Abstract
Nineteen isolates of rhizobacteria associated with sunflower (Helianthus annuus L.), collected from various locations in Pakistan, were screened for phosphate solubilization and indole-3-acetic acid (IAA) production. Two potential phosphate-solubilizing bacterial isolates with substantial IAA biosynthesis capacity, Ps-5 and Ss-2, were selected for further study. Based on 16S rRNA gene sequence analysis, isolate Ps-5 was identified as Bacillus sp. and Ss-2 as Alcaligenes faecalis. Both strains were found to be metabolically diverse in terms of the number and amount of different carbon substrates they utilized in the BIOLOG GN2/GP2 microplate assay. High-performance liquid chromatography analysis of the culture supernatant confirmed that Bacillus sp. Ps-5 produced considerable amounts of both lactic and tartaric acids, while A. faecalis Ss-2 secreted only lactic acid. There was a strong positive correlation between phosphate solubilization and organic acid production by both strains. Following inoculation, strain Ps-5 and Ss-2 were found to be good root colonizers and significantly (P ≤ 0.05) increased sunflower growth and phosphorus (P) uptake. However, inoculation had a non-significant (P ≤ 0.05) effect on sunflower yield parameters, including oil contents. Based on these results, we conclude that Ps-5 and Ss-2 are potent plant growth-promoting rhizobacteria strains with the ability to supplement the P requirements of sunflower crops. Further field inoculation studies are needed before these strains can be recommended as bio-inoculants. To the best of our knowledge, this is first report on the association and phytobeneficial potential of A. faecalis with sunflower.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
A microbial community that is closely affiliated with plant roots and densely populates the immediate environment of these plant roots is known as the rhizosphere microbiome or rhizo-microbiome (Chaparro et al. 2013). A proportion of bacteria within the dynamic environment of the rhizosphere are referred as plant growth-promoting rhizobacteria (PGPR). These microorganisms are an important biological asset for sustainable agriculture in that they promote plant growth directly by nitrogen fixation, macro- and/or-micro nutrient solubilization/mobilization, phytohormone production and/or indirectly by controlling phytopathogens through the production of siderophores, antibiotics, lytic enzymes, among others (Compant et al. 2005). PGPR are the most widely studied group of plant growth-promoting bacteria (PGPB) that are capable of establishing a dense population in rhizosphere, the soil adhering closely to root surfaces (Kloepper et al. 1989). The PGPB also found directly attached to roots are known as rhizoplanic bacteria, while those likely to penetrate into the root epidermal and cortical tissues are referred to as endophytes (Sylvia et al. 2005). PGPR are important biological tools for the effective release of insoluble phosphorus (P) from organic and inorganic pools through mineralization and solubilization processes, respectively, thereby helping plants obtain their nutrition from the soil (Hilda and Fraga 1999).
Although, P is the second most important nutrient for plant growth, only 1–5 % of total P in soil is in a form available to plants (Arcand and Schneider 2006; Zaidi et al. 2009). This has led to the excessive use of synthetic phosphate fertilizers becoming a normal routine for the farmers around the world. However, these mineral fertilizers form complexes with cations of calcium (Ca), aluminum (Al) and iron (Fe) in the soil and become insoluble (Sharan et al. 2008). In addition, synthetic mineral fertilizers have serious environmental and human health impacts and place a heavy economic burden on farmers. An alternative and environmentally friendly approach to achieve sustainable agriculture in a cost-effective manner would be to select and utilize those microorganisms and their biosystems with the mineral phosphate-solubilizing trait (Hameeda et al. 2006; Fankem et al. 2008; Park et al. 2010). Bacterial strains of genera Pseudomonas, Bacillus, Rhizobium and Enterobacter and fungal strains of genera Penicillium and Aspergillus are the most widely accepted P solubilizers (Whitelaw 1999; Shahid et al. 2012). The main phytobeneficial mechanism of mineral phosphate-solubilizing bacteria (PSB) is lowering of the pH of the surroundings through the production of oganic acids. These organic acids carry hydroxyl and carboxyl groups which chelate cations (Al, Fe, Ca) bound to soil phosphates, thereby making them soluble by detaching these cations (Mullen 2005; Trivedi and Sa 2008).
Of the organic acids synthesized by PSB, indole-3-acitic acid (IAA) is considered to be the most important. IAA is capable of playing a crucial role in cell growth and division and has the potential to be a major factor in root proliferation/elongation in plants (Seo and Park 2009). To date, three main IAA biosynthetic pathways have been reported in bacteria, namely, the indole-3-pyruvic acid, indole-3-acetamide and indole-3-acetonitrile pathways, respectively (Duca et al. 2014).
The association of PSB with sunflower roots and screening of the potential of these PSB for enhancing the growth and yield of sunflower plants has not as yet been completely studied. Recently, Ambrosini et al. (2012) reported the association between bacterial species/strains belonging to the genera of Enterobacter, Klebsiella, Grimontella, Novosphingobium, Microbacterium, Acinetobacter, Pantoea, Variovorax, Asticcacaulis, Chryseobacterium, Herbaspirillum, Mitsuaria, Moraxella, Serratia, Shinella, Sphingobium and Xanthomonas and the sunflower rhizosphere. Among these bacterial genera, members of the Enterobacter and Burkholderia were found to be the most abundant. In Pakistan there is a huge gap between edible oil production and market demand, resulting in sunflower oil being imported into Pakistan on a large scale. In the context of decreasing this gap, any improvement in the yield of sunflower crops is of keynote significance (Khan et al. 2000; Shah et al. 2013). Sunflower yield and yield components, including the oil yield, can be increased by the application of PSB (Ekin 2010). However, compared to studies on the rhizospheres of other plants, the isolation, characterization, identification and manipulation of potential PSB from the sunflower rhizosphere have been less explored worldwide, and to our knowledge no valid reports from Pakistan are available in this area of scientific research. Thus, the objective of this study was to characterize and identify promising PGPR strains which can supplement the P requirements of sunflower in a cost-effective manner without compromising crop yield and oil contents.
Materials and methods
Isolation and morphological studies
Sunflower (Helianthus annuus L.) plants at the flowering stage were uprooted with intact roots and adhering soil from various locations in Pakistan, including Faisalabad (30° 44′27.10 N, 72° 38′18.39 E, 154 m a.s.l.), Multan (30° 5′55.32 N, 71° 30′41.24 E, 126 m a.s.l.), Peshawar (34° 00′56.09 N, 71° 42′ 46.94 E, 304 m a.s.l.) and Tandojam (25° 25′ 50.72 N, 68° 33′ 46.81, E, 24 m a.s.l.). The shoots were excised using a sterilized knife, following which root portions were transferred to sterilized plastic bags (25 × 30 cm) and transported to the laboratory. Roots were shaken gently in sterile distilled water to remove the loosely adhering soil. One gram of the soil that remained tightly adhered to the roots was then removed from each sample and added in 9 mL of 0.85 % (w/v) NaCl solution; this solution was serially diluted as described by Somasegaran and Hoben (1994). A 100-μL aliquots from dilutions 10−4, 10−5 and 10−6 of each sample was spread on Luria-Bertani (LB) agar plates using a sterilized glass spreader and incubated at 28 ± 2 °C for 48 h. Bacterial isolates from each plate were selected based on prolific growth and colony appearance and streaked many times to achieve the maximum purity level. Twenty isolates purified in this manner were maintained on LB agar plates at 28 ± 2 °C for further studies, and five copies of each isolate were stored in 20 % (v/v) glycerol at −80 °C. Colony morphology, cell shape, motility and the Gram reaction were studied under the light microscope as described by Vincent (1970). The isolates from Faisalabad, Multan, Peshawar and Tandojam were designated Fs, Ms, Ps and Ss, respectively.
Phosphate solubilization and detection of organic acids
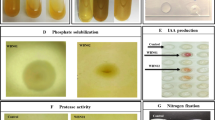
A single colony of each purified isolate was spotted on Pikovskaya’s agar (Pikovskaya 1948) plates, incubated at 28 ± 2 °C and observed for up to 240 h for halo-zone formation. For quantification of solubilized phosphate and detection of organic acids in the culture medium, we performed triplicate experiments in which 100-mL of Pikovskaya’s broth containing a single colony of each isolate was inoculated into a 500-mL Erlenmeyer flask and incubated at 28 ± 2 °C for 240 h in an orbital shaker at 150 rpm. A 20-mL sample of bacterial culture from each flask was harvested after 168 h (for all isolates) and after 120, 168 and 240 h (for two isolates) and centrifuged at 13,000 g for 10 min to collect the supernatant. The quantitative measurement of phosphate solubilization was performed according to phosphomolybdate blue colour method (Murphy and Riley 1962) using a spectrophotometer (λ = 882 nm). To determine the identity and concentration of organic acids, the supernatant of two selected isolates was first filtered through 0.2-μm nylon filters (Millipore, USA). Then, 20-μL samples of each supernatant were analysed using a high-performance liquid chromatography (HPLC) system equipped with Turbochrom software (Perkin Elmer, USA) and a C18 column (length 150 mm, diameter 4 mm, pore size 120 Å). The mobile phase consisted of methanol:acetic acid (30:70, v/v), and a flow rate of 0.6 mL min−1 was set. Organic acids to be used as standards were purchased from Sigma (USA). The supernatant was analysed for the presence of gluconic acid, malic acid, lactic acid, acetic acid, citric acid, succinic acid and tartaric acid by spectrometry at a 210 nm wavelength.
Indole-3-acetic acid production
For the detection of IAA, a single colony of each purified isolate was inoculated into a 500-mL Erlenmeyer flask containing 100 mL of LB broth supplemented with tryptophan (100 mg L−1) and incubated at 28 ± 2 °C for 48 h in an orbital shaker at 150 rpm. The supernatant was collected in separate tubes after centrifugation at 13,000 g and acidified (pH 2.8) with hydrochloric acid. An equal volume of ethyl acetate was mixed with the supernatant in a separating funnel to extract the IAA (Tien et al. 1979). The upper layer of ethyl acetate containing the IAA was collected in a separate sterilized tubes, and excessive ethyl acetate was removed under vacuum at 45 °C, using a rotary evaporator. The recovered extract was dissolved in methanol (1 mL) and filtered through a 0.2-μm nylon filter (Millipore) prior to analysis on a HPLC (λ = 260 nm) system equipped with Turbochrom software (Perkin Elmer) and a C18 column at a flow rate of 0.5 mL min−1 using 30:70 (v/v) methanol:water as the mobile phase.
16S rRNA gene sequencing
Total genomic DNA of isolate Ps-5 and Ss-2 was isolated by the alkaline lysis method (Maniatis et al. 1982) and quantified on the Ultrospec™ 3100 pro UV/visible spectrophotometer (GE Healthcare Life Sciences, USA) at OD260, 260/280. The quantified DNA was then used to amplify the 16S rRNA gene with primers fD1 and rD1 as described by Weisburg et al. (1991), with the following modifications: 50 μL reaction volume in purified water, 5 μL of Taq buffer (Fermentas, Lithuania), 3 μL of 25 mM MgCl2 (Fermentas), 5 μL of 2 mM dNTPs (Fermentas), 0.5 μL of 100 % DMSO, 1.5 μL each of forward and reverse primer, 0.75 μL of 5 U μL−1 Taq DNA polymerase (Fermentas) and 40 ng of template DNA. Amplification by PCR was carried out in a thermal cycler (advanced Primus 96; PeQLab Biotechnologie, Germany) also under modified conditions of 30 cycles of 95 °C for 1 min, 55 °C for 30 s, and 72 °C for 1 min. The initial denaturation and final extension steps were performed at 95 °C for 5 min and 72 °C for 10 min, respectively. Amplified PCR products were purified using QIAquick PCR purification kit (Qiagen, USA) and sequenced directly on both sides by a commercial laboratory (Eurofins, Germany). The gene sequence was analysed using the Sequence Scanner software package; both ends were joined using Caps 3 assembly software and compared with others in the GenBank database using the NCBІ BLASTn tool. The final sequences were deposited in Genbank and accession numbers were obtained.
Phenotypic microarray analysis
For the phenotypic microarray analysis, 50-mL samples of culture media containing isolates Ps-5 and Ss-2 were grown in LB broth for 20 h in 250-mL Erlenmeyer flasks to obtain a population density of 108 CFU mL−1. A 30-mL sample of culture was then collected from each flask and centrifuged at 8,000 g for 10 min; the pellet was removed and dissolved in the same volume of phosphate buffer (pH 6.5). The target OD620 nm (0.34) which indicated a population density of 106 CFU mL−1 was obtained by diluting the suspension with the appropriate amount of phosphate buffer. The suspension was then incubated at room temperature for 3 h to starve the cells and deplete most of the energy reserve needed for bacterial cell growth. Each well of the Biolog GN2/GP2 micro-titer plate (Biolog, USA) was filled with 100-μL of suspension and the plates were incubated at 28 ± 2 °C for 72 h; the reaction was analyzed qualitatively for color development and quantitatively on the VERSA max micro-plate reader (Molecular Devices, USA) with SoftMax Pro software. The OD595 nm was determined after 24, 48 and 72 h of incubation.
Root colonization studies
In the pot experiment, both bacterial strains were recovered from the rhizosphere after 7, 15 and 30 days of transplanting on LB plates. To recover bacterial isolates, sunflower plants with intact roots were carefully uprooted and shaken gently in sterilized distilled water to remove the loosely adhering soil. Bacteria were then recovered by the dilution plating technique using 1 g of the soil which remained strictly adhering to the roots in 9-mL sterile water (Seldin et al. 1998). The bacterial colonies recovered were also examined under the stereoscope to confirm their morphology.
For the ultrastructure studies, sunflower (cv. FH-331) seeds were surface sterilized by dipping them in sodium hypochlorite (5 %, w/v) for 10 min, followed by five to six washings with sterilized water. The seeds were inoculated by immersion in the inoculum (1 × 108 CFU mL−1) of selected isolates for 30 min and allowed to germinate on 1.5 % (w/v) water agar plates. Root hairs of 10-day-old seedlings were cut into pieces (approx. 1–3 cm) and embedded in water agar again, followed by cutting out approximately 2- to 3-mm small agar cubes. The cubes were put in 1.5-mL tubes in the presence of 5 % gluteraldehyde as fixative (in 0.2 M PIPES buffer, pH 8.0). After 16–18 h, the fixative was replaced with 0.2 M PIPES buffer {0.58 g NaCl, 3 g PIPES [piperazine-N,N′ bis(2-ethanesulfonic acid)], 1 M NaOH, 0.2 g MgCl2·6H2O, pH 6.8} (Salema and Brandao 1973), and the samples were washed 2× 1 h in fresh buffer. The washed samples were treated with 0.2 % osmium tetraoxide made in PIPES buffer (0.2 M, pH 6.8) for 16–18 h and then washed again 2× 30 min with sterilized distilled water. After being treated with 5 % aqueous uranyl acetate for 16–18 h, the samples were washed with sterilized distilled water for 2× 30 min. The dehydration steps consisted of immersion in absolute alcohol for 2× 30 min, followed by immersion in propylene oxide (100 %) for 1× 30 min. Infiltration of samples was carried out with propylene oxide in a ratio of 1:1 for 24–48 h and then with spurr resin for a further 24–72 h. The accelerator benzyl di-methyl amine was used in all infiltration steps. The samples were transferred to flat embedding moulds and polymerized for 72 h at 65–70 °C, following which the polymerized resin blocks were removed from the oven and left at room temperature for at least 24 h before being cut into ultrathin sections (150–200 nm) on an ultra-microtome (RMC-7000; Boeckeler Instruments, USA). The sections were carefully placed on copper grids. All of the sections were double stained with uranyl acetate (30 min) and lead citrate (10 min). The grids were then washed with deionized water and observed under a transmission electron Microscope (TEM; JEOL 1010).
Pot experiment
A pot experiment was conducted in growth room by employing Completely Randomized Design (CRD). Surface-sterilized sunflower seeds (cv. FH-331) were germinated on 1.5 % water agar plates. For inoculum preparation, bacterial cultures were grown to a density of 1 × 109 CFU mL−1, centrifuged at 8,000 g and washed twice with saline solution (0.85 %, w/v). The cells were re-suspended in equal volumes of saline and diluted to 108 CFU mL−1. The growth room was fumigated before the start of the experiment to achieve maximum sterility, and growth conditions were set at 25 °C and 16/8 light/dark periods until harvesting of the plants. The pots (diameter 9 cm) were filled with 400 g of steam-sterilized sand. Tricalcium phosphate (TCP) was mixed in sand (200 mg kg−1 sand) as an insoluble inorganic P source, and 4-day-old agar-grown plants were inoculated by dipping the roots in inoculums for 30 min. In addition, 7 mL 100 g−1 inoculum was mixed in sand at the time of transplanting. Each pot was transplanted with one inoculated seedling, with one treatment left uninoculated. Pots were watered with Hoagland’s solution (Arnon and Hoagland 1940) without a P source (10 mL pot−1 daily). The data on various growth parameters, including P uptake by plants, was determined 30 days after transplanting. Root and shoot P content were determined by the vanadium phospho-molybdate yellow colour method (Yoshida et al. 1971).
Field experiment
Soil of the experimental field was chemically analyzed by the Department of Soil and Environmental Sciences, University of Poonch Rawalakot Azad Jammu & Kashmir prior to the start of the experiment. The field experiment was conducted at the experimental field area of the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, using two inoculated treatments of Bacillus sp. Ps-5 and Alcaligenes faecalis Ss-2 with a half dose of the recommended fertilizer. Three uninoculated treatments with a full and half dose of fertilizer and no fertilizer were also included for comprehensive comparison. The experiment was conducted in a randomized complete block design with three replications.The seed bed was prepared by ploughing the soil two to three times with a tractor-mounted cultivator followed by planking after each cultivation. Plot size was maintained at 3 × 2 m2. Sowing was done manually by the dibbling method, with three to four seeds per hill and a between-row and between-plant distance of 75 and 25 cm, respectively. Inoculum was prepared in the same way as described in the pot experiment, while seed inoculation was carried out by dipping sunflower seeds in inoculum for 30 min. Thinning was done when the crop attained the height of 10 cm, and an 8 kg ha−1 seed rate was used. Nitrogen (50 kg ha−1) was applied in the form of urea in two splits (at sowing and at first irrigation), and P (90 kg ha−1) in the form of triple superphosphate was incorporated in the soil at the time of sowing by means of single row drill. Plant protection measures were adopted to keep the crop free from weeds, insects, pests and diseases. All other agronomic practices were kept normal and uniform for all the treatments. After harvesting, the crop was left for 5 days to dry under the sun and then threshed by physical beating. Achene P contents were determined by the vanadium phospho-molybdate yellow colour method (Yoshida et al. 1971). Oil content analysis of sunflower achenes was carried out commercially by Oil Seed Brassica Lab, Plant Breeding & Genetics Division, Nuclear Institute for Food and Agriculture (NIFA), Peshawar, Pakistan, by gas chromatography as described by Erickson et al. (1980).
Measurements and data analysis
Data for the Biolog GN2/GP2 microplate assay were analyzed by principal component analysis (PCA) using SPSS 17 software (SPSS Inc., USA). Data for the pot and field experiments were analyzed statistically by analysis of variance(Steel et al. 1997) using Statistix (ver. 8.1) software. The least significant difference test (Fisher’s LSD) at 5 % probability was used to compare the differences among treatment means.
Results
Morphological and physiological characterization
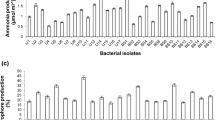
Most of the bacterial isolates were motile short rods, but some long rods and few cocci were also present. One isolate, Ps-3, was found to be oval shaped. Of the 19 isolates collected, 14 were Gram-negative and five, i.e. one each from Multan (Ms-4), Peshawar (Ps-5) and Tandojam (Ss-1) and two from Faisalabad (Fs-1 and Fs-4), were Gram-positive. Six isolates (Fs-6, Ms-3, Ps-2, Ps-3, Ps-5 and Ss-2) produced considerable halo-zones on TCP-supplemented agar medium. Analysis of the culture supernatants revealed that isolates Ps-5 and Ss-2 had the maximum P-solubilizing and IAA biosynthesis potentials, solubilizing 28.02 ± 1.85 and 28.91 ± 2.56 μg mL−1 TCP, respectively, and synthesizing 10.65 ± 2.08 and 2.27 ± 0.10 μg mL−1 IAA, respectively (Table 1).
Relationship between phosphate solubilization, pH change and organic acid synthesis
In the time-course studies, a trend towards increasing in vitro phosphate solubilization and organic acid synthesis was observed for both isolates with prolonged incubation time up to 240 h (Fig. 1a). Conversely, there was a decreasing trend in pH with increasing incubation time. The maximum amount of soluble phosphate in the culture supernatant after 240 h for isolate Ps-5 was estimated to be 57.40 μg mL−1, with a 3.93 drop in pH. For isolate Ss-2, the phosphate solubilization value was estimated to be 43.71 μg mL−1, with drop in pH of up to 5.2. After 240 h of incubation in vitro, isolate Ps-5 produced lactic and tartaric acids (22.09 and 1.31 μg mL−1, respectively), while Ss-2 synthesized lactic acid only (14.57 μg mL−1)(Fig. 1b). The amount of phosphate solubilized by Ps-5 was found to be significantly correlated (P ≤ 0.001) with its synthesis of both lactic acid (r = 0.99) and tartaric acid (r = 0.95)s. For Ss-2, a strong positive correlation (P ≤ 0.001) was found between phosphate solubilization and its production of lactic acid (r = 0.97).
Mean in vitro solubilization of insoluble phosphate with drop in pH (a) and production of organic acids (b) at different intervals of incubation by selected phosphate-solubilizing isolates from Peshawar (Ps-5) and Tandojam (Ss-2). The amount of soluble phosphorus (μg mL-1) calculated in the uninoculated control (negative control) was subtracted from the amount of soluble phosphate determined in the inoculated treatments. Data represent the mean ± standard deviation (SD)
Phenotypic microarray analysis
In the phenotypic microarray analysis, isolates Ps-5 and Ss-2 utilized 41 and 55 substrates, respectively, in the Biolog GP2/GN2 microtiter plates analysis. PCA of data based on absorbance values also verified the phenotypic results. The data retrieved 24, 48 and 72 h after inoculation demonstrated a clear divergence in carbon source utilization patterns between the two isolates (Fig. 2).
Identification through 16S rRNA gene sequence analysis
A 1,450-bp 16S rRNA gene sequence of isolate Ps-5, retrieved after commercial sequencing, established a 99 % sequence similarity with Bacillus cereus strain CA15 and a number of other Bacillus species, limiting identification to the species level. Thus, rhizotype Ps-5 was identified as Bacillus sp. (JQ248587). The 16S rRNA gene of isolate Ss-2 (1,318 bp) showed 99 % sequence identity with Alcaligenes faecalis strain B_IV_2L49 and a number of other strains of same species; hence, it was identified as A. faecalis (JQ268265).
Root colonization studies
In the growth room pot experiment, both strains were successfully recovered up to 30 days post-transplantion at a population density of around 106 CFU g−1 rhizosphere sand. There was a slight drop in the bacterial populations of both strain Ps-5 and strain Ss-2 at the 30-day post-transplantation time-point, with the density of Ps-2 dropping from 1.89 × 106 to 5.8 × 105 CFU g−1 rhizosphere sand and that of Ss-2 decreasing slightly to 3.4 × 105 CFU g−1 rhizosphere sand (Fig. 3). Ultrastructure studies also confirmed the colonization potential of both strains which were not only localized in the sunflower rhizosphere but also found inside root cortical cells (Fig. 4).
Mean population density of phosphate-solubilizing strains Ps-5 (Bacillus sp.) and Ss-2 (Alcaligenes faecalis) recovered from the sunflower (Helianthus annuus L.) rhizosphere at different time-points after transplantation. The slight decrease in the population density of both strains with increasing growth time was non-significant (P ≤ 0.05)
Effect of inoculation on sunflower growth and yield
In the pot experiment, inoculation of sunflower plants with Bacillus sp. Ps-5 significantly (P ≤ 0.05) enhanced sunflower root and shoot length, fresh and dry weight and P contents. The response of sunflower plants to inoculation with A. faecalis Ss-2 was significantly increased (P ≤ 0.05) in terms of shoot length, root and shoot fresh weight, root dry weight and plant P contents, as compared to uninoculated plants (Table 2). The soil of the field experiment was sandy loam which was slightly alkaline (pH 7.9) and low in organic matter content (0.68 %). Total nitrogen and available P of the soil was estimated at 0.55 g kg−1 and 4.6 mg kg−1, respectively. Under field conditions, the maximum growth, yield and physiological parameters of uninoculated sunflower plants receiving a full dose of the recommended fertilizer were measured as: plant height of 139 cm, a stem diameter of 3.25 cm, a head diameter of 38.33 cm, an achene yield of 2,902 kg ha−1, a biological yield of 9,792 kg ha−1, achene P contents of 14.71 g kg−1 and achene oil contents of 40.50 %. Inoculation with Bacillus sp. Ps-5 and A. faecalis Ss-2 significantly (P ≤ 0.05) increased sunflower plant height and P contents as compared to uninoculated plants receiving a half dose of recommended fertilizer. In contrast, the inoculation effect on yield and its attributed traits was found to be non-significant in comparison with both uninoculated treatments with a half and full dose of fertilizer, respectively (Table 3).
Discussion
In this study we purified and characterized 19 morphologically different bacterial isolates on the basis of their prolific growth. Gram-negative isolates dominated the overall collection, which is in accordance with well-documented reports of an abundance of Gram-negative bacteria in the soil and rhizosphere in general (Curl and Truelove 1986) and of the dominance of Gram-negative isolates (296/299) isolated from the sunflower rhizosphere (Ambrosini et al. 2012). The motility of bacteria is also considered to be an essential factor for successful root colonization (Bashan and Holguin 1994; Sakai et al. 1996). Although Broek and Vanderleyden (1995) and de Weert et al. (2002) suggested that non-motile bacteria also colonize plant roots as efficiently as motile bacteria, we found that all of the sunflower rhizo-types were motile, possibly due to motile bacteria having better access to root exudates in the rhizosphere area through chemotaxis. Of the 19 isolates tested, six (32 %) produced a halo-zone on the tricalcium phosphate-supplemented medium. The total inorganic phosphate solubilization range recorded in our study (11.29 ± 3.36 to 28.91 ± 2.56 μg mL−1) is in agreement with the findings of Oliveira et al. (2009) who determined the soluble phosphate range to be about 4–200 μg mL−1 in phosphate-solubilizing isolates from the maize rhizosphere. In terms of phytohormone production by PSB, IAA plays a major role in root growth and triggers cell division and growth (Vassilev et al. 2006; Seo and Park 2009). We detected bacterial in vitro IAA synthesis in the range of 0.06 ± 0.04 to 10.65 ± 2.08 μg mL−1, with the potential phosphate-solubilizing isolates Ps-5 and Ss-2 producing the highest amounts of the isolates tested. Many other PSB have also been reported to synthesize IAA and other phytohormones (Trivedi et al. 2011; Oliveira-Longatti et al. 2014). Thus, the substantial IAA production capacity combined with a good phosphate solubilization potential led us to screen isolates Ps-5 and Ss-2 for further studies. The hydroxyl and carboxyl groups of organic acids harbor secondary metabolites which chelate the cations bound to phosphates, transforming the latter to a soluble form (Kim et al. 1997; Sagoe et al. 1998). We found not only a significant relationship between phosphate solubilization and organic acid production in the cultures of isolates Ps-5 and Ss-2, but also a positive correlation between organic acid production and phosphate solubilization. There was an increasing trend in phosphate solubilization and a decreasing trend in pH in the in vitro culture experiments up to 240 h of incubation (Fig. 1a, b). Our findings are validated by similar results found in a number of earlier studies (Chen et al. 2006; Ma et al. 2009). A positive correlation between phosphate solubilization and organic acid production also validated the hypothesis that the inorganic phosphate solubilization ability of sunflower rhizosphere isolates was due to the production of organic acids. Both organic acid production and phosphate solubilization was directly proportional to the duration of the incubation, with a trend to increase during the passage of time. The increasing trend of phosphate solubilization with time has also been described earlier by Chen et al. (2006) and Ma et al. (2009). Studies by Trivedi and Sa (2008) and Park et al. (2010) revealed that the phosphate solubilization ability in wild-type and mutants of Pseudomonas corrugata (NRRL B-30409) and Burkholderia vietnamiensis, respectively, was related to the production of gluconic acid and 2-ketogluconic acids and ultimately with a decrease in the pH of the medium. A similar relationship between P solubilization and pH was found by Neijssel et al. (1983), Švitel and Šturdik (1995) and Hwangbo et al. (2003). Other organic acids, such as lactic acid, oxalic acid, tartaric acid, formic acid, malic acid, citric acid, succinic acid and propionic acid, have also been reported to be produced by bacteria in vitro (Chen et al. 2006; He and Sheng 2006; Gulati et al. 2010; Zhu et al. 2011). Some of these organic acids, especially 2-ketogluconic acid, citric acid, malic acid and oxalic acid, have been detected in the rhizosphere of various crops and vegetables (Jaeger III et al. 1999).
Stefanowicz (2006) developed and introduced Biolog Phenotype Microarray technique with microplates to determine the metabolic potential of microbial communities. As both of the screened isolates were isolated from soil samples collected at different locations, they utilized different numbers and amounts of the various substrates on the microtiter plates. This diversity in carbon source utilization pattern was also noted after the PCA (Fig. 2). Garland and Mills (1991), based on the results of a detailed Biolog study, reported that aquatic and soil bacteria, including those that associate with the rhizosphere, demonstrated a great diversity in their ability to metabolize carbon sources. The authors of a recent study conducted with fluorescent pseudomonads suggested that no specific group of substrates (carbon sources) in the Biolog microplate assay is responsible for the colonization ability of bacteria, rather that many substrates or substrate groups may lead to successful colonization (Oksinska et al. 2011). Based on our results, we also concluded that the screened isolates utilized many different substrate groups, indicating their diverse metabolic nature which might, possibly, contribute to their colonization potential.
16S rRNA gene amplification and sequencing was used to identify isolates Ps-5 and Ss-2 as this fragment is considered to be the most authentic taxonomic marker for bacterial identification at the genus and species level. Bacterial isolate Ps-5 was identified as Bacillus sp. (99 % sequence similarity with more than one Bacillus spp.; hence, it could not be identified to the species level), and Ss-2 was identified as Alcaligenes faecalis (99 % sequence identity with various strains of this species). Various strains of the genus Bacillus and Alcaligenes have been reported in the rhizosphere of various crops (Glick 1995; Joseph et al. 2007; Kaymak 2011) and have been isolated from sunflower roots and studied for their plant growth-promoting characteristics (Andreoli et al. 1993; Forchetti et al. 2007; Roberts et al. 2011; Ambrosini et al. 2012). Many Bacillus spp. and an Alcaligenes sp. SF2 strain have also been isolated as endophytes in sunflower (Forchetti et al. 2007). To the best of our knowledge, this is the first report of the isolation of an A. faecalis strain from the sunflower rhizosphere which has mineral phosphate-solubilizing ability.
In the inoculation experiment, the Bacillus sp. Ps-5 and A. faecalis Ss-2 strains had colonized the sunflower rhizosphere at a population density of 105–106 CFU g−1 of rhizosphere sand at 30 days post-transplantation (Fig. 3). These findings were consistent with those of a study conducted by Andreote et al. (2009) who successfully recovered the rifampicin resistant derivative of Pseudomonas putida strain P9R from the rhizoplane and endosphere of potato plants. Similar results were also described by Zinniel et al. (2002) and Shankar et al. (2011). The root colonization potential of the Bacillus sp. Ps-5 and A. faecalis Ss-2 strains was also determined by TEM, which is highly sensitive and reliable technique for ultra-structural root colonization studies. Both inoculated strains were successfully localized in the root surface area and even inside the root cortical cells, thereby demonstrating their endophytic nature (Fig. 4). These ultra-structural studies further validated that strains Ps-5 and Ss-2 were sunflower root-colonizing PGPR, which is of particular significance as PGPR strains can only stimulate plant growth when they have an optimum colonization potential. A number of earlier studies carried out with TEM and immunogold labeling techniques also revealed that PGPR strains can be localized in the rhizosphere, root cortical cells and nodules of various crops (Schloter et al. 1997; Hameed et al. 2005; Jeun et al. 2008; Yasmeen et al. 2012).
A significant (P ≤ 0.05) increase in sunflower growth parameters, including plant P contents, in inoculated plants after inoculation with Bacillus sp. Ps-5 and A. faecalis Ss-2 may possibly be attributed to the P-solubilizing, IAA-synthesizing and root-colonizing abilities of these strains (Table 2). As tricalcium phosphate was added as an insoluble P source in the pot experiment, the increase in sunflower P contents may be strongly related to tricalcium phosphate solubilization. In addition, the non-significant effect of strain Ss-2 on root fresh and dry weight may be due to its relatively lower ability to produce IAA. The inorganic phosphate solubilization and IAA production ability of PGPR are known to improve plant growth by increasing P-uptake from soil and its transport to plant shoots (Igual et al. 2001; Chen et al. 2006; Shirmardi et al. 2010). The promotion of root colonization and plant growth following inoculation with PGPR strains has also been described earlier (Andreote et al. 2009; Shankar et al. 2011).
Highly variable results were found under field conditions. A significant (P ≤ 0.05) increase in plant height and sunflower achene P contents were measured in inoculated plants treated with a half dose of the recommended fertilizer, while the effect on achene yield and oil contents was found to be non-significant when compared to uninoculated plants treated with half a dose of recommended fertilizer (Table 3). One possible explanation might be that the sterilized sand was used in the pot experiment and consequently there was no competition between inoculated bacteria and the indigenous micro-flora. However, in natural soil conditions, these selected strains were unable to compete with the indigenous micro-flora and could not adequately colonize sunflower roots. Soil is considered to be a highly unpredictable and heterogeneous medium, and it is very difficult to achieve the anticipated results in experiments using soil (Bashan 1998; Lucy et al. 2004). For comparison, we also included uninoculated treatment with full dose of fertilizer and recorded higher yield and yield-attributed traits in these uninoculated plants than inoculated plants with half fertilizer dose. Although inoculation alone was unable to compete with treatments of half and full doses of fertilizer, significantly higher achene P contents were observed in inoculated plants receiving a half dose of fertilizer than in uninoculated plants receiving a half dose of fertilizer, possibly due to P solubilization by the inoculated strains.
Conclusion
Based on these results, we conclude that Bacillus sp. Ps-5 and Alcaligenes faecalis Ss-2 are potential phosphate-solubilizing and plant-growth promoting strains which have the capacity to supplement the P requirements of sunflower. Both strains should be tested further under greenhouse and field conditions before applying them as commercial inoculants for improving sunflower crops.
References
Ambrosini A, Beneduzi A, Stefanski T, Pinheiro FG, Vargas LK, Passaglia LMP (2012) Screening of plant growth promoting rhizobacteria isolated from sunflower (Helianthus annuus L.). Plant Soil 356:245–264
Andreoli YE, Laich FS, Navarro CA (1993) In vitro control of Sclerotinia sclerotiorum and Gaeumannomyces graminis by bacteria of the fluorescent Pseudomonas group. Rev Argent Microbiol 25:70–79
Andreote FD, De Araújo WL, De Azevedo JL, Van Elsas JD, Da Rocha UN, Van Overbeek LS (2009) Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl Environ Microbiol 75:3396–3406
Arcand MM, Schneider KD (2006) Plant-and microbial-based mechanisms to improve the agronomic effectiveness of phosphate rock: a review. An Acad Bras Cienc 78:791–807
Arnon DI, Hoagland DR (1940) Crop production in artificial culture solution, sand and soil with special reference to factors influencing yield and absorption of inorganic nutrient. Soil Sci 50:463–483
Bashan Y (1998) Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol Adv 16:729–770
Bashan Y, Holguin G (1994) Root-to-root travel of the beneficial bacterium Azospirillum brasilense. Appl Environ Microbiol 60:2120–2131
Broek AV, Vanderleyden J (1995) The role of bacterial motility, chemotaxis and attachment in bacteria–plant interactions. Mol Plant-Microbe Interact 8:800–810
Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 8:e55731
Chen YP, Rekha PD, Arun AB, Shen FT, Lai WA, Young CC (2006) Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol 34:33–41
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959
Curl EA, Truelove B (1986) The rhizosphere. Springer, London
de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lungtenberg BJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant-Microbe Interact 15:1173–1180
Duca D, Lorv J, Patten CL, Rose D, Glick BR (2014) Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek 106:85–125
Ekin Z (2010) Performance of phosphate solubilizing bacteria for improving growth and yield of sunflower (Helianthus annuus L.) in the presence of phosphorus fertilizer. Afr J Biotechnol 9:3794–3800
Erickson DR, Pryde EH, Brekke OL, Mounts TL, Falb RA (1980) Handbook of soy oil processing and utilization. American Soybean Association/American Oil Chemists Society, St. Louis/Champaign
Fankem H, Ngo Nkot L, Deubel A, Quinn J, Merbach W, Etoa FX, Nwaga D (2008) Solubilization of inorganic phosphates and plant growth promotion by strains of Pseudomonas fluorescens isolated from acidic soils of Cameroon. Afr J Microbiol Res 2:171–178
Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl Microbiol Biotechnol 76:1145–1152
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117
Gulati A, Sharma N, Vyas P, Sood S, Rahi P, Pathania V, Prasad R (2010) Organic acid production and plant growth promotion as a function of phosphate solubilization by Acinetobacter rhizosphaerae strain BIHB 723 isolated from the cold deserts of the trans-Himalayas. Arch Microbiol 192:975–983
Hameed S, Mubeen F, Malik K, Hafeez F (2005) Nodule Co-occupancy of Agrobacterium and Bradyrhizobium with potential benefit to legume host. In: Wang YP (ed) Biological nitrogen fixation. Sustainable agriculture and the environment. Springer SBM, Dordrecht, pp 295–296
Hameeda B, Reddy YHK, Rupela O, Kumar G, Reddy G (2006) Effect of carbon substrates on rock phosphate solubilization by bacteria from composts and macrofauna. Curr Microbiol 53:298–302
He LY, Sheng XF (2006) Solubilization of potassium-bearing minerals by a wild-type strain of Bacillus edaphicus and its mutants and increased potassium uptake by wheat. Can J Microbiol 52:66–72
Hilda R, Fraga R (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17:319–359
Hwangbo H, Park RD, Kim YW, Rim YS, Park KH, Kim TH, Suh JS, Kim KY (2003) 2-Ketogluconic acid production and phosphate solubilization by Enterobacter intermedium. Curr Microbiol 47:87–92
Igual JM, Valverde A, Cervantes E, Velazquez E (2001) Phosphate-solubilizing bacteria as inoculants for agriculture: use of updated molecular techniques in their study. Agronomie 21:561–568
Jaeger CH III, Lindow SE, Miller W, Clark E, Firestone MK (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 65:2685–2690
Jeun YC, Lee YJ, Kim KW, Kim SJ, Lee SW (2008) Ultrastructures of Colletotrichum orbiculare in the leaves of cucumber plants expressing Induced systemic resistance mediated by glomus intraradices BEG110. Mycobiology 36:236–241
Joseph B, Patra RR, Lawrence R (2007) Characterization of plant growth promoting rhizobacteria associated with chickpea (Cicer arietinum L.). Int J Plant Prod 2:141–152
Kaymak HC (2011) Potential of PGPR in agricultural innovations. In: Maheshwari DK (ed) Plant growth and health promoting bacteria. Springer, Berlin, pp 45–79
Khan A, Iqbal M, Ahmad I, Iqbal N, Hussain M (2000) Effect of different water stress levels on yield and oil content of sunflower (Helianthus annuus L.) cultivators. Pak J Biol Sci 3:1632–1633
Kim KY, McDonald GA, Jordan D (1997) Solubilization of hydroxypatite by Enterobacter agglomerans and cloned Escherichia coli in culture medium. Biol Fertil Soils 24:347–352
Kloepper JW, Lifshitz R, Zablotowicz RM (1989) Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol 7:39–44
Lucy M, Reed E, Glick BR (2004) Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek 86:1–25
Ma Y, Rajkumar M, Freitas H (2009) Inoculation of plant growth promoting bacterium Achromobacter xylosoxidans strain Ax10 for the improvement of copper phytoextraction by Brassica juncea. J Environ Manag 90:831–837
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York
Mullen MD (2005) Phosphorus in soils: biological interactions. In: Hillel D (ed) Encyclopedia of soils in the environment. Elsevier, Oxford, pp 210–215
Murphy J, Riley JP (1962) Modified solution method for determination of phosphate in natural water. Anal Chim Acta 27:31–36
Neijssel OM, Tempest DW, Postma PW, Duine JA, Jzn JF (1983) Glucose metabolism by K+-limited Klebsiella aerogenes: Evidence for the involvement of a quinoprotein glucose dehydrogenase. FEMS Microbiol Lett 20:35–39
Oksinska MP, Wright SAI, Pietr SJ (2011) Colonization of wheat seedlings (Triticum aestivum L.) by strains of Pseudomonas spp. with respect to their nutrient utilization profiles. Eur J Soil Biol 47:364–373
Oliveira CA, Alves VMC, Marriel IE, Gomes EA, Scotti MR, Carneiro NP, Guimaraes CT, Schaffert RE, Sa NMH (2009) Phosphate solubilizing microorganisms isolated from rhizosphere of maize cultivated in an oxisol of the Brazilian Cerrado Biome. Soil Biol Biochem 41:1782–1787
Oliveira-Longatti SM, Mara LM, Soares BL, Bomfeti CA, Silva K, Ferreira PAA, Moreira FMS (2014) Bacteria isolated from soils of the western Amazon and from rehabilitated bauxite-mining areas have potential as plant growth promoters. World J Microbiol Biotechnol 30:1239–1250
Park KH, Lee QM, Jung HI, Jeong JH, Jeon YD, Hwang DY, Lee CY, Son HJ (2010) Rapid solubilization of insoluble phosphate by a novel environmental stress-tolerant Burkholderia vietnamiensis M6 isolated from ginseng rhizospheric soil. Appl Microbiol Biotechnol 86:947–955
Pikovskaya RI (1948) Metabolism of phosphorous in soil in connection with vital activity of some microbial species. Microbiologia 17:362–370
Roberts DP, Lohrke SM, McKenna L, Lakshman DK, Kong H, Lydon J (2011) Mutation of a degS homologue in Enterobacter cloacae decreases colonization and biological control of damping-off on cucumber. Phytopathology 101:271–280
Sagoe CI, Ando T, Kouno K, Nagaoka T (1998) Relative importance of protons and solution calcium concentration in phosphate rock dissolution by organic acids. Soil Sci Plant Nutr 44:617–625
Sakai M, Ozawa H, Futamata H, Matsuguchi T (1996) Effect of calcium ion on spinach root colonization by fluorescent pseudomonads through chemotaxis. Soil Sci Plant Nutr 42:323–331
Salema R, Brandao I (1973) The use of PIPES buffer in the fixation of plant cells, for electron microscopy. J Submicrosc Cytol 5:79–96
Schloter M, Wiehe W, Assmus B, Steindl H, Becke H, Höflich G, Hartmann A (1997) Root colonization of different plants by plant-growth-promoting Rhizobium leguminosarum bv. trifolii R39 studied with monospecific polyclonal antisera. Appl Environ Microbiol 63:2038–2046
Seldin L, Rosado AS, Da Cruz DW, Nobrega A, Van Elsas JD, Paiva E (1998) Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl Environ Microbiol 64:3860–3868
Seo PJ, Park CM (2009) Auxin homeostasis during lateral root development under drought condition. Plant Signal Behav 4:1002–1004
Shah NA, Aujla KM, Ishaq M, Farooq A (2013) Trends in sunflower production and its potential in increasing domestic edible oil production in Punjab, Pakistan. Sarhad J Agric 29:7–13
Shahid M, Hameed S, Imran A, Ali S, van Elsas JD (2012) Root colonization and growth promotion of sunflower (Helianthus annuus L.) by phosphate solubilizing Enterobacter sp. Fs-11. World J Microbiol Biotechnol 28:2749–2758
Shankar M, Ponraj P, Ilakkiam D, Gunasekaran P (2011) Root colonization of a rice growth promoting strain of Enterobacter cloacae. J Basic Microbiol 51:523–530
Sharan A, Darmwal NS, Gaur R (2008) Xanthomonas campestris, a novel stress tolerant, phosphate-solubilizing bacterial strain from saline–alkali soils. World J Microbiol Biotechnol 24:753–759
Shirmardi M, Savaghebi GR, Khavazi K, Akbarzedeh A, Farahbakhsh M, Rejali F, Sadat A (2010) Effect of microbial inoculants on uptake of nutrient elements in two cultivars of sunflower (Helianthus annuus L.) in saline soils. Not Sci Biol 2:57–66
Somasegaran P, Hoben HJ (1994) Handbook for rhizobia: methods in legume–rhizobium technology. Springer, Berlín
Steel RGD, Torrie JH, Dickey DA (1997) Principles and procedures of statistics, A biometrical approach. 3rd edn. McGraw Hill Book, New York
Stefanowicz A (2006) The Biolog plates technique as a tool in ecological studies of microbial communities. Pol J Environ Stud 15:669–676
Švitel J, Šturdik E (1995) 2-Ketogluconic acid production by Acetobacter pasteurianus. Appl Biochem Biotechnol 53:53–63
Sylvia DM, Fuhrmann JJ, Hartel P, Zuberer DA (2005) Principles and applications of soil microbiology. Pearson Prentice Hall, Upper Saddle River
Tien TM, Gaskins MH, Hubbell DH (1979) Plant growth substances produced by Azospirillum brazilense and their effect on the growth of pearl millet. Appl Environ Microbiol 37:1016–1024
Trivedi P, Sa T (2008) Pseudomonas corrugata (NRRL B-30409) mutants increased phosphate solubilization, organic acid production, and plant growth at lower temperatures. Curr Microbiol 56:140–144
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb Ecol 62:324–336
Vassilev N, Vassileva M, Nikolaeva I (2006) Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl Microbiol Biotechnol 71:137–144
Vincent JM (1970) A manual for the practical study of the root nodule bacteria. Burgess and Son Ltd., Leicester
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Whitelaw MA (1999) Growth promotion of plants inoculated with phosphate-solubilizing fungi. Adv Agron 69:99–151
Yasmeen T, Hameed S, Tariq M, Iqbal J (2012) Vigna radiata root associated mycorrhizae and their helping bacteria for improving crop productivity. Pak J Bot 44:87–94
Yoshida S, Forno DA, Cock JH (1971) Laboratory manual for physiological studies of rice. IRRI, Los Banos
Zaidi A, Khan M, Ahemad M, Oves M (2009) Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol Immunol Hung 56:263–284
Zhu F, Qu L, Hong X, Sun X (2011) Isolation and characterization of a phosphate-solubilizing halophilic bacterium Kushneria sp. YCWA18 from Daqiao Saltern on the coast of Yellow Sea of China. Evid Based Complement Alternat Med 2011:1–6
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru A, Arunakumari A, Barletta RG, Vidaver AK (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Acknowledgements
The authors are indebted to the Higher Education Commission (HEC) of Pakistan which provided the financial support for carrying out this research. We also thank Mr. Javed Iqbal for his technical help in the electron microscopy studies and Dr. Iftikhar Ali for his help in conducting the achene oil content analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shahid, M., Hameed, S., Tariq, M. et al. Characterization of mineral phosphate-solubilizing bacteria for enhanced sunflower growth and yield-attributing traits. Ann Microbiol 65, 1525–1536 (2015). https://doi.org/10.1007/s13213-014-0991-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0991-z