner M (2015) Rapid methods for quality assurance of foods: the next decade with polymerase chain reaction (PCR)-based food monitoring. Food Anal Methods 8:255–271. doi:10.1007/s12161-014-9915-6
Desfosses-Foucault E, Dussault-Lepage V, Le Boucher C, Savard P, La Pointe G, Roy D (2012) Assessment of probiotic viability during Cheddar cheese manufacture and ripening using propidiunn monoazide-PCR quantification. Fron Microbiol 3:350. doi:10.3389/fmicb.2012.00350
Elizaquivel P, Aznar R, Sanchez G (2014) Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J Appl Microbiol 116:1–13. doi:10.1111/jam.12365
Francesca N, Sannino C, Moschetti G, Settanni L (2013) Microbial characterisation of fermented meat products from the Sicilian swine breed "Suino Nero Dei Nebrodi". Ann Microbiol 63:53–62. doi:10.1007/s13213-012-0444-5
Furet JP, Quenee P, Tailliez P (2004) Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int J Food Microbiol 97:197–207. doi:10.1016/j.ijfoodmicro.2004.04.020
Garcia A, Kamara J, Vigre H, Hoorfar J, Josefsen M (2013) Direct quantification of Campylobacter jejuni in chicken fecal samples using real-time PCR: evaluation of six rapid DNA extraction methods. Food Anal Methods 6:1728–1738. doi:10.1007/s12161-013-9685-6
Herbel SR, Lauzat B, von Nickisch-Rosenegk M, Kuhn M, Murugaiyan J, Wieler LH, Guenther S (2013) Species-specific quantification of probiotic lactobacilli in yoghurt by quantitative real-time PCR. J Appl Microbiol 115:1402–1410. doi:10.1111/jam.12341
Huang C-H, Chang M-T, Huang L (2014) Use of highly variable gene (yycH) as DNA marker to resolve interspecific relationships within the Lactobacillus casei group and a target for developing novel species-specific PCR primers. Eur Food Res Technol 239:719–724. doi:10.1007/s00217-014-2278-9
Ilabaca C, Jara C, Romero J (2014) The rapid identification of lactic acid bacteria present in Chilean winemaking processes using culture-independent analysis. Ann Microbiol 64:1857–1859. doi:10.1007/s13213-014-0810-6
Ilha E, da Silva T, Lorenz J, de Oliveira Rocha G, Sant’Anna E (2014) Lactobacillus paracasei isolated from grape sourdough: acid, bile, salt, and heat tolerance after spray drying with skim milk and cheese whey. Eur Food Res Technol 240:977–984. doi:10.1007/s00217-014-2402-x
Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, Latorre A, Moya A (2011) Metatranscriptomic approach to analyze the functional human gut microbiota. Plos One 6(3):e17447. doi:10.1371/journal.pone.0017447
Kao Y-T, Liu Y-S, Shyu Y-T (2007) Identification of Lactobacillus spp. in probiotic products by real-time PCR and melting curve analysis. Food Res Int 40:71–79. doi:10.1016/j.foodres.2006.07.018
Ke L, Wang L, Li H, Lin H, Zhao L (2014) Molecular identification of lactic acid bacteria in Chinese rice wine using species-specific multiplex PCR. Eur Food Res Technol 239:59–65. doi:10.1007/s00217-014-2193-0
Kristo E, Biliaderis CG, Tzanetakis N (2003) Modelling of rheological, microbiological and acidification properties of a fermented milk product containing a probiotic strain of Lactobacillus paracasei. Int Dairy J 13:517–528. doi:10.1016/s0958-6946(03)00074-8
Li M, Zhao G, Liu J, Gao X, Zhang Q (2013) Effect of different heat treatments on the degradation of Salmonella nucleic acid. J Food Saf 33:536–544. doi:10.1111/jfs.12086
Lipp M, Brodmann P, Pietsch K, Pauwels J, Anklam E (1999) IUPAC collaborative trial study of a method to detect genetically modified soy beans and maize in dried powder. J Aoac Int 82:923–928
Machado SG, Soares Bazzolli DM, Dantas Vanetti MC (2013) Development of a PCR method for detecting proteolytic psychrotrophic bacteria in raw milk. Int Dairy J 29:8–14. doi:10.1016/j.idairyj.2012.09.007
Oliveira J, Cunha Â, Almeida A, Castilho F, Pereira M (2013) Comparison of methodologies for the extraction of bacterial DNA from mussels—relevance for food safety. Food Anal Methods 6:201–209. doi:10.1007/s12161-012-9419-1
Pereira TP, do Amaral FP, Dall'Asta P, Angonesi Brod FC, Maisonnave Arisi AC (2014) Real-Time PCR quantification of the plant growth promoting bacteria Herbaspirillum seropedicae strain SmR1 in maize roots. Mol Biotechnol 56:660–670. doi:10.1007/s12033-014-9742-4
Pirondini A, Bonas U, Maestri E, Visioli G, Marmiroli M, Marmiroli N (2010) Yield and amplificability of different DNA extraction procedures for traceability in the dairy food chain. Food Control 21:663–668. doi:10.1016/j.foodcont.2009.10.004
Poltronieri P, D'Urso OF, Blaiotta G, Morea M (2008) DNA arrays and membrane hybridization methods for screening of six Lactobacillus species common in food products. Food Anal Methods 1:171–180
Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD (2012) A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J Appl Microbiol 113:96–105. doi:10.1111/j.1365-2672.2012.05294.x
Ravula RR, Shah NP (1998) Selective enumeration of Lactobacillus casei from yogurts and fermented milk drinks. Biotechnol Tech 12:819–822
Reitschuler C, Lins P, Illmer P (2014) Primer evaluation and adaption for cost-efficient SYBR Green-based qPCR and its applicability for specific quantification of methanogens. World J Microbiol Biotechnol 30:293–304. doi:10.1007/s11274-013-1450-x
Rodríguez A, Córdoba J, Gordillo R, Córdoba M, Rodríguez M (2012) Development of Two Quantitative Real-Time PCR Methods Based on SYBR Green and TaqMan to Quantify Sterigmatocystin-Producing Molds in Foods. Food Anal Methods 5:1514–1525. doi:10.1007/s12161-012-9411-9
Rodriguez-Lazaro D, Lombard B, Smith H, Rzezutka A, D'Agostino M, Helmut R, Schroeter A, Burkhard M, Miko A, Guerra B, Davison J, Kobilinsky A, Hernandez M, Bertheau Y, Cook N (2007) Trends in analytical methodology in food safety and quality: monitoring microorganisms and genetically modified organisms. Trends Food Sci Technol 18:306–319. doi:10.1016/j.tifs.2007.01.009
Rodriguez-Lazaro D, Gonzalez-García P, Valero A, Hernandez M (2015) Application of the SureTect detection methods for Listeria monocytogenes and Listeria spp. in meat, dairy, fish, and vegetable products. Food Anal Methods 8:1-6. doi:10.1007/s12161-014-9970-z
Roy D (2005) Technological aspects related to the use of bifidobacteria in dairy products. Lait 85:39–56
Ruiz P, Sesena S, Llanos Palop M (2014) A comparative study of different PCR-based DNA fingerprinting techniques for typing of lactic acid bacteria. Eur Food Res Technol 239:87–98. doi:10.1007/s00217-014-2197-9
Rushdy AA, Gomaa EZ (2013) Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann Microbiol 63:81–90. doi:10.1007/s13213-012-0447-2
Tian W, Zhang Z, Liu D, Zhou T, Shen Q, Shen B (2013) An optimized DNA extraction and purification method from dairy manure compost for genetic diversity analysis. World J Microbiol Biotechnol 29:815–823. doi:10.1007/s11274-012-1236-6
Van de Casteele S, Vanheuverzwijn T, Ruyssen T, Van Assche P, Swings J, Huys G (2006) Evaluation of culture media for selective enumeration of probiotic strains of lactobacilli and bifidobacteria in combination with yoghurt or cheese starters. Int Dairy J 16:1470–1476. doi:10.1016/j.idairyj.2005.012.002
Villegas-Rivera G, Vargas-Cabrera Y, González-Silva N, Aguilera-García F, Gutiérrez-Vázquez E, Bravo-Patiño A, Cajero-Juárez M, Baizabal-Aguirre VM, Valdez-Alarcón JJ (2013) Evaluation of DNA extraction methods of rumen microbial populations. World J Microbiol Biotechnol 29:301–307. doi:10.1007/s11274-012-1183-2
Xue C, Zhang L, Fan R, Wang S, Li H, Luo X, Liu W, Song W (2015) Protective action of S-layer proteins from Lactobacillus paracasei M7 against Salmonella infection and mediated inhibition of Salmonella-induced apoptosis. Eur Food Res Technol 240:923–929. doi:10.1007/s00217-014-2396-4
Yu J, Sun Z, Liu W, Bao Q, Zhang J, Zhang H (2012) Phylogenetic study of Lactobacillus acidophilus group, L. casei group and L. plantarum group based on partial hsp60, pheS and tuf gene sequences. Eur Food Res Technol 234:927–934. doi:10.1007/s00217-012-1712-0
Conflict of Interest
Authors declare that they have no conflict of interest.
Compliance with Ethical Standards
This article does not contain any studies with human or animal subjects.
Informed consent was obtained from all individual participants included in the study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 172 kb)
Rights and permissions
About this article
Cite this article
Ilha, E.C., Scariot, M.C., Treml, D. et al. Comparison of real-time PCR assay and plate count for Lactobacillus paracasei enumeration in yoghurt. Ann Microbiol 66, 597–606 (2016). https://doi.org/10.1007/s13213-015-1137-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1137-7