Abstract
Two high-copy number vectors, pL97 and pL98, that contain the strong constitutive ermEp* from Saccharopolyspora erythraea and the ssrA promoter (ssrAp) from Streptomyces coelicolor, respectively, and an inducible high-copy vector, pL99, with the PnitA-NitR system from Rhodococcus rhodochrous, were constructed based on the pIJ101 replicon. These vectors have pUC18 and pIJ101 replication origins for high-copy plasmid number in Escherichia coli and Streptomyces, respectively, and the oriT (RK2) allows the efficient and convenient plasmid transfer from E. coli to Streptomyces. The transformants can be easily selected with apramycin. There is also a multiple cloning site (MCS) between promoter and terminator convenient for heterologous gene cloning. The enhanced green fluorescent protein (EGFP) gene and redD, a pathway-specific regulatory gene for the production of undecylprodigiosin in S. coelicolor, were inserted into these plasmids and could be over-expressed in S. coelicolor. Western blotting revealed that the expression of EGFP was dramatically increased compared to the cell with a single copy of EGFP. Furthermore, the production of undecylprodigiosin was also greatly enhanced in the cells constitutively over-expressing redD. Hence, the high-copy plasmids could be readily used to express foreign proteins and for production of secondary metabolites, especially the antibiotics, potentially for industrial applications.
Similar content being viewed by others
Introduction
Streptomyces are Gram-positive, soil-dwelling microorganisms, which can produce many kinds of antibiotics, enzymes, immuno-suppressives, etc., with commercial and academic value. Moreover, Streptomyces have also been developed as the hosts for the expression of heterogonous proteins, which might aggregate in the inclusion bodies of E. coli.
In order to maximize the productivity of Streptomyces, an easy and routine strategy is to over-express some pivotal genes in multiple-copy plasmids under strong promoters (Ann and van Mellaert 1993). pIJ101, a wide host range and multiple-copy plasmid from Streptomyces lividans ISP5434, can reach up to 100–200 copies per chromosome or 30% of the total DNA of a cell (Kieser et al. 1982, 2000). Several pIJ101 replicon-based over-expression vectors have previously been constructed, such as pIJ6021, pIJ4123 (Takano et al. 1995), and pHZ1272 (Yang et al. 1998), which all contain thiostrepton-inducible tipAp but do not have oriT. Recently, pTONA5 was reported as another E. coli–Streptomyces shuttle plasmid with a metalloendopeptidase promoter for hyperexpression of foreign proteins (Hatanaka et al. 2008).
ermEp*, a mutant promoter originating from erythromycin resistant gene of Saccharopolyspora erythraea, is a strong and constitutive promoter widely used in Streptomyces (Bibb et al. 1985; Janssen et al. 1989; Schmitt-John and Engels 1992). The ssrA gene, encoding the transfer-messenger RNA (tmRNA) for protein turn-over (Withey and Friedman 2003; Yang and Glover 2009), is also strongly expressed in Streptomyces, though a condition-dependent expression profile was observed (Paleckova et al. 2006, 2007; Yang and Glover 2009). Though this is a convenient method, there is the disadvantage that constitutively expressed proteins or secondary metabolites might be nocuous to cell growth, and have a severe effect on the production of the favored products. Accordingly, the introduction of an inducible expression system may be a preferable way. PnitA-NitR system has been demonstrated to be a powerful inducible expression system used in mycobacteria based on the positive feedback regulation of nitAp activity (Pandey et al. 2009). The expression of nitA, encoding nitrilase in Rhodococcus rhodochrous J1, is regulated by a positive regulator nitR. It has been reported that NitR coupling with inducer (ε-caprolactam) could bind to the specific site of the nitA promoter region to activate the expression of nitA (Komeda et al. 1996a, b).
In this paper, we present three multiple-copy plasmids in Streptomyces based on pIJ8668 backbone and pIJ101 replicon, two with strong and constitutive promoters and one with inducible system. Moreover, the egfp and redD were highly expressed in S. coelicolor to demonstrate the feasibility of these three over-expression vectors.
Materials and methods
Bacterial strains, plasmids and media
Escherichia coli TG1 was used as the host for plasmid construction and E. coli ET12567 (MacNeil et al. 1992) with the helper plasmid pUZ8002 was used for plasmid introduction into S. coelicolor by conjugation according to procedures previously described (Kieser et al. 2000). Escherichia coli strains were cultured in liquid or solid Luria–Bertani (LB) medium containing appropriate antibiotics at 37°C for plasmid propagation (Sambrook et al. 2000). Streptomyces coelicolor strains in this study are listed in Table 1. Wild-type strain M145 and its derivatives were maintained at 30°C on MS or R5 medium or in yeast extract–malt extract (YEME) liquid medium according to the manual (Kieser et al. 2000).
Plasmid construction
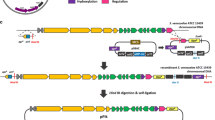
All primers and plasmids used in this study are listed in Tables 2 and 3, respectively. Promoter ermEp* was amplified by PCR using vector pLM1 (Mao et al. 2009a) as a template with primers 1 and 2, and then subcloned into pTA2 (Toyobo). Then, the ermEp* (0.2 kb) was obtained by BamHI digestion and ligated into the BglII site of pIJ8668 (5.0 kb) (Sun et al. 1999) resulting in the 5.2 kb of pIJ8668-ermEp. Subsequently, two synthetic oligonucleotides (primer 7 and 8) containing the multiple cloning sites (MCS) were heat denatured in buffer A (50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA) for 5 min and cooled down slowly to room temperature for renaturation. Then, the annealed double helix replaced the EGFP gene in pIJ8668-ermEp after digestion with NdeI-NotI to generate pIJ8668-ermEp-MCS. In parallel, both pIJ8600 (Sun et al. 1999) and pHZ1272 (Yang et al. 1998) were digested with XbaI-ClaI to get 6.3 kb of vector from pIJ8600 and 3.2 kb of pIJ101 replicon from pHZ1272, respectively, which were ligated for plasmid pIJ8600-101 ori (9.5 kb). Then, the resultant vector was digested with BamHI-BglII and self-ligated to create pIJ8600-101 ori-B/B. Finally, the 4.0-kb EcoRI-HindIII pIJ101 replicon-tfd from pIJ8600-101 ori-B/B was inserted into EcoRI-HindIII site of pIJ8668-ernEp-MCS to generate plasmid pL97. The construction of pL98 (pIJ8668-101 ori-ssrAp-MCS) is the same as described above except that S. coelicolor M145 genomic DNA served as a template for ssrAp amplification with primers 3 and 4.
The synthetic PnitA-NitR system (Shanghai Qinglan Biotech) cloned in pUC19 was digested with SphI-XbaI and then inserted into SphI-XbaI site of pIJ8600-101 ori to create pL99 (pIJ8600-101 ori-nit). The sequence of PnitA-nitR system was synthesized according to pNIT-1 (Genbank accession number FJ173069) (Pandey et al. 2009) with codon optimization for nitR according to Streptomyces codon usage (Kieser et al. 2000). The optimized sequence has been deposited in GenBank (accession number JN636796).
The KpnI-EcoRI egfp gene fragment from pIJ8668 was inserted into the KpnI-EcoRI site of pTA2 (Toyobo) generating pTA2-egfp. Then, the NdeI–BamHI egfp from pTA2-egfp was inserted into the three high-copy plasmids digested with NdeI-BglII, generating pL100, pL101, and pL102, respectively.
redD was amplified using primers 5 and 6, and cloned into pTA2 (Toyobo) after addition of dA. Then, the redD fragment digested by NdeI-BglII from pTA2-redD was inserted into pL97 and pL98 to generate pL103 and pL104, respectively.
SDS-PAGE and western blot analysis
Streptomyces coelicolor mycelia were collected after inoculation of spores in YEME liquid medium for the indicated time and resuspended in lysis buffer as described previously (Mao et al. 2009b). About 20 μg of total protein were separated in 12% SDS-PAGE followed by western blot with rabbit polyclonal anti-EGFP antibody (Proteintech, USA).
Antibiotic (undecylprodigiosin) production assay
For phenotype analysis, about 1010 spores were streaked on the R5 plate, incubated at 30°C for 30 h, and photographed at the bottom. Quantitative analysis of Red production has been previously described (Kieser et al. 2000; Mao et al. 2009b). Briefly, about 10 mg of mycelia collected from R5 medium overlaid with cellophanes at different developmental stages was extracted with acidified methanol overnight before vacuum freeze-drying, and the A530 of the supernatants was determined after centrifugation at 5,000 g for 5 min.
Results and discussion
Construction of novel multiple-copy vectors pL97, pL98 and pL99
Fragments harboring three different promoters, transcriptional terminators, selection markers, and the replicons for plasmid propagation were combined for a series of novel high-expression vectors (Figs. 1 and 2). The pIJ101 replicon required for the high copy plasmid number maintenance in S. lividans ISP5434 served as a replication element in Streptomyces (Kieser et al. 1982) and pUC18 ori was the replication origin in E. coli. The three plasmids also contained oriT origin for inter-species DNA transfer, which allowed convenient mobilization of shuttle–plasmid from E. coli to Streptomyces. The difference between the three vectors was mainly based on the strong promoters, among which ermEp* and ssrAp were constitutive while nitAp in PnitA-NitR system was inducible with the inducer ε-caprolactam. Additionally, the multiple cloning site (MCS) flanked by the strong promoter and transcriptional terminators tfd, which represented a major transcription terminator of bacteriophage fd, also allowed the convenient cloning of foreign genes.
Over-expression of EGFP in S. coelicolor
To evaluate the applicability of these vectors, the endogenous egfp gene from pIJ8668 (Sun et al. 1999) was cloned in these plasmids and expressed in S. coelicolor. The cell-free extract analysis by western blot revealed that the EGFP protein was expressed much more abundant in high-copy plasmid under ermEp* in comparison with the integrative single-copy plasmid (Fig. 3, lane 2 and 3). However, though much higher than that in the single copy plasmid (Fig. 3, lane 2 and 4), EGFP production under ssrAp was found to be slightly lower than that under ermEp* (Fig. 3, lane 3 and 4), suggesting that, though ssrAp was comparably a strong promoter, the foreign gene was transcribed more efficiently under ermEp*. Moreover, dramatically increased production of EGFP protein was observed when the inducer (ε-caprolactam) was added in the PnitA-NitR inducible system in multiple-copy plasmid, and 0.1% ε-caprolactam was sufficient for the full induction of PnitA-NitR system after 1 day (Fig. 3, lane 7–9). In addition, the PnitA-NitR system presented the highest expression level among the three promoters (Fig. 3, lane 3 and 4 and 8 and 9). However, the basal leaky expression was also observed without inducer (Fig. 3, lane 7), which might be caused by transcriptional read-through resulting from the inefficient synthetic short terminator.
Over-expression of EGFP. Strain L48, L49, and L50 were cultured in a 250-ml baffled flask containing 50 ml of YEME medium supplemented with 50 μg/ml apramycin and shaked at 210 rpm at 30°C to exponential phase. As for the PnitA-NitR inducible system, ε-caprolactam was added to the medium (final concentrations 0.1 and 1%) when cells grew to pre-exponential phase, and then further cultivation was carried out at 30°C for 24 h. Streptomyces coelicolor wide-type strain M145 without plasmid or with integrated plasmid pLM1 was cultured at the same condition. The cell extract was prepared and subjected to western blot with anti-EGFP antibody
Over-production of undecylprodigiosin by over-expression of redD in S. coelicolor
Streptomyces are the major producer of secondary metabolites including antibiotics and immunosuppressives, etc., especially for clinical applications. Then, the antibiotic undecylprodigiosin (Red) production from S. coelicolor was tested as a model for the production capacity of secondary metabolites after the positive regulator was over-expressed in these high-copy plasmids.
redD, the gene encoding the pathway specific transcription activator for Red production in S. coelicolor (Takano et al. 1992), was over-expressed in the multiple-copy vectors under ermEp* and ssrAp in strains L51 and L52, respectively, and L49 with ssrAp-egfp served as a control (Fig. 4). It was observed that after 30 h on the R5 plate, the lawn of cells over-expressing redD was much redder than the negative control, which only had the endogenously expressed redD (Fig. 4a). Consistent with the phenotypical result, quantitative measurement also showed that over-expression of redD in our constitutive multiple-copy plasmids could remarkably enhance the Red production at all developmental stages. Especially after 72 h, both strain L51 (redD under ermEp*) and L52 (redD under ssrAp) showed about 40 times higher than the control strain L49 (egfp under ssrAp) (Fig. 4b).
Undecylprodigiosin production assay. a L49, L51, and L52 were streaked on R5 medium, incubated at 30°C for 30 h and photographed from the bottom of the plate. b Quantitative measurement of Red production. The ratios of absorbance at A530 to mycelium dry weight of strains L51 (●), L52 (■), and L49 (▲) were calculated, respectively, and numbers in the graphs are the mean values of three independent experiments; bars standard deviations
One of the most effective strategies to improve the production of a particular protein or secondary metabolite in Streptomyces is to over-express the gene, the synthesis gene cluster, or the positive regulatory gene for this gene/gene cluster. In this paper, we constructed three high-copy expression vectors derived from pIJ8668 and pIJ101 to enlarge options for the choice of expression plasmids. The plasmids in our report were all E. coli–Streptomyces shuttle vectors, and the oriT in these vectors made the plasmid introduction into Streptomyces via conjugation very efficient and convenient, which was quite different from the classical pIJ101-derived vectors pIJ6021 and pIJ4123 (Takano et al. 1995). Moreover, the strong constitutive promoters ermEp* and ssrAp made it possible to permanently highly express foreign genes. However, especially for the expression of toxic compounds that would greatly affect the physiological processes of Streptomyces, the inducible system could be one of the best choices. After reaching the desired cell growth stage, the Streptomyces cells could be induced to express a large amount of toxic components in a comparably short time. The extensively used high-copy plasmids pIJ6021, pIJ4123, and pHZ1272 all contain the strong inducible promoter tipAp, whose activity could be induced by thiostrepton (Takano et al. 1995). However, thiostrepton is expensive, which limited it to the research area, and it could not be applied in the industrial field. In comparison, the inducer ε-caprolactam in PnitA-NitR system is quite cheap, and also the high expression level makes it possible for industrial production.
In this study, we presented two constitutive and one inducible high-expression plasmids based on pIJ101 replicon. These vectors could therefore be used to express heterogeneous genes in a high expression level, for the expression of biologically active foreign proteins that would be in the inclusion bodies of E. coli in a denatured form, and also some positive regulators that control the synthesis of secondary metabolites, such as antibiotics.
References
Ann J, van Mellaert L (1993) Streptomyces lividans as Host for heterologous protein production. FEMS Microbiol Lett 114:121–128
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bibb MJ, Janssen GR, Ward JM (1985) Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38:215–226
Hatanaka T, Onaka H, Arima J, Uraji M, Uesugi Y, Usuki H, Nishimoto Y, Iwabuchi M (2008) pTONA5: a hyperexpression vector in Streptomycetes. Protein Expr Purif 62:244–248
Janssen GR, Ward JM, Bibb MJ (1989) Unusual transcriptional and translational features of the aminoglycoside phosphotransferase gene (aph) from Streptomyces fradiae. Genes Dev 3:415–429
Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics: The John Innes Foundation.
Kieser T, Hopwood DA, Wright HM, Thompson CJ (1982) pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet 185:223–228
Komeda H, Hori Y, Kobayashi M, Shimizu S (1996a) Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Natl Acad Sci USA 93:10572–10577
Komeda H, Kobayashi M, Shimizu S (1996b) A novel gene cluster including the Rhodococcus rhodochrous J1 nhlBA genes encoding a low molecular mass nitrile hydratase (L-NHase) induced by its reaction product. J Biol Chem 271:15796–15802
MacNeil DJ, Occi JL, Gewain KM, MacNeil T, Gibbons PH, Ruby CL, Danis SJ (1992) Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115:119–125
Mao XM, Zhou Z, Cheng LY, Hou XP, Guan WJ, Li YQ (2009a) Involvement of SigT and RstA in the differentiation of Streptomyces coelicolor. FEBS Lett 583:3145–3150
Mao XM, Zhou Z, Hou XP, Guan WJ, Li YQ (2009b) Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J Bacteriol 191:6473–6481
Paleckova P, Bobek J, Felsberg J, Mikulik K (2006) Activity of translation system and abundance of tmRNA during development of Streptomyces aureofaciens producing tetracycline. Folia Microbiol 51:517–524
Paleckova P, Felsberg J, Bobek J, Mikulik K (2007) tmRNA abundance in Streptomyces aureofaciens, S. griseus and S. collinus under stress-inducing conditions. Folia Microbiol 52:463–470
Pandey AK, Raman S, Proff R, Joshi S, Kang CM, Rubin EJ, Husson RN, Sassetti CM (2009) Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb) 89:12–16
Sambrook J, MacCallum P, Russell D (2000) Molecular Cloning: A Laboratory Manual (Third Edition): Cold Spring Harbor Laboratory Press, NY
Schmitt-John T, Engels JW (1992) Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl Microbiol Biotechnol 36:493–498
Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ (1999) Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221–2227
Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ (1992) Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol Microbiol 6:2797–2804
Takano E, White J, Thompson CJ, Bibb MJ (1995) Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133–137
Withey JH, Friedman DI (2003) A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol 57:101–123
Yang C, Glover JR (2009) The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS One 4:e4459
Yang R, Hu Z, Deng Z, Li J (1998) Construction of Escherichia coli-Streptomyces shuttle expression vectors for gene expression in Streptomyces. Chin J Biotechnol 14:1–8
Acknowledgments
We gratefully thank Prof. Lin-Quan Bai in Shanghai Jiaotong University for providing the plasmid pHZ1272, and Prof. Mark J Buttner in John Innes Centre, Norwich, UK, for giving us the plasmids pIJ8600 and pIJ8668. This work was supported by the Science Foundation of Chinese University (No. 2009QNA6006) and the Zhejiang Provincial Natural Science Foundation of China (No. Y2100570) to Xu-Ming Mao.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, N., Wang, ZB., Wu, HP. et al. Construction of over-expression shuttle vectors in Streptomyces . Ann Microbiol 62, 1541–1546 (2012). https://doi.org/10.1007/s13213-011-0408-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-011-0408-1