Abstract
Pseudomonas aeruginosa often causes various acute and chronic infections in humans exploiting biofilm. Molecules interfering with microbial biofilm formation could be explored for the sustainable management of infections linked to biofilm. Towards this direction, the antimicrobial and antibiofilm activity of cuminaldehyde, an active ingredient of the essential oil of Cuminum cyminum was tested against Pseudomonas aeruginosa. In this regard, the minimum inhibitory concentration (MIC) of cuminaldehyde was found to be 150 μg/mL against the test organism. Experiments such as crystal violet assay, estimation of total biofilm protein, fluorescence microscopy and measurement of extracellular polymeric substances (EPS) indicated that the sub-MIC doses (up to 60 µg/mL) of cuminaldehyde demonstrated considerable antibiofilm activity without showing any antimicrobial activity to the test organism. Moreover, cuminaldehyde treatment resulted in substantial accumulation of cellular reactive oxygen species (ROS) that led to the inhibition of microbial biofilm formation. To this end, the exposure of ascorbic acid was found to restore the biofilm-forming ability of the cuminaldehyde-treated cells. Besides, a noticeable reduction in proteolytic activity was also observed when the organism was treated with cuminaldehyde. Taken together, the results demonstrated that cuminaldehyde could be used as a promising molecule to inhibit the biofilm formation of Pseudomonas aeruginosa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In nature, bacteria are found to switch between two distinct modes of existence. One is the free-living planktonic form while the other one is the biofilm (Gupta et al. 2016). Biofilm happens to be the most predominant form of bacterial adaptation in nature for the sustainable management of stress (Chakraborty et al. 2020). Biofilm is a highly complex structure of bacterial association that can originate either from the homogeneous or heterogeneous aggregation of microorganisms (Wimpenny et al. 2000). The existing literature revealed that the three-dimensional complex structure of biofilm gets stabilized by secreting extracellular polymeric substances (EPS) (Yang et al. 2011). EPS is composed of various biomolecules such as polysaccharides, proteins, lipids, and nucleic acids (Cortes et al. 2011). Furthermore, this matrix has been found to play a key role in amplifying the resistance of the bacterial biofilm to several antimicrobials (Roy et al. 2018). This in turn not only blocks the smooth diffusion of antimicrobials through biofilm, but also protects the sessile cells from bacteriophages, synthetic biocides, amoebae, disinfectants, UV radiation, pH stress, etc. (Chakraborty et al. 2018). Previous literature revealed that the biofilm cells are found to be ~ 10–1000 times more resistant than their planktonic counterpart in the context of drug resistance (Sharma et al. 2019). Besides, microorganisms in biofilm show an altered gene expression that helps them to evade the host immune system thereby causing various chronic and acute infections in humans (Gupta et al. 2016). According to the National Institute of Health (NIH), biofilm accounts for almost 80% of the microbial infections posing a serious threat to public healthcare (Khatoon et al. 2018).
Pseudomonas aeruginosa, a multi-drug resistant opportunistic pathogen has been found to cause an array of biofilm-linked diseases like endocarditis, periodontitis, kidney infections, cystic fibrosis, and so on (Gupta et al. 2016). Furthermore, most of the therapeutic approaches turned out to be quite challenging due to the emergence of drug resistance by the biofilm cells. Towards this direction, several molecules are being explored globally to manage this biofilm threat. However, natural molecules have been found to be an effective choice over synthetic molecules as they are cost-effective, abundant, and non-toxic to the human host (Mishra et al. 2020). Therefore, our current study has been aimed at investigating one such natural molecule, cuminaldehyde against microbial biofilm formation. This compound happens to be a major bioactive constituent of white cumin seeds (Cuminum cyminum) (Sowbhagya 2013). Moreover, a literature survey revealed that cuminaldehyde could show potential antibiofilm and antimicrobial activity against Staphylococcus aureus and Escherichia coli (Monteiro-Neto et al. 2020). However, the effect of cuminaldehyde on the biofilm inhibition of Pseudomonas aeruginosa is yet to be explored. Hence, efforts have been put together to bring forth the antimicrobial as well as antibiofilm activity of cuminaldehyde against Pseudomonas aeruginosa. Towards exploring the underlying mechanism of microbial biofilm inhibition, the existing literature recommended that the inhibition in biofilm formation could be regulated by the cellular accumulation of reactive oxygen species (ROS) (Gambino and Cappitelli 2016). ROS consists of diverse reactive chemical species (hydroxyl radical, superoxide, peroxide, and singlet oxygen) which get synthesized during aerobic metabolism (Imlay 2003). The accumulation of ROS has been found to exhibit diverse biological functions including microbial biofilm inhibition (Briviba et al. 1997). However, it has also been reported that the biofilm-forming ability of the microorganism could be restored under the influence of ascorbic acid, a known antioxidant agent (Chakraborty et al. 2018; Paul et al. 2021). Hence, in the present study, efforts have been given to understand whether ROS accumulation under the influence of cuminaldehyde exert any impact on the biofilm inhibition of Pseudomonas aeruginosa. The present report deciphered that the cuminaldehyde-mediated biofilm inhibition could be attributed to the cellular accumulation of ROS in the test organism.
Materials and methods
Microbial strain, growth media, and culture conditions
Pseudomonas aeruginosa MTCC 424, an efficient biofilm-forming bacteria was used as a test organism for the present study. In this study, the organism was cultivated in Luria Bertani (LB) medium at 37 °C for 24 h to experience its optimum growth. The test compound (cuminaldehyde) selected for the investigation was purchased from Sigma-Aldrich (product number: 135178; purity: 98%; density: 0.977 g/mL). In the present study, 5 µL cuminaldehyde was mixed with 995 µL of dimethyl sulfoxide (DMSO) (purchased from Merck Life Science Pvt Ltd) to prepare the stock solution (4.98 mg/mL). The stock solution was subsequently diluted in the growth media (LB) as per the requirements of the experiments.
Determination of the minimum inhibitory concentration (MIC) of cuminaldehyde against Pseudomonas aeruginosa
MIC of cuminaldehyde was determined against Pseudomonas aeruginosa by following the broth dilution system (BDS) (Clinical and Laboratory Standards Institute 2005; Chakraborty et al. 2021). In brief, to perform this experiment, 50 µL (1 × 105 CFU/mL) microbial culture was separately inoculated into several tubes containing 5 mL of sterile LB media. To it, different concentrations (50, 100, 150, 200 and 250 µg/mL) of cuminaldehyde were separately added. A control set was prepared in which the cells were grown without being exposed to cuminaldehyde. To test the effect of DMSO on the microbial growth profile, an equal number of cells were grown in LB in which only DMSO (5% v/v) was applied. Afterward, all the test tubes were incubated at 37 °C for 24 h in an incubator shaker. After the incubation, the turbidity of the growth medium was quantified by measuring the optical density (OD) at 600 nm. In this connection, the MIC of the compound was determined by considering the lowest concentration of cuminaldehyde that did not reveal any visible growth of the microorganism. In some cases, an equal number of cells (1 × 105 CFU/mL) were grown under different combinations of cuminaldehyde and ascorbic acid. After incubation at 37 °C for 24 h, the microbial growth in each combination was measured by recording OD at 600 nm.
Bacterial growth curve analysis
To understand the effect of the sub-MIC doses of cuminaldehyde on the growth profile of Pseudomonas aeruginosa, 50 μL microbial culture was inoculated into different conical flasks containing 100 mL of sterile LB media. To it, sub-MIC doses (20, 40, 60, and 80 μg/mL) of cuminaldehyde were added individually. In the control set, the microorganisms were allowed to grow in the absence of cuminaldehyde. Thereafter, all the conical flasks were grown at 37 °C. At regular time intervals (2 h), the microbial cultures were recovered from each growth media including the control. Thereafter, the turbidity, an indicator of the microbial growth was recorded by measuring the OD at 600 nm.
Estimation of the biofilm mass by crystal violet (CV) assay
Crystal violet (CV) assay was performed to determine the degree of biofilm mass of Pseudomonas aeruginosa under different conditions (Chakraborty et al. 2020). To examine the same, 50 µL (1 × 105 CFU/mL) of microbial culture was inoculated into 5-mL sterile LB media in different tubes. After that, various sub-MIC (20, 40 and 60 µg/mL) doses of cuminaldehyde were applied to the inoculated growth media. However, in some cases, ascorbic acid (25 and 50 µg/mL) was immediately added into those tubes in which the cells were exposed to cuminaldehyde. A control set was also made in which the microbial culture of Pseudomonas aeruginosa was neither exposed to cuminaldehyde nor ascorbic acid. All the glass test tubes were then incubated at 37 °C for 24 h. After the incubation, planktonic cells were discarded from each tube and gently washed with sterile double distilled water. Then, the tubes were allowed to dry for 10 min and subsequently stained with 5 mL of 0.4% CV solution. All the tubes were further incubated for 30 min to allow effective staining of the biofilm cells developed over the glass surfaces. Thereafter, the CV solution was discarded from all the test tubes and rinsed with sterile double distilled water to wash the excess CV (if any). Then, 33% glacial acetic acid was added to each test tube to dissolve the biofilm cells bound to CV. Finally, the intensity of the color generated in each tube was estimated by measuring the OD at 630 nm.
Estimation of total biofilm protein
Estimation of total biofilm protein has been found to be a popular method of measuring the abundance of microbial populations in a biofilm (Paul et al. 2021). Hence, to determine the same, an equal number of cells (1 × 105 CFU/mL) were either challenged with sub-MIC concentrations (20, 40 and 60 µg/mL) of cuminaldehyde or left untreated. Then, all the test tubes were incubated at 37 °C for 24 h. Post incubation, the planktonic cells were removed followed by washing the tubes with sterile double distilled water. Then, 5 mL of 0.3 (M) NaOH solution was added to each test tube and subsequently boiled for 15 min at 100 °C. Thereafter, the cellular suspensions were centrifuged at 8000 rpm for 8 min. Finally, the supernatant was collected and the protein present in it was determined using the Folin-Lowry method (Lowry et al. 1951).
Estimation of extracellular polymeric substances (EPS)
The production of EPS by Pseudomonas aeruginosa under the influence of cuminaldehyde was determined by the EPS extraction assay as described by Tribedi and Sil (2014). In brief, 50 µL (1 × 105 CFU/mL) microbial culture was separately inoculated in sterile LB media. To it, various sub-MIC doses (20, 40 and 60 µg/mL) of cuminaldehyde were separately added. A control set was also prepared wherein the organisms were allowed to grow in sterile LB in the absence of cuminaldehyde. Then, all the experimental sets were incubated at 37 °C for 24 h. After the incubation, planktonic cells were removed from all the experimental sets including the control one. The biofilm cells were then suspended in sterile double distilled water and thoroughly vortexed for proper mixing. Then, the suspensions were centrifuged at 6000 rpm for 10 min. The pellet was collected and further treated with 10 mM EDTA followed by vortexing for another 10 min. Thereafter, the samples were again centrifuged at 6000 rpm for 10 min. After that, the supernatants collected from the two rounds of centrifugation were mixed and treated with 2.2 volumes of chilled absolute ethanol followed by incubating at − 20 °C for 1 h. After the incubation, the samples were centrifuged again at 6000 rpm for 20 min at 4 °C. The pellets were then collected and suspended in sterile double distilled water. Finally, the extent of EPS present in the prepared suspension was estimated using the phenol–sulphuric acid method (Dubois et al. 1956).
Analysis of biofilm formation by fluorescence microscopy
To analyze the change in microbial biofilm formation under the exposure of cuminaldehyde, a fluorescene microscopic study was performed. Firstly, 50 μL (1 × 105 CFU/mL) culture of Pseudomonas aeruginosa was separately inoculated into different test tubes containing 5 mL of sterile LB media. To it, sub-MIC doses (20, 40, and 60 μg/mL) of cuminaldehyde were added. As expected, a control set was also prepared where the organisms were allowed to grow in the absence of cuminaldehyde. Sterile glass coverslips were added to all the test tubes including the control set under aseptic conditions. All the experimental sets were then incubated for 24 h at 37 °C. Post incubation, the respective coverslips were recovered from each test tube and stained with acridine orange solution (4 μg/mL) to observe the microbial biofilm population developed over the coverslips. After that, the coverslips were gently washed to remove the excess stain (if any) and viewed under a fluorescence microscope (FITC filter) at excitation and emission wavelength of 491 nm and 516 nm, respectively (Chakraborty et al. 2018; Paul et al. 2021).
Determination of cellular accumulation of reactive oxygen species (ROS)
The microbial production of ROS under the influence of cuminaldehyde was measured by 2′-7′-dichlorofluorescein diacetate (DCFDA) based ROS detection assay kit (ab113851) purchased from Abcam. An equal number of microorganisms (1 × 108 CFU/mL) were separately collected from both cuminaldehyde treated (20, 40, 60 and 80 µg/mL) and untreated growth media. DCFDA (5 µM) was added to the collected cells in 0.9% saline. Then, the DCFDA-treated cells were further incubated at 37 °C for another 4 h (in dark) for the deacetylation of DCFDA by microbial enzymes. The accumulation of ROS (if any) under the influence of cuminaldehyde could oxidize the deacetylated DCFDA (non-fluorescent) to a fluorogenic molecule DCF (2′,7′-dichlorofluorescein). The fluorescence emission of DCF was then measured by a fluorescence spectrophotometer at an excitation and emission wavelength of 488 nm and 535 nm, respectively (Chakraborty et al. 2021; Paul et al. 2021).
Microbial metabolic activity measurement
To analyze the metabolic activity of Pseudomonas aeruginosa under the influence of cuminaldehyde, the azo-casein degrading assay was performed by following the protocol described in Chakraborty et al. (2018). In brief, 50 μL microbial culture (1 × 105 CFU/mL) of Pseudomonas aeruginosa was separately inoculated into fresh test tubes containing 5 mL of sterile LB media. Then, the cells were either exposed to the sub-MIC doses (20, 40 and 60 μg/mL) of cuminaldehyde or left untreated. Then, all the test tubes including the control were incubated for 24 h at 37 °C. Post incubation, microbial cultures collected from each set was centrifuged at 10,000 rpm for 15 min. After that, supernatant (2 mL) was collected from each experimental set and mixed with 500 μL of azo-casein (0.3%) solution followed by incubating them at 37 °C for 1 h. Then, 10% trichloroacetic acid was added to stop the reaction. Afterward, the reaction mixture was centrifuged at 10,000 rpm for 5 min. Post centrifugation, the supernatant was collected and the amount of color developed in the supernatant was measured by recording the OD at 440 nm (Chakraborty et al. 2018).
Statistical analysis
One-way analysis of variance (ANOVA) was used in this study to determine the statistical difference of the results. Each experiment was repeated thrice and the average was taken for each set. P values were estimated to determine the degree of significant difference between the cuminaldehyde treated and untreated sets. P values < 0.05 were marked with (*), P values < 0.01 were marked with (**), and P values < 0.001 were marked with (***). P values which happened to be more than 0.05 were marked as N.S. (no statistical difference).
Results and discussion
Cuminaldehyde showed efficient antimicrobial activity against Pseudomonas aeruginosa
Cuminaldehyde, a major component of the essential oil of Cuminum cyminum has been reported to exhibit diverse biological functions including antimicrobial activity (Chen et al. 2011; Tariq et al. 2019). In this context, cuminaldehyde was recently reported to show considerable antibacterial activity against Staphylococcus aureus wherein the MIC of the compound was found to be 12 mg/mL (Monteiro-Neto et al. 2020). It was also documented that cuminaldehyde could enhance the antimicrobial action of ciprofloxacin against Staphylococcus aureus and Escherichia coli-induced infections when it was co-incubated with the antibiotic (Monteiro-Neto et al. 2020). In this study, efforts were put together to understand the antimicrobial activity of cuminaldehyde (if any) against the test organism (Pseudomonas aeruginosa). In this regard, we found that with the rise of the concentrations of cuminaldehyde, the microbial growth of the test organism got decreased considerably (Fig. 1A). The microbial growth profile indicated that cuminaldehyde at a concentration of 150 μg/mL prevented the visible microbial growth of the test organism (Fig. 1A). Hence, the minimum inhibitory concentration (MIC) of cuminaldehyde could be considered as 150 μg/mL (Fig. 1A). Since DMSO was used to increase the solubility of cuminaldehyde, we had investigated whether DMSO could show any antimicrobial property against the test organism. To test the same, DMSO (5% v/v) was applied alone to the organisms growing in sterile LB. The result revealed that DMSO did not exhibit any antimicrobial activity against Pseudomonas aeruginosa (data not shown). Thus, the observations suggested that the antimicrobial action was exhibited by cuminaldehyde only. To gain further confidence, the extent of microbial growth under the presence and absence of cuminaldehyde was quantified by recording the optical density (OD) at 600 nm. The observation conveyed that with the gradual increase in the concentration of cuminaldehyde, the degree of microbial growth got reduced significantly (Fig. 1B). Thus, the results demonstrated that cuminaldehyde exhibited promising antimicrobial activity against Pseudomonas aeruginosa.
Cuminaldehyde showed considerable antimicrobial activity against Pseudomonas aeruginosa. A MIC determination. The pattern of the microbial growth of the test organism was analysed under the presence and absence of cuminaldehyde. The MIC dose of cuminaldehyde against the test organism was marked with an asterisk (*) symbol. B Measurement of microbial growth. The degree of microbial growth of Pseudomonas aeruginosa was determined by measuring the OD of the microbial cultures collected from both cuminaldehyde treated and untreated growth media. Each experiment was conducted three times and the average of the three was considered for the present study. The error bars indicated the standard error of the mean. To denote the statistical difference between the control and treated sets, P values were calculated using ANOVA. P values < 0.01 and < 0.001 were denoted with (**) and (***), respectively. P values > 0.05 did not show any significant difference with the control, hence marked as N.S (no statistical difference)
Effect of the sub-MIC doses of cuminaldehyde on the growth profile of Pseudomonas aeruginosa
Since cuminaldehyde showed promising antimicrobial activity against Pseudomonas aeruginosa (MIC: 150 μg/mL), sub-MIC doses (up to 80 µg/mL) of cuminaldehyde were screened for the antibiofilm studies. Hence, various sub-MIC doses (up to 80 µg/mL) of cuminaldehyde were selected to test whether the tested concentrations of the compound could affect the growth profile of Pseudomonas aeruginosa. The microbial growth profile showed that the sub-MIC doses (up to 60 µg/mL) of cuminaldehyde did not affect the microbial growth profile of the test organism (Fig. 2A). However, the extent of microbial growth got reduced significantly when the cells were exposed to a higher concentration (80 µg/mL) of cuminaldehyde (Fig. 2A). To gain further confidence, the microbial growth curve of Pseudomonas aeruginosa was analyzed under the presence and absence of cuminaldehyde. In this regard, we observed that the microbial growth curve remained similar between the cuminaldehyde treated (up to 60 µg/mL) and untreated cells (Fig. 2B). However, a sharp decline in growth curve was spotted when the cells were exposed to a higher concentration (80 µg/mL) of cuminaldehyde (Fig. 2B). Therefore, the result demonstrated that the sub-MIC doses (up to 60 µg/mL) of cuminaldehyde did not reveal antimicrobial activity against the test organism. Hence, in the present study, cuminaldehyde concentrations up to 60 µg/mL were considered for the biofilm inhibition studies. Towards this direction, several literatures reported that antimicrobial molecules could be used for the biofilm inhibition studies when they were applied in sub-MIC doses (Das et al. 2016; Chakraborty et al. 2021; Paul et al. 2021). In this connection, it was reported that thymoquinone could show both antimicrobial (MIC dose: 20 μg/mL) as well as antibiofilm activities (Sub-MIC doses: 5 and 10 μg/mL) against Pseudomonas aeruginosa (Chakraborty et al. 2021). In a recent report, it was documented that 1,4-naphthoquinone exhibited antimicrobial activity against Staphylococcus aureus at a concentration of 100 μg/mL (Paul et al. 2021). However, the lower concentrations (5 and 10 μg/mL) of the same molecule showed efficient antibiofilm activity against the same organism (Paul et al. 2021). Thus, the reported observations suggested that the concentrations selected for the biofilm inhibition studies did not exhibit any antimicrobial activity to the test organism (Chakraborty et al. 2021; Paul et al. 2021). Hence, in the present report, we tested the effect of the screened sub-MIC doses (up to 60 μg/mL) of cuminaldehyde on the microbial biofilm formation as the said concentrations (up to 60 μg/mL) did not exhibit any antimicrobial activity.
Sub-MIC doses of cuminaldehyde did not show any antimicrobial activity against Pseudomonas aeruginosa. Cells were inoculated in sterile LB media supplemented with different concentrations (sub-MIC doses) of cuminaldehyde. A separate set was marked as control in which cells were grown in the absence of cuminaldehyde. A Growth profile. Post incubation, microbial culture was separately collected from both cuminaldehyde treated and untreated growth media followed by measuring the OD at 600 nm. Each experiment was repeated thrice. The result happened to be the average of three independent observations. The error bars indicated the standard error of the mean. P value < 0.05 (denoted with *) indicated the significant difference in results between the cuminaldehyde treated and untreated microbial cultures. B Growth curve analysis. Microbial cultures were collected from both cuminaldehyde treated and untreated growth media at different time gap followed by recording the OD of the same at 600 nm. Each experiment was repeated thrice. The result happened to be the average of three independent observations. The error bars suggested the standard error of the mean
Cuminaldehyde showed efficient antibiofilm activity against Pseudomonas aeruginosa
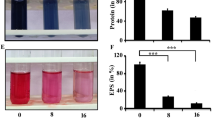
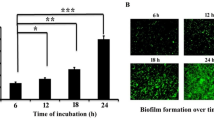
The exploration of molecules in attenuating microbial biofilm formation has been found to draw paramount attention in recent times as molecules interfering with biofilm formation could compromise microbial pathogenesis remarkably (Gupta et al. 2016). Since cuminaldehyde exhibits various biological activities, in this study, the antibiofilm activity of cuminaldehyde was tested against Pseudomonas aeruginosa. The existing literature documented that CV staining assay could be used effectively to address the antibiofilm activity of a compound (Mukherjee et al. 2013; Paul et al. 2021). Hence, in the current study, CV assay was used to understand the antibiofilm activity of cuminaldehyde (if any) against Pseudomonas aeruginosa. The CV assay result indicated that the tube in which the cells were not exposed to cuminaldehyde retained the maximum CV stain (Fig. 3A). However, the intensity of CV staining got reduced considerably when the cells were treated with cuminaldehyde (Fig. 3A). The result further explained that the maximum biofilm formation took place in the control set in which the cells were not exposed to cuminaldehyde (Fig. 3B). However, the maximum inhibition (~ 75%) in biofilm formation occurred when the cells were exposed to cuminaldehyde at a concentration of 60 µg/mL (Fig. 3B). Since DMSO was used as a vehicle to increase the solubility of cuminaldehyde, we had tested whether DMSO (1.2% v/v) could show any biofilm-attenuating activity against Pseudomonas aeruginosa. The result of the CV staining assay revealed that DMSO did not show any antibiofilm activity against the test organism (data not shown). Thus, the observations of the CV staining assay revealed that cuminaldehyde could show promising antibiofilm activity against Pseudomonas aeruginosa. To validate the observations of the CV assay, total biofilm protein was measured under the presence and absence of cuminaldehyde as the determination of the total biofilm protein could give an indirect estimation about the extent of microbial population developed in a biofilm (Tribedi and Sil 2014; Das et al. 2016; Chakraborty et al. 2021). In this regard, we observed a considerable variation in the measurement of the total biofilm protein between the cuminaldehyde treated and untreated cells (Fig. 3C, D). The result of the protein assay manifested that the highest biofilm protein was recovered from the cuminaldehyde untreated cells (Fig. 3C, D). However, the degree of total biofilm protein got reduced by ~ 15%, ~ 21% and ~ 38% under the exposure of cuminaldehyde at a concentration of 20, 40 and 60 µg/mL, respectively (Fig. 3C, D). It is noteworthy to mention that the CV staining assay and total biofilm protein measurement assay followed the same pattern validating the antibiofilm activity of cuminaldehyde (Fig. 3B, D). Furthermore, efforts were put together to measure the production of EPS by Pseudomonas aeruginosa under different conditions. EPS, being an effective indicator of biofilm development plays an important role in holding the integrity of the Pseudomonas biofilm structure (Gupta et al. 2016). The exisiting literature revealed that interference in EPS production could inhibit the microbial biofilm formation considerably (Tribedi and Sil 2014; Bhattacharyya et al. 2018). Hence, we investigated whether cuminaldehyde could decrease the microbial production of EPS. The EPS secretion by the test organism was found to be the highest when the cells were not exposed to cuminaldehyde (Fig. 3E). However, the microbial EPS production got decreased by ~ 7%, ~ 23% and ~ 40% when the cells were treated with cuminaldehyde at a concentration of 20, 40 and 60 µg/mL, respectively (Fig. 3E). Thus, the results indicated that cuminaldehyde reduced the production of EPS that resulted in the inhibition of microbial biofilm formation. Furthermore, the antibiofilm activity of cuminaldehyde was confirmed by observing the cuminaldehyde treated and untreated cells under a fluorescence microscope. The result indicated that there was a remarkable variation in the degree of microbial biofilm formation between the cuminaldehyde treated and untreated cells (Fig. 3F). In the control set, the cells showed prominent clusters resembling efficient biofilm development over the surface. However, the degree of such clusters got reduced substantially when the cells were exposed to cuminaldehyde (Fig. 3F). Hence, the results demonstrated that cuminaldehyde showed promising biofilm-attenuating activity against Pseudomonas aeruginosa.
The sub-MIC doses of cuminaldehyde showed efficient antibiofilm activity against Pseudomonas aeruginosa. Pseudomonas aeruginosa was challenged with the sub-MIC doses of cuminaldehyde while growing in sterile LB. In the control set, cells were grown in the absence of cuminaldehyde. Post incubation, CV assay, estimation of total biofilm protein and measurement of EPS were carried out to examine the degree of microbial biofilm formation of the test organism under different conditions. A CV staining profile. The degree of microbial biofilm formed over the glass tubes were initially stained with CV and subsequently dissolved into glacial acetic acid. B CV assay. The intensity of violet color developed in the test tubes were measured by determining the OD at 630 nm. C Total biofilm protein profile. Post incubation, the extent of microbial biofilm developed over the glass tubes was analyzed by performing Lowry assay. D Determination of the total biofilm protein. The intensity of the color developed in the tubes of both cuminaldehyde treated and untreated sets was quantified by recording their OD at 750 nm. E EPS profile. The amount of EPS produced by the test organism under different conditions were measured by the phenol–sulphuric acid method. Each experimental set was repeated thrice. The error bars indicated the standard error of the mean. To denote the statistical difference between the control and treated sets, P values were calculated using ANOVA software. The P values < 0.001, < 0.01, and < 0.05 were denoted with (***) and (**) and (*), respectively. F Fluorescence microscopic image analysis. Glass coverslips were taken out from both cuminaldehyde treated and untreated growth media and viewed under a fluorescence microscope after staining with acridine orange. The result happened to be a representative of the three independent experiments
Antibiofilm activity of cuminaldehyde could be attributed to the accumulation of ROS in Pseudomonas aeruginosa
Previous literature revealed that the cellular accumulation of ROS could play an important role in the inhibition of microbial biofilm formation (Dwivedi et al. 2014; Paul et al. 2021). It was reported that the 3,6-di(pyridin-2-yl)-1,2,4,5-tetrazine (pytz)-capped silver nanoparticles (TzAgNPs) were found to generate ROS in microbial cells that resulted in the inhibition of biofilm formation (Chakraborty et al. 2018). In a recent report, we noticed that 1,4-naphthoquinone also exhibited biofilm inhibition by generating ROS in microorganisms (Paul et al. 2021). It is noteworthy to mention that one phytochemical namely thymoquinone also inhibited Pseudomonas biofilm by accumulating ROS (Chakraborty et al. 2021). Hence, in the present study, we hypothesized that cuminaldehyde-mediated biofilm inhibition could be attributed to the ROS generation in Pseudomonas aeruginosa. Thus, efforts were concentrated to study the effect of cuminaldehyde on the ROS profile of the test organism. It was observed that with the gradual increase in the concentrations of the test compound, the DCF signal got increased considerably (Fig. 4A). This result indicated that the increasing concentrations (up to 60 µg/mL) of cuminaldehyde could significantly increase the generation of ROS in the test organism (Fig. 4A). It was also noted that the maximum generation of ROS took place when the cells were exposed to cuminaldehyde at a concentration of 80 µg/mL (Fig. 4A). However, this concentration (80 µg/mL) of cuminaldehyde was not selected for the biofilm inhibition studies as the said concentration (80 µg/mL) affected the microbial growth profile considerably (see Fig. 2A). Hence, microbial biofilm inhibition studies were conducted with cuminaldehyde at or below the concentrations of 60 µg/mL as the mentioned concentrations did not show any antimicrobial activity. To understand the influence of ROS accumulation on biofilm inhibition, we had exposed an equal number of cells to varying concentrations of cuminaldehyde and ascorbic acid. After the incubation, the extent of biofilm formation of Pseudomonas aeruginosa under different conditions was determined by performing CV assay. The result of the CV assay mentioned that the maximum biofilm mass was observed in the control set in which the cells were neither treated with cuminaldehyde nor ascorbic acid (Fig. 4B). We also observed that the highest microbial biofilm inhibition (~ 75%) took place when the cells were treated with cuminaldehyde at a concentration of 60 µg/mL (Fig. 4B). However, when the cuminaldehyde-treated cells were further exposed to different concentrations (25 and 50 µg/mL) of ascorbic acid, the potential of biofilm-forming ability of the test organism got increased by ~ 45% and ~ 58%, respectively (Fig. 4B). Thus, the result revealed that ascorbic acid could efficiently restore the biofilm-forming ability of the cuminaldehyde-treated organisms. In this context, we also investigated whether ascorbic acid alone could make any impact on the biofilm profile of the organism. Towards this direction, an equal number of cells were grown either in the presence (25 and 50 µg/mL) or absence of ascorbic acid. However, cuminaldehyde was not added to it. The result of the CV assay revealed no significant difference in the biofilm mass between the ascorbic acid (25 and 50 µg/mL) treated and untreated cells (Fig. 4B). Hence, the observation demonstrated that ascorbic acid alone could not make any impact on microbial biofilm formation (Fig. 4B). Therefore, the result suggested that ROS accumulation was found to reduce the microbial biofilm formation considerably. Taken together, the results indicated that the cuminaldehyde treatment was found to generate ROS in Pseudomonas aeruginosa that resulted in the inhibition of microbial biofilm formation.
Cellular accumulation of ROS efficiently inhibited the microbial biofilm formation of Pseudomonas aeruginosa. A Cellular ROS profile. Cells were collected from both cuminaldehyde treated and untreated growth media followed by determining the cellular accumulation of ROS using DCFDA method. B Microbial biofilm profile. Cells were independently grown in autoclaved LB supplemented with different combinations of cuminaldehyde and ascorbic acid. Post incubation, planktonic cells were removed and the degree of biofilm mass formed over the test tubes were measured by following the CV assay. The result demonstrated the average of three independent observations. Error bars indicated the standard error of the mean. To denote the statistical difference between the control and treated sets, P values were calculated using ANOVA software. P values < 0.001, < 0.01, and < 0.05 were denoted with (***) and (**) and (*), respectively
Ascorbic acid exhibited growth restoration of cuminaldehyde-treated organisms
The results so far revealed that lower concentrations of cuminaldehyde (up to 60 µg/mL) did not exhibit antimicrobial activity whereas the higher concentration (150 µg/mL) of the same compound exhibited substantial antimicrobial activity against Pseudomonas aeruginosa. To understand the underlying cause of the antimicrobial activity of cuminaldehyde against Pseudomonas aeruginosa, a similar number of cells were grown in LB supplemented with different combinations of cuminaldehyde and ascorbic acid. Post incubation, the extent of microbial growth was analyzed in each tube wherein we observed no difference in the turbidity of the microbial cultures between the cuminaldehyde treated (60 µg/mL) and untreated growth media (Fig. 5A). However, the turbidity of the microbial cultures was found to be the lowest when the cells were incubated with the MIC dose (150 µg/mL) of the compound (Fig. 5A). Interestingly, when ascorbic acid was added to this set, the turbidity or microbial growth was seen to re-establish again (Fig. 5A). Furthermore, the extent of microbial growth in each tube was measured by recording the OD of the microbial cultures at 600 nm. The result revealed no significant difference in OD between the cuminaldehyde treated (60 µg/mL) and untreated growth media (Fig. 5B). As expected, almost no microbial growth was observed when the cells were exposed to the MIC dose (150 µg/mL) of the compound. However, when the cuminaldehyde (150 µg/mL)-treated cells were exposed to ascorbic acid, the microbial growth was seen to be restored by ~ 58% (Fig. 5B). Previous literature reported that ascorbic acid could be used as an antioxidant agent to scavenge ROS in microorganisms leading to the restoration of microbial growth under oxidative stress (Chakraborty et al. 2018). Thus, the result suggested that the treatment of ascorbic acid was found to restore the growth of cuminaldehyde-treated cells. Hence, the present report indicated that the antimicrobial activity of cuminaldehyde could be attributed to the accumulation of ROS in Pseudomonas aeruginosa.
Ascorbic acid exhibited growth restoration of cuminaldehyde-treated cells. A Microbial growth profile. The pattern of microbial growth of Pseudomonas aeruginosa was analysed under the influence of various combinations of cuminaldehyde and ascorbic acid. B Determination of the microbial growth. Post incubation, microbial culture was separately collected from each growth media followed by measuring the OD at 600 nm. Each experiment was repeated three times. The average value was taken from three independent experiments. Error bars indicated the standard error of mean. To denote the statistical difference between the control and treated sets, P values were calculated using ANOVA software. P values < 0.01 and < 0.001 were denoted with (**) and (***), respectively. P values > 0.05 did not show any significant difference with the control, hence marked as N.S (no statistical difference)
Cuminaldehyde exhibited a reduction in the secretion of virulence factor from Pseudomonas aeruginosa
Biofilm cells were reported to secrete various virulence factors including protease enzymes during microbial pathogenesis (Adonizio et al. 2008; Chakraborty et al. 2020). Protease enzymes belong to hydrolytic enzymes that hydrolyze the host proteins and thereby augment microbial invasion and pathogenesis (Das et al. 2016). Hence, in the present study, we aimed to examine whether cuminaldehyde could reduce the production of protease enzymes from the test organism. To do the test, the azo-casein degrading assay was performed wherein the amount of azo dye released after the enzymatic hydrolysis of the casein was measured. The result showed that the highest protease production was seen when the cells were not exposed to cuminaldehyde (Fig. 6). However, the azo-casein degrading activity got decreased by ~ 40% when the cells were exposed to cuminaldehyde at a concentration of 60 µg/mL (Fig. 6). The current results demonstrated that cuminaldehyde not only prevents biofilm formation, but also inhibits the production of protease enzymes from microorganisms.
Cuminaldehyde showed a considerable inhibition in the proteolytic activity of Pseudomonas aeruginosa. The proteolytic activity of Pseudomonas aeruginosa under the presence and absence of cuminaldehyde was determined using azo-casein as a substrate. Each experiment was conducted three times and their average was presented. The error bars indicated the standard error of the mean. To denote the statistical difference between the control and treated sets, P values were calculated using ANOVA. P values < 0.05 and < 0.01 were denoted with (*) and (**), respectively
Conclusion
The emergence of biofilm-mediated drug resistance has been found to be a major concern in the context of sustainable management of public healthcare. Towards this direction, cuminaldehyde, a natural phytochemical identified in Cuminum cyminum exhibited promising antibiofilm activity against Pseudomonas aeruginosa by generating ROS. We also noticed that the compound did not display any antimicrobial activity while exhibiting antibiofilm activity. Hence, the compound could be used alone or in combination with other bioactive agents to curb the biofilm-linked infections caused by Pseudomonas aeruginosa.
Availability of data and material
All data generated or analysed during this study are included in this article. The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Adonizio A, Kong KF, Mathee K (2008) Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob Agents Chemother 52:198–203
Bhattacharyya P, Agarwal B, Goswami M, Maiti D, Baruah S, Tribedi P (2018) Zinc oxide nanoparticle inhibits the biofilm formation of Streptococcus pneumoniae. Antonie Van Leeuwenhoek 111:89–99
Briviba K, Klotz LO, Sies H (1997) Toxic and signaling effects of photochemically or chemically generated singlet oxygen in biological systems. Biol Chem 378:1259–1265
Chakraborty P, Joardar S, Ray S, Biswas P, Maiti D, Tribedi P (2018) 3, 6-Di (pyridin-2-yl)-1, 2, 4, 5-tetrazine (pytz)-capped silver nanoparticles (TzAgNPs) inhibit biofilm formation of Pseudomonas aeruginosa: a potential approach toward breaking the wall of biofilm through reactive oxygen species (ROS) generation. Folia Microbiol 63:763–772
Chakraborty P, Dastidar DG, Paul P, Dutta S, Basu D, Sharma SR, Basu S, Sarker RK, Sen A, Sarkar A, Tribedi P (2020) Inhibition of biofilm formation of Pseudomonas aeruginosa by caffeine: a potential approach for sustainable management of biofilm. Arch Microbiol 202:623–635
Chakraborty P, Paul P, Kumari M, Bhattacharjee S, Singh M, Maiti D, Dastidar DG, Akhter Y, Kundu T, Das A, Tribedi P (2021) Attenuation of Pseudomonas aeruginosa biofilm by thymoquinone: an individual and combinatorial study with tetrazine-capped silver nanoparticles and tryptophan. Folia Microbiol (praha) 66:255–271
Chen Q, Hu X, Li J, Liu P, Yang Y, Ni Y (2011) Preparative isolation and purification of cuminaldehyde and p-menta-1,4-dien-7-al from the essential oil of Cuminum cyminum L. by high-speed counter-current chromatography. Anal Chim Acta 689:149–154
Clinical and Laboratory Standards Institute (2005) Performance standards for antimicrobial susceptibility testing: fifteenth informational supplement M100-S15. CLSI, Wayne
Cortes ME, Bonilla JC, Sinisterra RD (2011) Biofilm formation, control and novel strategies for eradication. Sci against Microbial Pathog Commun Curr Res Technol Adv 2:896–905
Das MC, Paul S, Gupta P, Tribedi P, Sarkar S, Manna D, Bhattacharjee S (2016) 3-Amino-4-aminoximidofurazan derivatives: small molecules possessing antimicrobial and antibiofilm activity against Staphylococcus aureus and Pseudomonas aeruginosa. J Appl Microbiol 120:842–859
Dubois M, Gilles KA, Hamilton JK, Rebers PT, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Dwivedi S, Wahab R, Khan F, Mishra YK, Musarrat J, Al-Khedhairy AA (2014) Reactive oxygen species-mediated bacterial biofilm inhibition via zinc oxide nanoparticles and their statistical determination. PLoS ONE 9:e111289
Gambino M, Cappitelli F (2016) Mini-review: biofilm responses to oxidative stress. Biofouling 32:167–178
Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribedi P (2016) Biofilm, pathogenesis and prevention-a journey to break the wall: a review. Arch Microbiol 198:1–15
Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57:395–418
Khatoon Z, McTiernan CD, Suuronen EJ, Mah TF, Alarcon EI (2018) Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 4(12):e01067
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Mishra R, Panda AK, De Mandal S, Shakeel M, Bisht SS, Khan J (2020) Natural anti-biofilm agents: strategies to control biofilm-forming pathogens. Front Microbiol 11:566325
Monteiro-Neto V, de Souza CD, Gonzaga LF, da Silveira BC, Sousa NCF, Pontes JP, Santos DM, Martins WC, Pessoa JFV, Júnior ARC, Almeida VSS, de Oliveira NMT, de Araújo TS, Maria-Ferreira D, Mendes SJF, Ferro TAF, Fernandes ES (2020) Cuminaldehyde potentiates the antimicrobial actions of ciprofloxacin against Staphylococcus aureus and Escherichia coli. PLoS ONE 15:e0232987
Mukherjee K, Tribedi P, Mukhopadhyay B, Sil AK (2013) Antibacterial activity of long-chain fatty alcohols against mycobacteria. FEMS Microbiol Lett 338:177–183
Paul P, Chakraborty P, Chatterjee A, Sarker RK, Dastidar DG, Kundu T, Sarkar N, Das A, Tribedi P (2021) 1, 4-Naphthoquinone accumulates reactive oxygen species in Staphylococcus aureus: a promising approach towards effective management of biofilm threat. Arch Microbiol 203:1183–1193
Roy R, Tiwari M, Donelli G, Tiwari V (2018) Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9:522–554
Sharma D, Misba L, Khan AU (2019) Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control 8:76
Sowbhagya HB (2013) Chemistry, technology, and nutraceutical functions of cumin (Cuminum cyminum L): an overview. Crit Rev Food Sci Nutr 53:1–10
Tariq S, Wani S, Rasool W, Shafi K, Bhat MA, Prabhakar A, Shalla AH, Rather MA (2019) A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens. Microb Pathog 134:103580
Tribedi P, Sil AK (2014) Cell surface hydrophobicity: a key component in the degradation of polyethylene succinate by Pseudomonas sp. AKS2. J Appl Microbiol 116:295–303
Wimpenny J, Manz W, Szewzyk U (2000) Heterogeneity in biofilms. FEMS Microbiol Rev 24:661–671
Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S (2011) Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol 13:1705–1717
Acknowledgements
The authors would like to thank Ms. Ahana Chatterjee for critically reading the manuscript. We are extremely grateful to Dr. Debasish Maity (Professor, Department of Human Physiology, Tripura University, India) for sharing the organism with us to carry out our research activities.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
SC, PP, PC, SD, RKS performed the experiments and wrote the manuscript. SS analysed the results and wrote the manuscript. AD and PT conceived the idea, designed the experiments, analysed the results and edited the manuscript critically.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that they do not have any conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
About this article
Cite this article
Chatterjee, S., Paul, P., Chakraborty, P. et al. Cuminaldehyde exhibits potential antibiofilm activity against Pseudomonas aeruginosa involving reactive oxygen species (ROS) accumulation: a way forward towards sustainable biofilm management. 3 Biotech 11, 485 (2021). https://doi.org/10.1007/s13205-021-03013-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-03013-1