Abstract
Probiotics are live microorganisms that when administered in adequate amounts, confer health benefits on the host. In this study, 13 strains of Bifidobacterium were isolated from three samples of breast-fed infant feces. The isolates were identified based on conservative gene sequencing and phylogenetic analysis. In vitro tests included survival under simulated gastrointestinal tract conditions, aggregation, hydrophobicity, intestinal epithelial cell adhesion, antimicrobial activity, and antibiotic resistance according to international guidelines for probiotics. The results suggest that B. bifidum, B. adolescentis, and B. breve had high adhesive ability compared with B. longum and B. catenulatum/B. pseudocatenulatum group strains. In particular, B. bifidum IF3-211 has a highest adhesion index (8273 ± 247 and 18,009 ± 1476 adhering bacteria per 100 HT-29 and Caco-2 cells, respectively), far higher than the two reference strains, B. lactis Bb12 and B. longum BBMN68. B. adolescentis IF1-11 showed highest autoaggregation (82.52 ± 0.24 %) and coaggregation (45.59 ± 4.16 %) with L. monocytogenes among isolates. In conclusion, B. bifidum IF3-211 and B. adolescentis IF1-11 showed promising characteristics as probiotic candidates that have good potential for application in food industry.
Similar content being viewed by others
Introduction
Bifidobacteria represent one of the dominant groups of microorganisms colonizing the human intestine, constituting >1 % of the intestinal population in adults and representing up to 90 % of the fecal anaerobic bacteria in breast-fed infants (Mueller et al. 2006; Penders et al. 2006). Formation of the human gut microbiota starts at birth and is established after the first year of life; however, the composition and temporal patterns of the microbial communities vary largely among babies (Palmer et al. 2007). Thus, infant feces is a good source for the collection of wild bifidobacteria. Several selective techniques have been developed for the enumeration and isolation of bifidobacteria (Muñoa and Pares 1988; Beerens 1990; Hartemink et al. 1996; Silvi et al. 1996; Ferraris et al. 2010; Miranda et al. 2014). The United Nations Food and Agriculture Organization and the World Health Organization (FAO/WHO) define probiotics as “live micro-organisms, which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO 2006). Certain members of the genus Bifidobacterium are the most frequently used human probiotics because of their health-promoting properties (Russell et al. 2011), such as improvement of the intestinal microbial balance of the host, lowering the risk of gastrointestinal diseases, assimilation of cholesterol, and immunomodulatory effects (Saavedra et al. 1994; Pereira and Gibson 2002; Fukuda et al. 2011; Fanning et al. 2012).
To have functional effects in the intestine, probiotics have to survive the transit through the gastrointestinal tract (GIT). Thus, it is critical for bacteria to withstand the different challenges found along the GIT, mainly acidic pH and gastric enzymes in the stomach, and bile, pancreatin, and other intestinal enzymes in the small intestine (Sánchez et al. 2013). In addition, probiotics should adhere to the intestinal mucosa and significantly inhibit the adhesion of a variety of enteropathogenic bacteria (Del Re et al. 2000; Ouwehand et al. 2002). Adhesion, hydrophobicity and autoaggregation of bifidobacterial strains and Lactobacillus have been found to be strongly related (Del Re et al. 2000; Pan et al. 2006; Rahman et al. 2008), but with the exception in Lactobacillus (Tuo et al. 2013; García-Cayuela et al. 2014). Hydrophobicity and autoaggregation are based mainly on the proteins, glycoproteins, teichoic, and lipoteichoic acids on the cell-wall surface of bacteria, and secreted factors (Goh and Klaenhammer 2010).
As one of the most widely used probiotic bacteria, bifidobacteria are included in many functional foods and dietary supplements (Candela et al. 2008). However, few bifidobacterial strains have been commercialized due to their high sensitivity to environmental stresses (Scheller and O’Sullivan 2011). The commercial bifidobacteria strains in fermented milk products are mainly Bifidobacterium animalis (Raeisi et al. 2013), which is reported to be one of the most tolerant to environmental stresses (Sánchez et al. 2008), such as the widely used probiotic B. animalis subsp. lactis Bb12 (Garrigues et al. 2010). In contrast, the wild-type strains are generally sensitive to acid, bile salts, and oxygen (Simpson et al. 2005; Andriantsoanirina et al. 2013). In addition, probiotic safety issues such as virulence and transfer of antibiotic resistance need to be addressed (Saarela et al. 2000). The beneficial properties of probiotics and the increased human consumption of these products have augmented efforts to identify potential probiotic strains (Muňoz-Quezada et al. 2013). Selection of probiotics for food products should be based on their safety and technological and functional properties (Prasanna et al. 2014).
The number of formula-fed infants is on the rise in China and other countries (Blanchard et al. 2013; Tang et al. 2014). Harmful microorganisms and maturation of the intestinal immune system have an important influence on the infant's intestinal microbiome, and the number and diversity of bifidobacteria in formula-fed infants are generally low compared to those in breast-fed infants (Harmsen et al. 2000; Roger et al. 2010). Thus, supplementation of formula with probiotics is an important field of research (Braegger et al. 2011). However, most of the current commercial probiotic bifidobacterial strains are from limited species with less functional properties. In this study, 13 strains of Bifidobacterium were isolated from breast-fed infant feces collected from remote rural areas in China and were screened for desirable probiotic traits such as tolerance to simulated gastrointestinal juice, intestinal epithelial cell adhesion, aggregation activity, and antimicrobial activity, as well as antibiotic-resistance profiles. The candidate probiotic strains have potential for use as novel probiotic strains in the dairy industry, for example, incorporated into infant formulas.
Material and methods
Strain isolation and cultivation
Three samples of fresh feces from breast-fed, healthy infants (vaginally delivered) were collected—two from a 4-month-old infant in the rural area of Xinjiang Uygur autonomous region, China, and one from a 4-month-old infant in the rural area of Shandong province, China. The samples were diluted and plated on de Man, Rogosa, and Sharpe (MRS) medium supplemented with 0.05 % (w/v) L-cysteine hydrochloride, 3 g/L lithium chloride, and 5 mL/L propionic acid (MRScPL) (Hartemink et al. 1996; Silvi et al. 1996). The plates were incubated for 48 h at 37 °C under anaerobic (10 % H2, 10 % CO2, 80 % N2) conditions. The colonies were incubated in MRS supplemented with 0.05 % L-cysteine hydrochloride (MRSc). After centrifugation, the cells were suspended in sterile 10 % (w/v) reconstituted skim milk, frozen in liquid nitrogen and stored at −80 °C until further use. To characterize the properties of the bifidobacterial isolates, the probiotics B. lactis Bb12 and B. longum BBMN68, cultured under the same conditions, were used as reference strains.
Bifidobacterial strain identification
Isolates were suggested to be bifidobacteria on the basis of their anaerobic requirement, cellular morphology, and Gram staining. For molecular biological identification of the isolates, total DNA was extracted by a previously described method (Zuo et al. 2013). Genus-specific PCR was performed to confirm strain assignment to the genus Bifidobacterium using the primers rpoB-F: 5’-AACATCGGTCTGATCGGCTC-3’ and rpoB-R: 5’-GCTGCATGTTGGTACCCATC-3’ (to detect the rpoB gene) (Kim et al. 2010). Species identification was performed by PCR amplification of the partial 16S rRNA gene using primers Bif164-F: 5’-GGGTGGTAATGCCGGATG-3’ and Bif662: 5’-CCACCGTTACACCGGGAA-3’ (Langendijk et al. 1995), and the partial transaldolase gene using primers ForTal: 5’-CGTCGCCTTCTTCTTCGTCTC-3’ and RevTal: 5’-CTTCTCCGGCATGGTGTTGAC-3’ (Requena et al. 2002). The PCR product was partially sequenced and compared to an all-nucleotide database using Blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree was constructed using the neighbor-joining distance method of MEGA program version 4.0.
Survival under conditions simulating the human GIT
Assays of tolerance to low pH or bile salts
All methods were based on Arboleya et al. (2011). A 5-mL aliquot of bacterial cells from overnight (16 h) culture was harvested by centrifugation (6000 g, 10 min, 4 °C), washed twice with 0.85 % NaCl and resuspened in 500 μL of the same buffer. A 100-μL aliquot of the bacterial suspension was added to 900 μL simulated gastric juice (125 mM NaCl, 7 mM KCl, 45 mM NaHCO3 , and 3 g/L pepsin [Sigma], adjusted to pH 2.5 with HCl) or bile juice (45 mM NaCl, 1 g/L pancreatin [Sigma] and 3 g/L Oxgall [Sigma], adjusted to pH 8.0 with NaOH). Suspensions were then incubated under anaerobic conditions for 1 h. Plate counts in MRSc were performed at time 0 and after incubation, and results are presented as percent survival.
Bile salt hydrolysis assay
Fresh bacterial cultures were dropped onto MRS agar containing 0.5 % (w/v) taurodeoxycholic acid (Sigma T0875), then anaerobically incubated at 37 °C for 48 h. Strains with bile salt hydrolase activity were surrounded by a halo of precipitated deconjugated bile salts (Jones et al. 2008).
Autoaggregation and coaggregation assays
Autoaggregation assays were performed according to Del Re et al. (2000) with some modifications. Bifidobacterial strains were grown for 16 h at 37 °C in MRSc broth, the cells were harvested by centrifugation at 6000 g for 10 min, and the pellets were washed twice and suspended in phosphate buffered saline (PBS, pH 7.4) to yield an optical density at 600 nm (OD600) of 1.0. After incubation at 37 °C for 2 h, 0.1 mL of the upper suspension was transferred to another tube with 1.9 mL PBS and OD600 was measured. Percent autoaggregation was expressed as 1 - (OD600 of upper suspension/OD600 of total bacterial suspension) × 100.
Preparation of cell suspensions for coaggregation was the same as for the autoaggregation analysis. Equal volumes (1 mL) of the cell suspensions of a bifidobacterial strain and the pathogen strain Listeria monocytogenes were mixed in a cuvette, and the OD600 was immediately measured (designated A0). After incubation of the mixture at 37 °C for 2 h, the OD600 was measured again (designated At). Percent coaggregation was calculated using the equation of Nagaoka et al. (2007): coaggregation % = (A0 - At)/A0 × 100.
Hydrophobicity assay
Hydrophobicity of the bifidobacterial strains was determined by xylene extraction according to Pablo et al. (1998) and Pan et al. (2006). After growth in MRSc broth for 24 h, bacterial cells were harvested by centrifugation at 6000 g for 5 min, and washed twice with 50 mM K2HPO4 (pH 6.5) buffer. Absorbance at 600 nm (A600) was adjusted to 0.5 ± 0.05, then 0.6 mL xylene was added to 3 mL of bacterial suspension and vortexed for 180 s. The aqueous phase was removed after 1 h of incubation at room temperature and its A600 was measured. Affinity to hydrocarbons was reported as adhesion percentage according to the formula [(A0 - A)/A0] × 100, where A0 and A are the absorbance before and after extraction with organic solvents, respectively.
Adhesion to HT-29 and Caco-2 cells
The adhesive activity of the bifidobacterial strains was assessed using HT-29 and Caco-2 cells as an intestinal epithelial cell model according to Tuo et al. (2013) with slight modifications. HT-29 and Caco-2 cells were seeded in 24-well cell culture plates at a concentration of 5 × 105 cells per well. The plates were cultured at 37 °C in a humidified atmosphere of 5 % CO2 and 95 %. After about 24 h of incubation, a confluent monolayer was obtained.
For adhesion assay, HT-29 and Caco-2 cell monolayers on the 24-well plates were washed twice with PBS (pH 7.4). Overnight-grown bifidobacterial strains were harvested by centrifugation at 6000 g for 5 min at 4 °C and washed twice with PBS (pH 7.4) and then resuspended in Dulbecco’s Modified Eagle Medium (antibiotic-free, fetal bovine serum-free). Bacterial suspension (1 mL of 1 × 108 CFU/mL) was added to the 24-well plates and incubated for 1 h at 37 °C in a 5 % CO2 atmosphere. After incubation, each well was washed six times with PBS (pH 7.4) to remove free, unattached bacterial cells. The monolayers were fixed in methanol, Gram-stained and examined microscopically under an oil-immersion lens. Adhesion was evaluated in 20 random microscopic fields and the mean ± standard deviation of adhering bacteria per 100 epithelial cells was determined.
Sensitivity to antibiotics
Bifidobacterial susceptibility to antibiotics was analyzed by minimum inhibitory concentration (MIC) assay. The different bifidobacterial strains were cultured in MRSc broth supplemented with various concentrations (0.125 to 1024 μg/mL) of antibiotics (including ampicillin, chloramphenicol, kanamycin, streptomycin, tetracycline, erythromycin, rifampicin, vancomycin) and examined in triplicate for growth in a microplate reader (OD600) following a 24-h incubation period at 37 °C.
Antimicrobial activity against pathogens
The capacity of the strains to inhibit intestinal pathogens was determined by the agar-well diffusion method (Touré et al. 2003). Fresh overnight bifidobacterial MRSc culture supernatants were collected by centrifugation (12,000 g, 15 min, 4 °C). The cell-free supernatant (CFS) was divided into two aliquots, one adjusted to pH 6.5, and the other left unadjusted.
An initial inoculum of approximately 106 CFU/mL of the pathogen strain was incorporated into soft agar (1 %, w/v) plates with the appropriate medium for the target strain (BHI agar medium for Staphylococcus aureus ATCC25923 [Collado et al. 2005]; nutrient agar medium for Salmonella enterica ATCC13076 [Cheikhyoussef et al. 2009]; LB medium for Escherichia coli ATCC 8099). CFS (100 μL) was transferred in an Oxford cup on the surface of the agar. The plates were incubated at 37 °C for 12 h, and the diameter of the inhibition zone was measured. Tetracycline (10 μg/mL) was used as a positive control.
Statistical analysis
Data are presented as means per group ± standard errors of the means (SEM). Differences were considered significant at p < 0.05.
Results
Strain isolation and identification
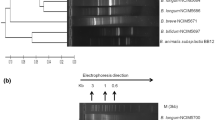
Nearly 200 colonies were isolated from three feces samples collected from breast-fed infants. Of these, 30 were suggested to be bifidobacteria based on cellular morphology, rpoB gene amplification, and Gram staining. Their 16S rRNA and transaldolase genes were partially sequenced, resulting in the identification of five different species that had greater than 99 % sequence identity to B. adolescentis, B. longum, B. breve, B. catenulatum/B. pseudocatenulatum group, and B. bifidum, respectively. Among them, strains IF1-03, IF1-04, IF1-11, and IF1-12 were regarded as B. adolescentis, IF3-31, IF3-53, and IF3-111 as B. longum, IF2-141, IF2-191, and IF3-131 as B. catenulatum/B. pseudocatenulatum group strains, IF2-173, IF2-174 as B. breve, and IF3-211 as B. bifidum (Table 1). The phylogenetic tree of the identified bacteria based on partial 16S rRNA gene and transaldolase gene sequences provided the relative positions of the isolates (Fig. 1).
The partial 16S rRNA and transaldolase gene sequences of bifidobacteria were deposited in GenBank with accession numbers KP256207 to KP256219 and KP256220 to KP256232, respectively.
Resistance to simulated conditions of human GIT
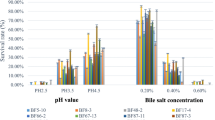
Under the simulated GIT conditions, strains isolated from the breast-fed infants showed varied resistance to acid and bile salts. Tolerance to low pH was highly variable among strains, but all strains showed lower survival in simulated gastric juice after 1 h exposure than the two reference strains, B. lactis Bb12 and B. longum BBMN68 (Table 2). B. adolescentis IF1-12 displayed the highest survival, while B. catenulatum/B. pseudocatenulatum group and B. breve strains were more sensitive to low pH (Table 2). However, the isolated bifidobacterial strains were extremely sensitive to simulated bile juice. The survival rates after exposure for 1 h to bile juice were lower than 0.0001 % for all isolates, whereas the reference strain B. lactis Bb12 showed 0.46 % survival after bile juice challenge.
Aggregation property
All of the isolated bifidobacterial strains exhibited autoaggregation after 2 h incubation at 37 °C (Table 3). The strain B. adolescentis IF1-11 showed the highest autoaggregation activity, far higher than the two reference strains (B. lactis Bb12 and B. longum BBMN68; Table 3). After 16 h static cultivation in MRSc at 37 °C, B. adolescentis IF1-11 and B. bifidum IF3-211 cells aggregated and sank to the bottom of the Hungate tube (Supplementary Fig. 2).
The coaggregation ratios between bifidobacterial strains and L. monocytogenes are shown in Table 3. B. adolescentis strain IF1-11 showed the highest coaggregation ability with L. monocytogenes. All four B. adolescentis strains, three B. longum strains and the B. bifidum isolate showed significantly higher coaggregation ability than those of the two reference strains (Table 3).
Hydrophobicity distribution
Cell-surface hydrophobicity showed big differences, as measured by xylene extraction, among the bifidobacterial strains (Table 3). All of the isolated strains showed lower hydrophobicity than B. lactis Bb12 (Table 3). However, most of the strains showed significantly higher hydrophobicity than B. longum BBMN68 (Table 3), especially B. bifidum IF3-211, which presented hydrophobicity approaching that of B. lactis Bb12.
Adhesion to intestinal epithelial cells
Adhesion of the bifidobacterial strains to HT-29 and Caco-2 cells was evaluated and the results are presented in Table 3. The strains did not exhibit similar adhesion abilities, despite being in the same genus. In general, B. longum and B. catenulatum/B. pseudocatenulatum group strains had low adhesive ability compared with B. bifidum, B. adolescentis, and B. breve. The most adhesive strain was B. bifidum IF3-211, showing significantly higher adhesion than the two reference strains (Table 3). This was followed by B. breve IF2-173 and B. adolescentis IF1-11 cells (Table 3).
Antibiotic-resistance profiles
Table 4 shows the MICs of the tested bifidobacterial strains against antibiotics of different groups: RNA-synthesis inhibitor (rifampicin), cell-wall inhibitors (ampicillin and vancomycin), and protein-synthesis inhibitors (kanamycin, streptomycin, tetracycline, erythromycin, and chloramphenicol). Strains were considered resistant when they showed MIC values higher than the MIC breakpoints established by the European Food Safety Authority (EFSA 2008). Accordingly, all of the isolated bifidobacterial strains could be characterized as susceptible to ampicillin and chloramphenicol, but resistant to kanamycin, streptomycin, and vancomycin (Table 4).
Antimicrobial activity
All of the bifidobacterial strains were tested for antimicrobial activity against selected pathogens in well-diffusion assays. As indicated in Table 5, CFS of all of the Bifidobacterium strains inhibited Salmonella enterica ATCC13076, and CFS of B. adolescentis and B. breve weakly inhibited Escherichia coli and Staphylococcus aureus ATCC25923. However, no inhibition was observed for any of the CFS in which the pH had been neutralized (data not shown).
Discussion
The wide use of infant formula in China carries potential health-risk concerns (Tang et al. 2014); infant formula is less efficient at assisting in the development of the gut microbiota and the immune system than breast milk (Harmsen et al. 2000). Increasing bifidobacterial levels is considered a target in infant formula development through, for example, supplementation with live Bifidobacteria (Braegger et al. 2011). Thus, isolation of bifidobacterial strains from breast-fed infants is of great importance. In China, however, wild bifidobacterial strains have been only scarcely isolated and characterized from human feces (Pan et al. 2006; Liu et al. 2013). Here we confirmed the identity of 13 Bifidobacterium strains isolated from breast-fed infant feces by partial sequencing of the 16S rRNA and transaldolase genes; these included strains of B. adolescentis, B. catenulatum/B. pseudocatenulatum group, B. longum, B. breve, and B. bifidum. Differentiation of B. catenulatum and B. pseudocatenulatum is difficult; strains IF2-141 and IF2-191 were regarded as B. catenulatum, IF3-131 was regarded as B. pseudocatenulatum according to phylogenetic tree based on partial transaldolase gene (Fig. 1b). In spite of this, a much better method to discriminate them, such as multi-gene-based analysis, is needed (Kim et al. 2010).
To test the potential of these wild bifidobacterial strains for use as probiotics, in vitro functional characterization and safety assessment were carried out according to the guidelines established by the FAO/WHO working group (FAO/WHO 2006). Bifidobacteria of human intestinal origin have been proposed to be more suitable for probiotic applications, but they are sensitive to environmental stresses such as low pH and bile salts as they enter the GIT (Scheller and O’Sullivan 2011). Survival under GIT conditions is crucial for probiotic strains. In this study, all of the isolated strains showed high sensitivity to bile salts, albeit generally lower sensitivity than reported in previous studies (Arboleya et al. 2011; Andriantsoanirina et al. 2013). This suggests that resistance to bile salts is highly strain- and species-dependent, although all isolates showed bile salt hydrolase activity. It should be noted that there are many methods to improve the viability of wild bifidobacteria for use in the food industry, such as stress adaptation and microencapsulation (Hansen et al. 2004; Noriega et al. 2004; Picot and Lacroix 2004; Sánchez et al. 2007).
In analyses of cell-surface properties, B. adolescentis IF1-11 exhibited the highest autoaggregation values and highest coaggregation with L. monocytogenes. B. catenulatum/B. pseudocatenulatumgroup strain IF3-131 and B. bifidum IF3-211 had the highest hydrophobicity values, with the latter value being close to that of the reference strain B. lactis Bb12. In most cases, hydrophobicity and autoaggregation abilities are strongly related to the adhesion properties of bifidobacteria and lactobacilli (Del Re et al. 2000; Tuo et al. 2013). Adhesion to the intestinal mucosa is considered one of the main criteria for the selection of potential probiotics, as it may increase their persistence in the intestine, giving the probiotic time to exert its effects (Kolida et al. 2006). B. bifidum IF3-211 showed the highest adhesion levels to HT-29 and Caco-2 cells, far exceeding those of B. lactis Bb12 and B. longum BBMN68; this corresponds to the high hydrophobicity of B. bifidum IF3-211, but not to its low autoaggregation. However, B. bifidum IF3-211 did exhibit high autoaggregation when cells were grown to the stationary phase (Fig. 2) or in low pH conditions (data not shown), suggesting that the autoaggregation characteristic of B. bifidum IF3-211 is dependent on environmental conditions, mainly pH, as described previously (Canzi et al. 2005; Guglielmetti et al. 2009). Most of the B. adolescentis strains and B. breve IF2-173 showed relatively higher adhesion ability than B. longum BBMN68. Except for B. adolescentis IF1-11, which had high autoaggregation values, the other three B. adolescentis strains had relatively low autoaggregation and hydrophobicity. But B. adolescentis IF1-11 showed lower adhesion ability to HT-29 cells and higher adhesion ability to Caco-2 cells than the other B. adolescentis strains. B. breve IF2-173 showed far higher adhesion ability to HT-29 cells than B. breve IF2-174, but its hydrophobicity was lower. We could, therefore, hypothesize that the aggregation and hydrophobicity phenotypes are not always correlated with adhesion abilities, and they are not the only mechanism involved in adhesion. For example, adhesion of Lactobacillus rhamnosus GG and Lactococcus lactis TIL448 was found to be mediated by pili, structures known to mediate the adhesion of many pathogens (Kankainen et al. 2009; Meyrand et al. 2013). Remarkably, the tight adherence (Tad) pili and sortase-dependent pili are responsible for the adhesion of B. breve UCC2003 and B. bifidum PRL2010 to the intestinal epithelium, respectively (Motherway et al. 2011; Turroni et al. 2013). In addition, the moonlighting protein transaldolase has been shown to play a role in the autoaggregation and adhesion of B. bifidum to mucin (Gonzalez-Rodriguez et al. 2012). However, B. longum and B. catenulatum/B. pseudocatenulatum group strains showed relatively lower adhesion ability. In fact, the adhesion properties of these two bifidobacteria are generally poorer than those of other species such as B. breve and B. bifidum (Del Re et al. 2000; He et al. 2001). Thus, to select new potential probiotic strains, a case-by-case assessment is required. Other mechanisms related to adhesion and pathogen-exclusion properties of these wild bifidobacterial strains need to be further investigated.
The production of antimicrobial compounds against pathogens by bifidobacterial strains was determined by agar diffusion assay. All of the selected bifidobacteria's supernatants could inhibit the foodborne pathogen Salmonella enterica, and inhibition of E. coli and S. aureus was found with some bifidobacterial secretions. However, no inhibition was observed for any of the CFS when the pH was neutralized, indicating that the inhibition effect is mainly due to organic acids, such as short-chain fatty acids, lactic acid, and acetic acid (Midtvedt and Midtvedt 1992; Makras and De Vuyst 2006). Bifidobacterial antimicrobial activity is due to a number of metabolites, organic acids, and most importantly, bacteriocins, although only limited classes of bacteriocins produced by bifidobacteria have been characterized in depth (Martinz et al. 2013). The capacity to produce antimicrobial compounds is one of the critical characteristics of bifidobacteria in terms of effectively and competitively excluding pathogens in the intestine, and exerting their probiotic effect on the host (Ouwehand and Salminen 1998).
It is important to determine the safety of wild bifidobacterial strains, particularly their antibiotic resistance profiles (Sanders et al. 2010). Antibiotic MIC assay suggested that all of the isolated bifidobacterial strains are susceptible to ampicillin and chloramphenicol, but resistant to kanamycin, streptomycin, and vancomycin. B. lactis Bb12 is resistant to kanamycin, streptomycin, and vancomycin (Zhou et al. 2005), and some other studies suggest that bifidobacteria were resistant to kanamycin and streptomycin (Sharma et al. 2014). Kiwaki and Sato (2009) suggest that bifidobacteria resistance to streptomycin might be caused by mutations in the rpsL gene, but sequencing of rpsL genes from all the isolates plus the two reference strains didn't find any mutations at nucleotide position 128 (data not shown). Thus, the rpsL gene was not responsible for streptomycin resistance in these strains. Testing them for the presence of the enterococcal vanA and vanB genes by PCR using the primers provided by Klein et al. (2000) gave a negative result (data not shown), similar with the previously reports (Klein et al. 2000; Zhou et al. 2005). Some strains of B. animalis subsp. lactis and B. bifidum have shown acquisition of the tetracycline resistance gene tet(W) (Meile et al. 2008), and some commercial probiotic strains, including B. longum, B. bifidum, and B. thermophilum, are also resistant to the antibiotics erythromycin, streptomycin, and chloramphenicol (Sato and Iino 2009; Mayrhofer et al. 2011; Wei et al. 2012). Although none of the Bifidobacterium species with qualified presumption of safety (QPS) status, including B. adolescentis, B. animalis, B. bifidum, B. breve, and B. longum (EFSA 2012), have been associated with human clinical disease, their antibiotic resistance phenotypes should be thoroughly characterized to prevent the potential transfer of antibiotic resistance genes to other bacteria, especially pathogens, in the intestinal habitat (Ammor et al. 2008).
The main objective of this study was to find new probiotic candidates for use in functional fermented food, such as infant formula. In summary, we characterized 13 bifidobacterial strains isolated from breast-fed infant feces, phenotypically and genotypically, according to international guidelines for probiotics. In addition, in vitro tests were performed to assess the probiotic potential of these strains. Our results suggest that some of the strains isolated from breast-fed infant feces, notably B. bifidum IF3-211 and B. adolescentis IF1-11, may have valuable probiotic potential in functional food products, althrough further safety evaluation and human studies would be needed.
References
Ammor MS, Flórez AB, Van Hoek AHAM, de los Reyes-Gavilán CG, Aarts HJM, Margolles A, Mayo B (2008) Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J Mol Microb Biotech 14:6–15
Andriantsoanirina V, Allano S, Butel MJ, Aires J (2013) Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe 21:39–42
Arboleya S, Ruas-Madiedo P, Margolles A, Solís G, Salminen S, de los Reyes-Gavilán CG, Gueimonde M (2011) Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int J Food Microbol 149:28–36
Beerens H (1990) An elective and selective isolation medium for Bifidobacterium spp. Lett Appl Microbiol 11:155–175
Braegger C et al (2011) Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by the ESPGHAN committee on nutrition. J Pediatr Gastroenter Nutr 52:238–250
Blanchard E, Zhu P, Schuck P (2013) Infant formula powders. In: Bhandari B, Bansal N, Zhang M, Schuck P (eds) Handbook of food powders: Processes and properties. Vol. no. 255. Woodhead Publishing, Cambridge, UK, pp 465–480
Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, Campieri M, Brigidi P (2008) Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 125:286–292
Canzi E, Guglielmetti S, Mora D, Tamagnini I, Parini C (2005) Conditions affecting cell surface properties of human intestinal bifidobacteria. Anton Leeuw Int J G 88:207–219
Cheikhyoussef A, Pogori N, Chen HQ, Tian FW, Chen W, Tang J, Zhang H (2009) Antimicrobial activity and partial characterization of bacteriocin-like inhibitory substances (BLIS) produced by Bifidobacterium infantis BCRC 14602. Food Control 20:553–559
Collado MC, Hernández M, Sanz Y (2005) Production of bacteriocin-like inhibitory compounds by human fecal Bifidobacterium strains. J Food Protect 68:1034–1040
Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31:438–442
EFSA (2008) Technical guidance prepared by the Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) on the update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 732, 1e15 (output obsolete)
EFSA (2012) Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed. EFSA J 10:3020. doi:10.2903/j.efsa.2011.2497
Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, van Sinderen D, Dougan G (2012) Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109:2108–2113
FAO/WHO (2006) Probiotics in food. Health and nutritional properties and guidelines for evaluation. FAO Food and Nutrition paper 85. ISBN: 92-5-105513-0
Ferraris L, Aires J, Waligora-Dupriet AJ, Butel MJ (2010) New selective medium for selection of bifidobacteria from human feces. Anaerobe 16:469–471
Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H (2011) Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547
García-Cayuela T, Korany AM, Bustos I, Gómez de Cadiñanos LP, Requena T, Peláez C, Martínez-Cuesta MC (2014) Adhesion abilities of dairy Lactobacillus plamtarum strains showing an aggregation phenotype. Food Res Int 57:44–50
Garrigues C, Johansen E, Pedersen MB (2010) Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a wildly consumed probiotic strain. J Bacteriol 192:2467–2468
Goh YJ, Klaenhammer TR (2010) Functional roles of aggregation-promoting factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 76:5005–5012
Gonzalez-Rodriguez I, Sanchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A (2012) Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol 78:3992–3998
Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D (2009) Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr Microbiol 59:167–172
Hansen LT, Allan-Wojtas PM, Jin Y-L, Paulson AT (2004) Survival of Ca-alginate microencapsulated Bifidobacterium spp. in milk and simulated gastrointestinal conditions. Food Microbiol 19:35–45
Hao YL, Huang DW, Guo HY, Xiao M, An HR, Zhao L, Zuo FL, Zhang B, Hu SN, Song SH, Chen SW, Ren FZ (2011) Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy Chinese centenarian. J Bacteriol 193:787–788
Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW (2000) Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenter Nutr 30:61–67
Hartemink R, Kok BJ, Weenk GH, Rombouts FM (1996) Raffinose-Bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J Microbiol Meth 27:33–43
He F, Ouwehand AC, Hashimoto H, Isolauri E, Benno Y, Salminen S (2001) Adhesion of Bifidobacterium spp. to human intestinal mucus. Microbiol Immunol 45:259–262
Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR (2008) Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585
Kankainen M, Paulin L, Tynkkynen S, Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, de Vos WM (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198
Kim BJ, Kim HY, Yun YJ, Kim BJ, Kook YH (2010) Differentiation of Bifidobacterium species using partial RNA polymerase β-subunit (rpoB) gene sequences. Int J Syst Evol Micr 60:2697–2704
Kiwaki M, Sato T (2009) Antimicrobial susceptibility of Bifidobacterium breve strains and genetic analysis of streptomycin resistance of probiotic B. breve strain Yakult. Int J Food Microbiol 134:211–215
Klein G, Hallmann C, Casas IA, Abad J, Louwers J, Reuter G (2000) Exclusion of vanA, vanB and vanC type glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridization methods. J Appl Microbiol 89:815–824
Kolida S, Saulnier DM, Gibson GR (2006) Gastrointestinal microflora: probiotics. Adv Appl Microbiol 59:187–219
Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW (1995) Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-dpecific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 61:3069–3075
Liu WJ, Chen YF, Kwok LY, Li MH, Sun T, Sun CL, Wang XN, Dan T, Menghebilige ZHP, Sun TS (2013) Preliminary selection for potential probiotic Bifidobacterium isolated from subjects of different Chinese ethnic groups and evalution of their fermentation and storage characteristics in bovine milk. J Dairy Sci 96:6807–6817
Makras L, De Vuyst L (2006) The in vitro inhibition of Gram-negative pathogenic bacteria by bifidobacteria is caused by the production of organic acids. Int Dairy J 16:1049–1057
Martinz FAC, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP (2013) Bacteriocin production by Bifidobacterium spp.: A review. Biotechnol Adv 13:482–488
Mayrhofer S, Mair C, Kneifel W, Domig KJ (2011) Susceptibility of bifidobacteria of animal origin to selected antimicrobial agents. Chemot Res Practi 989520
Meile L, Le Blay G, Thierry A (2008) Safety assessment of dairy microorganisms: Propionibacterium and Bifidobacterium. Int J Food Microbiol 126:316–320
Meyrand M, Guillot A, Goin M, Furlan S, Armalyte J, Kulakauskas S, Cortes-Perez NG, Thomas G, Chat S, Péchoux C, Dupres V, Hols P, Dufrêne YF, Trugnan G, Chapot-Chartier M-P (2013) Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol Cell Proteomics 12:3935–3947
Midtvedt A-C, Midtvedt T (1992) Production of short chain fatty acids by the intestinal microflora during the first 2 years of human life. J Pediatr Gastr Nutr 15:395–403
Miranda RO, de Carvalho AF, Nero LA (2014) Development of a selective sulture medium for bifidobacteria, Raffinose-Propionate Lithium Mupirocin (RP-MUP) and assessment of its usage with PetrifilmTM Aerobic Count plates. Food Microbiol 39:96–102
Motherway MOC, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JAM, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D (2011) Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved hostcolonization factor. Proc Natl Acad Sci U S A 108:11217–11222
Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, Clavel T, Koebnick C, Zunft H-JF, Doré J, Blaut M (2006) Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 72:1027–1033
Muñoa FJ, Pares R (1988) Selective medium for isolation and enumeration of Bifidobacterium spp. Appl Environ Microbiol 54:1715–1718
Muňoz-Quezada S, Chenoll E, Vieites JM, Bermúdez-Brito SM, Gomez-Llorente C, Matencio E, Bernal MJ, Romero F, Suárez A, Ramón D, Gil A (2013) Isolation, identification and characterisation of three novel probiotic strains (Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036) from the faeces of exclusively breast-fed infants. Brit J Nutr 109:S51–S62
Nagaoka S, Hojo K, Murate S, Mori T, Ohshima T, Maeda N (2007) Interactions between salivary Bifidobacterium adolescentis and other oral bacteria: in vitro coaggregation and coadhesion assays. FEMS Microbiol Lett 281:183–189
Noriega L, Gueimonde M, Sánchez B, Margolles A, de los Reyes-Gavilán CG (2004) Effect of the adaptation to high bile salts concentration on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int J Food Microbiol 94:79–86
Ouwehand AC, Salminen S (1998) The health effects of cultured milk products with viable and non viable bacteria. Int Dairy J 8:749–758
Ouwehand AC, Salminen S, Isolauri E (2002) Probiotics: an overview of beneficial effects. Anton Leeuw Int J G 82:279–289
Pablo FP, Yessica M, Disalvo EA, De Antoni GL (1998) Surface properties of bifidobacterial strains of human origin. Appl Environ Microbiol 64:21–26
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO (2007) Development of the human infant intestinal microbiota. Plos Biol 5:1556–1573
Pan WH, Li PL, Liu ZY (2006) The correlation between surface hydrophobicity and adherence of Bifidobacterium strains from centenarians’ faeces. Anaerobe 12:148–152
Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–21
Pereira DIA, Gibson GR (2002) Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl Environ Microbiol 68:4689–4693
Picot A, Lacroix C (2004) Encapsulation of bifidobacteria in whey protein-based microcapsules and survival in simulated gastrointestinal conditions and in yoghurt. Int Dairy J 14:505–515
Prasanna PHP, Grandison AS, Charalampopoulos D (2014) Bifidobacteria in milk products: an overview of physiological and biochemical properties, exopolysaccharide production, selection criteria of milk products and health benefits. Food Res Int 55:247–262
Raeisi SN, Ouoba LII, Farahmand N, Sutherland J, Ghoddusi HB (2013) Variation, viability and validity of bifidobacteria in fermented milk products. Food Con 34:691–697
Rahman MM, Kim WS, Kumura H, Shimazaki KI (2008) Autoaggregation and surface hydrophobicity of bifidobacteria. World J Microbiol Biotechnol 24:1593–1598
Requena T, Burton J, Matsuki T, Munro K, Simon MC, Tanaka R, Watanabe K, Tannock GW (2002) Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl Environ Microbiol 68:2420–2427
Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL (2010) Examination of faecal Bifidobacteterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341
Russell DA, Ross RP, Fitzgerald GF, Stanton C (2011) Metabolic activities and probiotic potential of bifidobacteria. Int J Food Microbiol 149:88–105
Saarela M, Mogensen G, Fondėn R, Mättö J, Mattila-Sandholm T (2000) Probiotic bacteria: safety, functional and technological properties. J Biotechnol 84:197–215
Saavedra JM, Bauman NA, Perman JA, Yolken RH, Oung I (1994) Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. The Lancet 344:1046–1049
Sánchez B, Champomier-Vergés MC, Collado MC, Anglade P, Baraige F, Sanz Y, de los Reyes-Gavilán CG, Margolles A, Zagorec M (2007) Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl Environ Microbiol 73:6450–6459
Sánchez B, Ruiz L, de los Reyes-Gavilan CG, Margolles A (2008) Proteomics of stress response in Bifidobacterium. Front Biosci 13:6905–6919
Sánchez B, Ruiz L, Gueimonde M, Ruas-Madiedo P, Margolles A (2013) Adaptation of bifidobacteria to the gastrointestinal tract and functional consequences. Pharmacol Res 69:127–136
Sanders ME, Akkermans LMA, Haller D, Hammerman C, Heimbach JT, Hörmannsperger G, Huys G (2010) Safety assessment of probiotics for human use. Gut Microbes 1:164–185
Sato T, Iino T (2009) Genetic analyses of the antibiotic resistance of Bifidobacterium bifidum strain Yakult YIT 4007. Int J Food Microbiol 137:254–258
Scheller M, O’Sullivan DJ (2011) Comparative analysis of an intestinal strain of Bifidobacterium longum and a strain of Bifidobacterium animalis subspecies lactis in chrddar cheese. J Dairy Sci 94:1122–1131
Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R (2014) Antibiotic resistance among commercially available probiotics. Food Res Int 57:176–195
Silvi S, Rumney CJ, Rowland IR (1996) An assessment of three selective media for bifidobacteria in faeces. J Appl Bacteriol 81:561–564
Simpson PJ, Stanton C, Fitzgerald GF, Ross RP (2005) Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J Appl Microbiol 99:493–501
Tang L, Lee AH, Binns CW, Yang YX, Wu Y, Li YX, Qiu LQ (2014) Widespread usage of infant formula in China: a major public health problem. Birth 41:339–343
Touré R, Kheadr E, Lacroix C, Moroni O, Fliss I (2003) Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes. J Appl Microbiol 95:1058–1069
Tuo YF, Yu YL, Ai LZ, Wu ZG, Guo BH, Chen W (2013) Aggregation and adhesion properties of 22 Lactobacillus strains. J Dairy Sci 96:4252–4257
Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sánchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Cuglielmetti S, van Sinderen D, Ventura M (2013) Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156
Wei YX, Zhang ZY, Liu C, Malakar PK, Guo XK (2012) Safety assessment of Bifidobacterium longum JDM301 based on complete genome sequences. World J Gastroentero 18:479–488
Zhou JS, Pillidge CJ, Gopal PK, Gill HS (2005) Antibiotic susceptibility profiles of new probiotic Lactobacillus ans Bifidobacterium strains. Int J Food Microbiol 98:211–217
Zuo FL, Feng XJ, Sun XF, Du C, Chen SW (2013) Characterization of plasmid pML21 of Enterococcus faecalis ML21 from koumiss. Curr Microbiol 66:103–105
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31071507), the National High Technology Research and Development Program (“863” Program, No. 2008AA10Z310), and the National Science and Technology Support Program, Ministry of Science and Technology of China (2011BAD09B03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors had a conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(JPG 5546 kb)
Rights and permissions
About this article
Cite this article
Zuo, F., Yu, R., Feng, X. et al. Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast-fed infant feces. Ann Microbiol 66, 1027–1037 (2016). https://doi.org/10.1007/s13213-015-1187-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1187-x