Abstract
Gram-positive and Gram-negative bacteria often develop biofilm through different mechanisms in promoting pathogenicity. Hence, the antibiofilm molecule needs to be examined separately on both organisms to manage the biofilm threat. Since the antibiofilm activity of piperine against Staphylococcus aureus was already reported; here, we aimed to examine the antibiofilm activity of it against Pseudomonas aeruginosa. P. aeruginosa is an opportunistic Gram-negative pathogen that can cause several healthcare-associated infections by exploiting biofilm. Several experiments like crystal violet assay, estimation of total protein, measurement of extracellular polymeric substance, and microscopic analysis confirmed that lower concentrations (8 and 16 µg/mL) of piperine could inhibit the microbial biofilm formation considerably. Besides, it could also reduce the secretion of virulence factors from P. aeruginosa. Further investigation showed that the cell surface hydrophobicity and microbial motility of the test organism got reduced under the influence of piperine. Piperine exposure was found to increase the accumulation of reactive oxygen species (ROS) that resulted in the inhibition of biofilm formation. Furthermore, the molecular simulation studies suggested that piperine could affect the quorum sensing network of P. aeruginosa. Towards this direction, we noticed that piperine treatment could decrease the expression of the quorum sensing gene (lasI) that resulted in the inhibition of biofilm formation. Besides biofilm inhibition, piperine was also found to disintegrate the pre-existing biofilm of P. aeruginosa without showing any antimicrobial property to the test organism. Thus, piperine could be used for the sustainable protection of public-healthcare by compromising the biofilm assembly of P. aeruginosa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic human pathogen which is listed as priority pathogen in Research and Development by the World Health Organization in 2017 [1]. It causes statistically striking chronic diseases every year including chronic wound infections, urinary tract infections, cystic fibrosis, endocarditis, osteomyelitis etc. which are mainly attributed to its inherent resistance to antimicrobials [2]. Literature survey revealed that many factors including biofilm formation have been found to play a major role in exhibiting drug resistance [2]. Biofilm is an agglomeration of microorganisms that often get stabilized by the self-secreted extracellular polymeric substances (EPS) comprising of polysaccharides, proteins, lipids, eDNA etc. [3]. Approximately, 85–90% of biofilm mass is composed of such EPS matrix thereby making the biofilm cells impervious to external challenges and therapeutics [4]. P. aeruginosa has been found to promote the secretion of several virulence factors such as rhamnolipid, pyocyanin, and proteolytic enzymes during biofilm development. Microbial biofilm allows bacteria to spread pathogenesis either by activating the virulence profile or offering resistance to a wide range of antibiotics [2]. Towards this direction, several natural molecules are being investigated to disarm and eradicate such biofilm-associated challenges. In this respect, we have considered one such natural molecule named as piperine which is an alkaloid of black pepper. According to literature, piperine shows various health benefits such as anti-diabetic, anti-inflammatory, anti-cancerous, anti-arthritic, and anti-parasitic effects [5, 6]. Besides, piperine was reported to be an excellent bio-active compound which can enhance the bio-availability of several antibiotics such as ampicillin and nor-floxacin [7]. It was documented that piperine could show antimicrobial as well as antibiofilm activity against several biofilm-forming organisms such as Staphylococcus aureus and Klebsiella pneumoniae [8, 9]. Furthermore, piperine was also found to enhance the antibiofilm activity of streptomycin, kanamycin, and amikacin against Salmonella enterica [10]. The existing literature revealed that an antibiofilm molecule could behave differently with the change in the test organism [11,12,13,14]. In this connection, we have found that L-tryptophan, a natural amino acid, could efficiently inhibit the biofilm formation of a Gram-negative organism, P. aeruginosa [12]. However, we have also noticed that instead of biofilm inhibition, the same amino acid was found to promote the biofilm formation of a Gram-positive organism, Staphylococcus aureus [13]. Literature report also stated that molecule named as maipomycin A showed efficient antibiofilm activity against Gram-negative bacteria but failed to show such effect against the microbial biofilm formation of Gram-positive bacteria [14]. Besides, molecule named as Aryl rhodanines inhibited microbial biofilm formation of Gram-positive bacteria but remained ineffective against biofilm formation of Gram-negative bacteria [11]. Towards this direction, we observed the antibiofilm activity of piperine against a Gram-positive S. aureus [8]. However, the antibiofilm property of the same against a Gram-negative P. aeruginosa remains unexplored. Therefore, in the present report, efforts have been targeted to examine the antibiofilm activity of piperine (if any) against P. aeruginosa. Further investigation suggested that piperine could show inhibition in biofilm formation as well as disintegration of the pre-existing biofilm of P. aeruginosa.
Materials and Methods
Microbial Strain, Growth Media, and Culture Conditions
In the current report, the Gram-negative bacteria, P. aeruginosa (MTCC 424) was used as the test organism. The bacterium was cultivated in Luria Bertani (LB) media purchased from Himedia. The test organism was grown at 37 °C for 24 h to achieve its optimum growth. Piperine, the test compound used in the present report was dissolved in an organic solvent, dimethyl sulfoxide (DMSO) to prepare the stock solution.
Determination of the Antibiofilm Property of Piperine Against P. aeruginosa
Crystal violet (CV) assay happens to be a well acknowledged method that can be used to measure the extent of microbial biofilm formation under different conditions [15]. To understand the effect of piperine on microbial biofilm formation, an overnight culture of P. aeruginosa (1 × 105 CFU/mL) was inoculated into several tubes containing 5 mL of sterile LB media. Thereafter, the tested concentrations (8 and 16 µg/mL) of piperine were added to it and the tubes were subsequently incubated for 24 h at 37 °C. After the incubation, the non-adherent planktonic cells were discarded from each tube, rinsed with sterile double distilled water, and air-dried. Then, the dried tubes were stained with 5 mL CV solution (0.4%) for 30 min. After that, the CV solution was discarded and the tubes were then rinsed with sterile double distilled water. Finally, to measure the intensity of adhered biofilm cells on the glass surface, 33% glacial acetic acid was added to each tube and the optical density (OD) of the same was recorded at 630 nm.
Estimation of Total Biofilm Protein of P. aeruginosa
The extent of microbial biofilm formation could be measured indirectly by estimating the total biofilm protein as a directly proportional relationship prevails between protein estimation and microbial abundance [16]. To understand the influence of piperine on the biofilm association, overnight culture (1 × 105 CFU/mL) of the test bacteria was inoculated into several tubes containing 5 mL of sterile LB media. Subsequently, the respective culture tubes were challenged with the tested concentrations (8 and 16 µg/mL) of piperine. In this connection, a control set was prepared in which the cells were grown without being exposed to piperine. All the tubes were then incubated at 37 °C for 24 h. Post incubation, the LB media containing the planktonic cells were discarded and washed with sterile double distilled water. Thereafter, 5 mL of 0.3 (N) NaOH was added to all the tubes and boiled for 30 min at 100 °C. The prepared suspensions were then centrifuged at 8000 rpm for 10 min and the respective supernatant was subjected to protein estimation by following Lowry method [17].
Determination of EPS
EPS happens to be a common indicator of microbial biofilm assembly [18]. To determine the amount of EPS produced by the test organism under the presence and absence of piperine, an equal number of cells (1 × 105 CFU/mL) from an overnight culture were added in sterile LB media. To it, the respective concentrations (8 and 16 µg/mL) of piperine were added to the test tubes and all the tubes were further incubated for 24 h at 37 °C. Post incubation, the non-adherent planktonic cells were discarded from each tube and subsequently rinsed with sterile phosphate buffer saline. Then, 5 mL Congo red (1% w/v) solution was added to the tubes and incubated for 30 min at dark. Thereafter, the adhered cells were washed and DMSO was subsequently added to the tubes to record the absorbance of the same at 490 nm.
Analysis of Microbial Colonization Under Fluorescence Microscope
Fluorescence microscopic analysis was carried out to observe the effect of piperine on the microbial biofilm formation [19]. In this direction, overnight inoculum (1 × 105 CFU/mL) of the test organism was inoculated into several tubes containing sterile LB media. To it, sterile cover slips were added so that microorganisms could develop biofilm over the surface. The tubes were later incorporated with the tested concentrations (8 and 16 µg/mL) of piperine. A control set was also prepared wherein the test organism was grown in the absence of piperine. After an incubation of 24 h at 37 °C, the cover slips were recovered aseptically and stained with acridine orange (4 µg/mL). Then, the stained cover slips were observed under a fluorescence microscope (under green FITC filter) to analyze the pattern of microbial colonization over the cover slips.
Measurement of Protease Activity
Microbial biofilm has often been found to enhance the production of several virulence factors including the release of protease enzyme [2]. To determine the effect of piperine on the production of protease enzyme from the test organism, equal number of the test organism (1 × 105 CFU/mL) was allowed to grow into glass tubes containing 5 mL of autoclaved LB media. Thereafter, the tubes were supplemented with various concentrations (8 and 16 µg/mL) of piperine. Alongside, another set was also kept where the bacterial inoculum was not treated with any concentrations of piperine. All the experimental sets were then incubated for 24 h at 37 °C. After that, all the experimental sets were centrifuged at 10,000 rpm for 15 min. Afterward, 2 mL of supernatant from each set was mixed with 500 μL of azo-casein (0.3%) solution followed by incubating them at 37 °C for 1 h. Then, the reaction of the same was stopped by adding 10% trichloroacetic acid. Afterward, the solution of each tube was centrifuged at 10,000 rpm for 5 min and the absorbance of the resulting supernatant was measured at 440 nm.
Measurement of Rhamnolipid Production
The amount of rhamnolipid produced by the test organism under the presence and absence of piperine was measured by following the orcinol-sulfuric acid assay as described in Cheng et al. (2017) [20]. To perform this assay, an overnight grown culture of P. aeruginosa was inoculated in several tubes containing sterile LB media. To this, the tested concentrations (8 and 16 µg/mL) of piperine were added to the respective tubes. Alongside, a control set was also prepared in which the cells were grown in the absence of piperine. All the tubes were then incubated for 48 h at 37 °C. Thereafter, the planktonic cells were decanted and the adhered biofilm cells were collected followed by dissolving them in sterile phosphate buffer saline (PBS) solution. Then, 1 mL biofilm cell suspension was mixed with 9 mL of orcinol (0.19%) in 53% sulfuric acid solution. After that, the mixture was incubated at 80 °C for 30 min followed by cooling at room temperature for 15 min. Thereafter, the absorbance of the same was measured at 420 nm.
Measurement of Pyocyanin Production
The amount of pyocyanin produced by the test organism under the presence and absence of piperine was determined by following the protocol described in Chatterjee et al. (2020) [21]. In brief, equal inoculums (1 × 105 CFU/mL) of the test organism were inoculated in sterile LB media. To it, different concentrations (0, 8 and 16 µg/mL) of piperine were added. The tubes were then incubated for 4 days at 37 °C. Post incubation, each culture was centrifuged at 10,000 rpm for 10 min. Then, the supernatant was separately collected and mixed with chloroform in the ratio 5:3. After vigorous vortexing, 1.5 mL of 0.2 M HCl was added to each tube. Thereafter, the upper pink layer was collected and its absorbance was recorded at 520 nm.
Measurement of Cell Surface Hydrophobicity
Cell surface hydrophobicity of the test organism under the presence and absence of piperine was determined by bacterial adherence to hydrocarbon (BATH) assay as described by Rosenberg et al. (1981) [22] with minor modification. At first, test organism (1 × 105 CFU/mL) was inoculated in sterile glass tubes having 5 mL of autoclaved LB broth. Selected concentrations (8 and 16 µg/mL) of piperine were added to it. In the control set, no piperine was added. After overnight incubation of all the test tubes at 37 °C, planktonic cells were discarded from each tube and subsequently washed with sterile double distilled water. Following this step, 5 mL of sterile phosphate buffer (pH 8.0) was added to each tube and vortexed vigorously. After that, the suspension collected from each tube was centrifuged at 6000 rpm for 10 min. Afterward, pellet was collected, suspended in 3.4 mL of sterile double distilled water and OD of each suspension was measured at 420 nm. Then, 0.6 mL of chloroform was mixed with the suspension. Following adequate shaking, all the tubes were kept aside for 30 min to allow the phase separation. Finally, the aqueous phase from each tube was collected separately and OD was measured again. The following formula could be used to understand the cell surface hydrophobicity of the test organism:
Measurement of Swarming Motility
To understand the effect of piperine on the swarming motility of the test organism, equal number (1 × 105 CFU/mL) of cells were inoculated in sterile LB media supplemented with selected concentrations (8 and 16 µg/mL) of piperine and was incubated at 37 °C for 24 h. One control set of the test organism was also incubated in similar condition where no piperine was added. After the incubation, an equal number of cells were separately spotted onto the middle of the semi-solid tryptone soy agar media. Then, all the plates were kept in aseptic condition at 37 °C for 24 h. Thereafter, the diameter (in mm) of bacterial motility was measured from the respective spot.
Measurement of the Generation of Reactive Oxygen Species
To estimate the amount of ROS generated by the test organism under the influence of piperine, a DCFDA (2′, 7′-dichlorofluorescein diacetate)-dependent ROS measurement assay kit (ab113851) was used for the same. Cellular esterase enzyme has been reported to deacetylate DCFDA into a non-fluorescent DCF which gets further oxidized to fluorescent DCF under high ROS profile. The amount of fluorescent DCF produced by the organism (if any) under the influence of piperine was recorded through a fluorescence spectrophotometer.
To do the test, overnight grown cells (1 × 108 CFU/mL) were treated with different concentrations (8 and 16 µg/mL) of piperine. A control set was also prepared wherein the cells were not exposed to piperine. Then, all the tubes including the control were incubated at 37 °C for 30 min. Post incubation, the DCFDA exposed cells were centrifuged at 10,000 rpm for 8 min. Thereafter, the supernatant was discarded and cell pellets were collected followed by washing them with sterile Luria broth. Then, the cell pellets were further exposed to the selected concentrations (8 and 16 μg/mL) of piperine. In this connection, some piperine exposed cells were also challenged with ascorbic acid (50 µg/mL). To validate the observations, a negative control was also made in which microbial cells were neither exposed to piperine nor ascorbic acid. Finally, the amount of DCF produced by the test organism under piperine exposure was measured by a fluorescence spectrophotometer [23].
In silicoDocking Study of LasR with Piperine
The PDB file of the quorum-sensing signaling protein LasR (PDB id: 3JPU) of P. aeruginosa was downloaded from the RCSB PDB database (last accessed on 26th December 2021). After the removal of non-standard residues, the protein molecule was prepared with the Quick Prep module of Molecular Operating Environment (MOE) 2019. The Protonated 3D module of MOE 2019 was used to add hydrogen and assign the ionization state of amino acid residues. It was followed by energy minimization to a 0.1-unit RMS gradient to get the final structure of the receptor protein. The DOCK application of MOE 2019 was used for the receptor-ligand docking studies. Initially, the receptor was considered ‘rigid’ and the ‘Triangle Matcher’ replacement method was used for the generation of 30 poses that were screened on the basis of London dG score. It was followed by the refinement of poses with the ‘Induced Fit’ method to get the top five poses based on GBVI/WSA dG score.
Molecular Dynamics Simulations
The molecular dynamics (MD) simulations of 3JPU, and its complex with piperine was performed using NAMD (Nanoscale Molecular Dynamics) program; v 2.8. Amber10: EHT force field was applied and a generalized Born Implicit solvent model was followed for MD simulation. The complex was initially subjected to a restraint energy minimization process where the ligand atoms were kept restrained. It was followed by constant pressure and temperature discrete-time Langevin molecular dynamics [24] for 120 ns. At first, the system was heated to 310 K from 300 K at a time period of 100 ps, and equilibrated at 310 K for 100 ps. Then, the MD simulation was continued for 120 ns at 310 K temperature and 1-atm pressure. Finally, the system was cooled to 300 K at a time period of 100 ps. The equation of motion was discretized at a 2-fs interval. Frames were assembled at 5-ps intervals. A total of 24,060 frames were collected per simulation. The trajectory files were studied with VMD [25]. The VEGA ZZ 3.2.1 program [26] was considered for constructing the data of RMSD, RMSF, gyration radius, and polar surface area (PSA).
Measurement of Quorum Sensing Gene expression by Real-time PCR
The expression of a quorum sensing gene (lasI) under the influence of piperine was measured by real-time PCR as per the protocol published by Paul et al. [27]. To carry out this experiment, an equal number of organisms were allowed to grow separately under the selected concentrations (8 and 16 µg/mL) of piperine. In the control set, similar numbers of cells were grown in sterile LB without piperine. All the experimental sets were incubated at 37 °C for 24 h. After the incubation, similar numbers of biofilm cells were collected from both piperine treated and untreated set. The RNA was extracted from them using the Trizol reagent. The purity of the collected RNA was measured by a spectrophotometer. Next, the entire RNA from each sample was allowed to get transcribed into the first strand of cDNA using the reverse transcriptase enzyme of M-MLV (Takara). Then, the same amount of cDNA (1 mg) was taken from each experimental set and subsequently real-time-amplified using the SYBR Premix kit (Applied Biosystems). In the negative control, a reaction mixture was prepared in which cDNA was not used. The expression level of the quorum sensing gene (lasI) was calculated under the influence of piperine using the comparative threshold cycle method (2 − ΔΔCt) with 16S rRNA as the control gene. The gene-specific primers along with the sequences were mentioned in the supplementary table 1.
Determination of Biofilm-Disintegrating Property of Piperine
To determine whether the tested concentrations (8 and 16 µg/mL) of piperine could disintegrate the pre-formed biofilm of P. aeruginosa, overnight grown inoculum (1 × 105 CFU/mL) of the test organism was allowed to grow in several test tubes having sterile LB media. Sterile cover slips were separately added to the tube for the development of biofilm over it. After an incubation of 24 h at 37 °C, the developed biofilm was challenged with the selected concentrations (8 and 16 µg/mL) of piperine followed by incubating them for another 4 h at 37 °C. One control set was also taken into consideration where no piperine was exposed to pre-developed biofilm but incubated for the same extent of time. Then, the degree of disintegration of pre-existing biofilm under the influence of piperine was measured by conducting CV assay [3]. The total protein recovery assay was also conducted to estimate the extent of biofilm disintegration of the test organism under the exposure of piperine. Besides, the cover slips were separately recovered from the respective tubes and stained with acridine orange (4 µg/mL) to observe the extent of disintegration of the pre-existing biofilm under a fluorescence microscope.
The eDNA Extraction and Quantification
The eDNA happens to be an important component that offers considerable stability to the biofilm [28]. Therefore, to observe the effect of piperine on the eDNA of microbial biofilm, eDNA was extracted from the cells by following the protocol described in Sahu et al. [29]. In brief, similar numbers (1 × 105 CFU/mL) of cells were inoculated into glass petri-plates having 20 mL of sterile LB. All the plates were then incubated for 24 h at 37 °C. After the incubation, the developed biofilm cells were either treated (8 and 16 µg/mL) or untreated with piperine for 4 h at 37 °C. Post incubation, biofilm cells were scrapped from the bottom of the petridish followed by centrifuging them at 8000 rpm for 8 min. After the centrifugation, supernatant was collected as it carries the EPS part. To extract the cell-bound EPS, cell pellets were re-suspended in 500 µL of EDTA (10 mM) and centrifuged again at 8000 rpm for 8 min. Then, the supernatant was collected and pooled with the previous supernatant. The pooled fraction was then mixed with 2.2 volume of chilled absolute ethanol and incubated for 1 h at 8 °C. Afterward, the suspension was centrifuged at 8000 rpm for 8 min wherein the pellet was collected as the source of EPS. The pellet was suspended in 500 µL of Tris–EDTA buffer and 150 µL of ice-cold isopropanol. The suspension was incubated at 4 °C for 3 h followed by centrifugating at 12,000 rpm for 15 min. Then, the pellet was collected, mixed with Tris–EDTA buffer (500 µL), treated with 10 µL of Proteinase K (10 mg/mL) and incubated for another 1 h at 37 °C. After the incubation, 150 µL ice-cold isopropanol was added to it. Then, the suspension was centrifuged at 8000 rpm for 8 min. Finally, the pellet was collected and re-suspended in 50 µL of Tris-buffer. The concentrations and purity of the extracted eDNA was subsequently measured through a spectrophotometer.
Determination of the Antimicrobial Property of the Selected Concentrations of Piperine
To test whether the selected concentrations (8 and 16 µg/mL) of piperine could show any microbial growth inhibitory property, a series of experiments were conducted. In this regard, organisms (1 × 105 CFU/mL) were allowed to grow in sterile LB media (100 mL) for 24 h at 37 °C. After that, various concentrations (8 and 16 µg/mL) of piperine were added to the media. A control set was also prepared in which cells were grown without being exposed to piperine. During the incubation period, microbial cultures were recovered at different time points (2-h interval) followed by recording the OD of the same at 600 nm. After that, the viable microbial counts were determined in both piperine treated and untreated cultures. In this connection, 1 mL microbial culture was collected from each conical flask and serially diluted (10−1 to 10−5) with sterile 0.85% NaCl solution. Thereafter, 100 µL samples were transferred from each dilution to an autoclaved LB agar plate and subsequently spread over the plate. After an overnight incubation at 37 °C, the number of colony forming unit (CFU) of each plate was calculated [3]. In addition, spot assay and clear zone assay were also performed to find out the antimicrobial activity of the selected concentrations (8 and 16 µg/mL) of piperine. To perform the spot assay, 10 µL samples was collected from each dilution and subsequently spotted on autoclaved Luria agar plate followed by incubating the same for 24 h at 37 °C. Post incubation, the developed spot of the respective dilutions was analyzed. Besides, to perform the clear zone assay, 100 µL of P. aeruginosa was spread over different LB agar plates to form bacterial lawn. Then, 3 wells were punched in each plate wherein desired concentrations (0, 8, and 16 µg/mL) of piperine were added. After an overnight incubation at 37ºC, clear zone (if any) around the well was measured.
Analysis of the Effect of Piperine Removal on the Formation of Microbial Biofilm
To examine whether the antibiofilm effect of piperine persists even after its exclusion from the growth media, cells (1 × 105 CFU/mL) were allowed to grow at 37 °C for 24 h in the presence (8 and 16 µg/mL) and absence of piperine. Post incubation, microbial cultures were centrifuged at 8000 rpm for 10 min. After that, supernatants were discarded to remove piperine from the growth media. The cell pellets collected from both piperine treated and untreated set were suspended again in sterile LB media and incubated at 37 °C for another 6 h, 12 h, 18 h, and 24 h. After the incubation, CV assay was conducted to understand the degree of biofilm formation of the test organism under different conditions.
Measurement of Antibiofilm Property of DMSO
As piperine is insoluble in water, DMSO was used as a solvent to prepare the stock solution of the same. Hence, we examined whether DMSO alone could show any antibiofilm activity against the same organism. In this connection, an equal number (1 × 105 CFU/mL) of P. aeruginosa was allowed to grow in sterile LB media (5 mL) for 24 h at 37 °C. After that, instead of piperine, various concentrations (0, 0.08, and 0.16% in v/v) of DMSO were added to the cells. A control set was also incubated under the same condition where DMSO was not introduced. Post incubation, CV assay was performed to understand the antibiofilm effect of DMSO (if any) on the test organism.
Statistical Analysis
Each experiment was repeated three times to achieve statistical confidence. One-way analysis of variance (ANOVA) was used to perform the significance test. The degree of significance was incorporated as p value < 0.05 (*), p value < 0.01 (**), and p value < 0.001 (***) in contrast to control. The p values with more than 0.05 indicated that there were no significant difference and hence presented as N.S. (no statistical difference). A trial version of Minitab 19 was used for the present study.
Results
Piperine Exhibited Effective Antibiofilm Property Against P. aeruginosa
To assess the antibiofilm effect of piperine against P. aeruginosa, a similar number of the bacteria were inoculated in several tubes containing sterile LB media. Thereafter, different concentrations (0, 8, 16, 24, and 32 µg/mL) of piperine were independently added into in. Post incubation, the degree of biofilm formation of the test organism was examined under the absence and presence of piperine by following the CV assay [15]. The result showed that the intensity of CV stain was found to be the highest in the control set whereas the intensity of the stain got decreased considerably under the influence of piperine (Fig. 1A). It was noted that the level of biofilm formation got decreased by ~ 45% and ~ 55% when the cells were exposed to 8 and 16 µg/mL concentrations of piperine, respectively (Fig. 1B). Beyond this concentration (16 µg/mL), no such change in biofilm inhibition was observed (Fig. 1B). Since piperine was dissolved in DMSO, the effect of DMSO on the microbial biofilm formation was also taken into account. The result indicated that the cells treated and untreated with DMSO revealed the similar extent of biofilm formation (Supplementary Fig. 1). Thus, it appears that instead of DMSO, piperine could show the biofilm inhibiting property. To further confirm the antibiofilm effect of piperine, total biofilm protein was estimated under the absence and presence of piperine by following the Lowry assay [17]. The result showed that the intensity of color corresponding to the availability of protein was found to vary between the piperine treated and untreated conditions (Fig. 1C). We observed that the highest amount of protein was measured in the control set where piperine was not added (Fig. 1D). However, with the increase of the concentrations of piperine, the estimation of total biofilm protein got reduced significantly (Fig. 1D). The level of protein recovery got decreased by ~ 39% and ~ 53% when the cells were treated with piperine at 8 and 16 µg/mL concentrations, respectively (Fig. 1D). To gain further confidence, the effect of piperine on EPS production from the test organism was measured under the absence and presence of piperine by following Congo red assay. Congo red has been reported to interact with the polysaccharide part of EPS [30]. The result showed that the maximum color was developed in the control set (Fig. 1E). However, the intensity of the same got reduced considerably under the influence of piperine (Fig. 1E). Furthermore, the highest reduction (~ 88%) in the EPS production was noticed when the test bacterium was treated with piperine at a concentration of 16 µg/mL (Fig. 1F). Thus, the result suggested that piperine could inhibit the formation of biofilm by inhibiting the production of EPS. To gain more confidence, the change in the degree of biofilm formation of the test organism under the influence of piperine was analyzed through a fluorescence microscope. The microscopic image analysis suggested that the maximum number of biofilm clusters were spotted in the control set which were free from piperine (Fig. 1G). However, on increasing the concentrations of the test compound, the clusters were seen to decrease considerably (Fig. 1G). Thus, the results demonstrated the antibiofilm effect of piperine against P. aeruginosa. To comprehend the transient or lasting role of piperine on the biofilm of P. aeruginosa, cells were either treated or untreated with piperine for 24 h at 37 °C. Post incubation, cells were harvested from both piperine treated and untreated growth media. After that, the cells were re-inoculated in sterile LB media and incubated further up to 24 h. In this connection, we observed that with the advancement of time, the biofilm-forming pattern remained almost same for the cells which were collected from both piperine treated and untreated growth media (Supplementary Fig. 2). Thus, the result indicated that the effect of piperine on microbial biofilm inhibition was found to be transient.
Piperine showed efficient biofilm inhibitory activity against P. aeruginosa. A similar number of cells of P. aeruginosa were grown in sterile LB media under the presence and absence of the selected concentrations of piperine. After the desired period of incubation, planktonic cells were discarded followed by measuring the antibiofilm activity of piperine against the test organism. A Biofilm profile. The CV staining profile of both piperine treated and untreated cells. B Estimation of biofilm formation. Crystal violet assay was conducted to estimate the percentage of microbial biofilm formed on the glass surface under the presence and absence of piperine. C Total biofilm protein profile. Total biofilm protein profile of both piperine treated and untreated cells. D Estimation of total biofilm protein. Total biofilm protein on the glass surface under the influence of piperine was measured by Lowry method. E EPS profile. Congo red staining profile of both piperine treated and untreated cells. F Measurement of EPS. Congo red assay was conducted to quantify the extent of EPS produced by P. aeruginosa under the piperine exposed and unexposed conditions. Each experiment was performed three times. Error bars represented standard error of the mean. The p values < 0.001 were marked with (***) to show the statistical difference among the observations. G. Fluorescence microscopic image analysis. The cover slips were recovered from both piperine treated and untreated culture media, stained with acridine orange and observed under a fluorescence microscope. The figure is representative image of 20 different fields and from three independent experiments
Piperine Curbed the Secretion of Virulence Factors from P. aeruginosa Biofilm
The existing literature revealed that microorganisms associated with biofilm could secrete a number of virulence factors thereby promoting pathogenicity considerably [31]. Hence, in the present study, besides biofilm inhibition, efforts were put through to understand the effect of piperine on the production of several virulence factors namely protease secretion, rhamnolipid and pyocyanin synthesis from the test organism. In this connection, we noticed that the highest level of protease activity was obtained in the control set which was not exposed to piperine (Fig. 2A). However, the extent of protease activity was found to be reduced by ~ 24% when the cells were treated with piperine at a concentration of 16 µg/mL (Fig. 2A). The extent of the production of rhamnolipid and pyocyanin were also measured under the presence and absence of the test compound. As expected, the result revealed that the amount of rhamnolipid production was found to be the highest in the cells which were not treated with piperine (Fig. 2B). On contrary, the rhamnolipid production got reduced by ~ 35% when the cells were exposed to piperine at a concentration of 16 µg/mL (Fig. 2B). The pyocyanin production by the test organism followed the same trend wherein we observed that the exposure of piperine could reduce the pyocyanin production by ~ 46% (Fig. 2C). Hence, the results suggested that the tested concentrations of piperine could considerably curb the virulence factors production from P. aeruginosa.
Piperine attenuated the secretion of various virulence factors from P. aeruginosa. An equal number of the test organism was inoculated into the sterile LB media either challenged with piperine or left untreated with the same. After an incubation of 24 h at 37 °C, protease activity, rhamnolipid and pyocyanin production was measured in both piperine treated and untreated growth media. A Determination of protease activity. Protease activity of both piperine exposed and unexposed cells were quantified by performing azo-casein assay. B Rhamnolipid measurement. The percentage of rhamnolipid secreted by P. aeruginosa under the influence of piperine was determined by orcinol-sulfuric acid assay. C Pyocyanin measurement. The percentage of pyocyanin produced by the test organism under the presence and absence of piperine was determined. Each experiment was repeated thrice. The result represented the average of the three independent observations. Error bars represented standard error of the mean. The p values < 0.01 were marked with (**), and p values < 0.001 were marked with (***) to show the statistical difference among the observations
Elucidation of the Underlying Mechanism of Microbial Biofilm Inhibition Under the Influence of Piperine
Existing literature revealed that cell surface hydrophobicity could play an important role in the formation of microbial biofilm [32]. In the present study, efforts were given to understand the effect of piperine on the cell surface hydrophobicity of the test organism. Hence, cells were grown under the absence and presence of piperine followed by measuring the cell surface hydrophobicity of the test organism. The result showed that the cell surface hydrophobicity was found to be the highest in the cells which were not exposed to piperine (Fig. 3A). The cell surface hydrophobicity got decreased by ~ 20% when the cells were treated with piperine at a concentration of 16 µg/mL (Fig. 3A). Therefore, the result stated that the selected concentrations of piperine could reduce the cell surface hydrophobicity of the test organism. Furthermore, the contour plot confirmed that the increase in piperine concentration affected the cell surface hydrophobicity of the organism that resulted in the inhibition of microbial biofilm formation (Supplementary Fig. 3). Besides, efforts were put together to test whether the selected concentrations of piperine could compromise microbial motility (swarming) of P. aeruginosa. The result exhibited that the highest motility was noticed in the semi-solid agar plate which was not challenged with piperine (Fig. 3B). On treatment with piperine, the bacterial cells were observed to impede its motility (Fig. 3B). To confirm this result, the diameter (in mm) of microbial motility was measured from the center of inoculation under the absence and presence of piperine. It was observed that the microbial motility got decreased by ~ 56% when the cells were treated with piperine at a concentration of 16 µg/mL (Fig. 3C). Thus, it could be stated that the selected concentrations of piperine effectively inhibited the swarming motility of the P. aeruginosa. Since swarming motility has been found to promote microbial biofilm formation, the interference in swarming motility under the influence of piperine could inhibit the biofilm formation considerably. Further literature survey revealed that reactive oxygen species (ROS) could inhibit microbial biofilm formation considerably [8, 19]. To understand the effect of piperine on the accumulation of ROS, similar numbers of cells were allowed to grow under the absence and presence of piperine. Post incubation, cells were collected followed by measuring the accumulation of ROS using DCFDA (2′, 7′-dichlorofluorescein diacetate)-dependent ROS measurement assay kit (ab113851). The result showed that the maximum (5-folds increase from control) ROS accumulation took place when the cells were treated with piperine at a concentration of 16 µg/mL (Fig. 3D). However, the cells showed a twofolds decrease in ROS accumulation when the cells were treated with both piperine and ascorbic acid (Fig. 3D). Furthermore, by following the CV assay, it was found that the bacterial cells which were not exposed to either piperine or ascorbic acid, showed the maximum biofilm formation (Fig. 3E). However, on exposing the cells to piperine, inhibition in biofilm formation was noticed significantly (Fig. 3E). On adding ascorbic acid to the piperine treated cells, it was found that the biofilm-forming ability of the test organism got restored efficiently (Fig. 3E). Hence, the results indicated that piperine enhanced the accumulation of ROS in the bacterial cells that could affect the biofilm-forming ability of the test organism.
Piperine showed antibiofilm activity by targeting cell surface hydrophobicity, microbial motility and ROS profile of the test organism. A similar number of test organisms were either challenged with piperine or left untreated with same. All the cultures were subsequently incubated at 37 °C for 24 h. Post incubation, different mechanisms were explored to observe the antibiofilm activity of piperine against the test organism. A Cell surface hydrophobicity profile. The percentage of cell surface hydrophobicity of the test organism under the presence and absence of piperine was measured by BATH assay. B Microbial motility profile. Piperine treated and untreated culture was spotted individually on semi-solid TSB agar plates followed by incubating the same at 37 °C for 24 h. Post incubation, the microbial motility was observed under the presence and absence of piperine. C Measurement of colony diameter. The zone of diameter of each microbial colony either treated or untreated with piperine was measured (in mm) independently. D. ROS profile. DCFDA-based assay kit was followed to measure the accumulated ROS in both piperine exposed and unexposed cells. E The effect of ROS on biofilm profile. CV assay was taken into account to analyze the effect of ROS accumulation on the degree of biofilm formation. Each experiment was repeated thrice. Error bars represented the standard error of the mean. The p values < 0.05 were marked with (*), p values < 0.01 were marked with (**), and p values < 0.001 were marked with (***) to show the statistical difference among the observations
Docking Analysis Revealed that Piperine Could Interfere with Quorum Sensing Property of P. aeruginosa
The literature report stated that quorum sensing could play an important role in the development of biofilm over a surface [2]. Hence, in the present study, efforts were targeted to understand whether piperine could affect the quorum sensing profile of the test organism. In this connection, the docking studies has been performed to indicate the putative interaction/s between piperine and LasR (PDB Id: 3JPU) protein. The three-dimensional image indicated prominent interactions between piperine and 3JPU (Fig. 4A). The two-dimensional image also confirmed the interactions between piperine and 3JPU (Fig. 4B). The binding energy of the interactions between piperine and 3JPU was found to be -7.92 kcal/mole. Piperine formed alkyl-alkyl interactions with TRP 88 and LEU 110 residue of 3JPU (Fig. 4B). The pi-alkyl interaction was observed between piperine with ALA 127, ALA 50, LEU 40, TYR 47, and VAL 76 residue of 3JPU (Fig. 4B). The pi-electron-rich benzene ring of piperine was found to show pi-pi T-shaped interaction and pi-anion interaction with TYR 56 and ASP 73 resides of 3JPU, respectively (Fig. 4B). We also observed a carbon-hydrogen bond between VAL 76 residue of 3JPU and the carbonyl O-atom of piperine (Fig. 4B). All these favorable interactions might contribute to the exhibition of high affinity of piperine binding to 3JPU. To gain further confidence of the interaction between piperine and 3JPU, molecular dynamics study was carried out. At first, the system was heated to 310 K from 300 K for a time period of 100 ps, and equilibrated at 310 K for 100 ps. Then, the MD simulation was continued for 120 ns at 310 K temperature and 1-atm pressure. Finally, the system was cooled to 300 K for a time period of 100 ps. The RMSD plot of free 3JPU and 3JPU-piperine complex were shown in Fig. 5A. The average RMSD value of free 3JPU (2.80 ± 0.09 Å) and 3JPU-piperine complex (3.41 ± 0.21 Å) were found to be almost similar. Since there was no abrupt change in RMSD value, it was suggested that a stable complex has been formed between 3JPU and piperine [33]. Besides RMSD, the RMSF plot was also prepared and presented in Fig. 5B. The RMSF value of 3JPU (0.9632 ± 0.425 Å) and 3JPU-piperine complex (0.8736 ± 0.315 Å) was found be close suggesting no substantial difference between them. The reduction in RMSF values of different amino acid residues within 1 to 60 could be attributed to the interactions between piperine and some amino acid residues of 3JPU. Though there was an increase in RMSF values of amino acid residues within 95 to 105 and 132 to 145, the stability of the complex has not been compromised as the plot of Radius of Gyration (Fig. 5C) and PSA (Fig. 5D) showed overlapping graph for free 3JPU and 3JPU-piperine complex. The average values of Gyration Radius of 3JPU (14.65 ± 0.07 Å) alone and 3JPU-piperine complex (14.66 ± 0.06 Å) were found to be similar. There was no considerable difference observed in the PSA values of 3JPU (1826.63 ± 76.62 Å) and 3JPU-piperine complex (1891.71 ± 69.34 Å). The existing literature supported that the obtained results confirmed the stability of 3JPU-piperine complex [34]. To understand the effect of piperine on the quorum sensing linked gene expression, we selected lasI gene as it comes under the regulation of LasR protein [27]. In this regard, we found that the expression of lasI gene got reduced considerably with the increasing concentrations of piperine (Fig. 5E). To gain confidence, we also measured the expression of 16S rRNA gene under the presence and absence of piperine as this gene (16S rRNA) does not offer any function to quorum sensing property of the test organism. The result indicated that piperine treated and untreated cells showed a similar level of 16S rRNA gene expression profile (Fig. 5E). Hence, the results demonstrated that piperine was found to affect the quorum sensing property that could lead to compromise the microbial biofilm formation.
Molecular dynamics study of the interactions between piperine and 3JPU complex. A RMSD plot. B RMSF plot of backbone residues. C Plot for Gyration radius at the different time points of study. D Plot surface area (PSA) analysis at different time points of study. E Real-time PCR profile of lasI gene. Equal number of cells was allowed to grow at 37 °C for 24 h under the presence and absence of piperine. Post incubation, RNA was isolated from both piperine exposed and unexposed cells and the expression of the quorum sensing gene (lasI gene) was estimated by real-time PCR. Each experiment was repeated thrice. Error bars represented standard error of the mean. The p values < 0.01 were marked with (**) to show the statistical difference among the observations
Piperine Could Disintegrate the Pre-existing Biofilm of P. aeruginosa
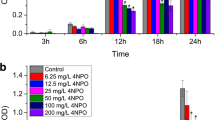
To test the potential of piperine as a biofilm-disintegrating agent, efforts were given to develop biofilm first followed by challenging the same with piperine. In this connection, an overnight grown culture of P. aeruginosa was allowed to grow in sterile LB media at 37 °C for 24 h. The extent of biofilm formed by the test organism at different time intervals (6, 12, 18, 24 h) was measured by following the CV assay (Fig. 6A). The result indicated that with the increase in the time of incubation, biofilm formation of the test bacteria gradually increased (Fig. 6A). The maximum biofilm formation took place when the cells were incubated for 24 h (Fig. 6A). Beyond this time, no further increase in biofilm development was observed (data not shown). To reassure this observation, fluorescence microscopic analysis was carried out. To do the same, sterile LB media was prepared in several tubes in which sterile cover slips were incorporated. All the tubes were then inoculated with the test organism and further incubated at 37 °C for 24 h. At different time points (6, 12, 18, 24 h), the respective cover slips were recovered, stained with acridine orange and viewed under a fluorescence microscope. It was noticed that the biofilm clusters gradually got increased with the progression of time (Fig. 6B). Thus, all the results suggested that the bacteria could develop substantial biofilm after an incubation of 24 h. Thereafter, efforts were directed to understand whether piperine could disintegrate the pre-existing biofilm of the test organism. The concentrations of piperine selected for this study was 8 and 16 µg/mL as these concentrations showed effective biofilm inhibition against this bacterium. In this context, the developed biofilm cells were exposed to piperine (8 and 16 µg/mL) for another 4 h at 37 °C. Post incubation, CV assay was performed in which we observed the maximum staining for the biofilm cells which were not challenged with piperine (Fig. 7A). However, the intensity of CV staining got reduced considerably when the pre-existing biofilm was challenged with piperine (Fig. 7A). Further analysis revealed that piperine at a concentration of 16 µg/mL showed ~ 40% disintegration of pre-existing biofilm (Fig. 7B). To confirm the results of CV assay, protein assay was also performed wherein we observed the presence of maximum biofilm protein in the control set (Fig. 7C). However, the amount of the same got reduced under the influence of piperine (Fig. 7C). Further studies revealed that piperine (16 µg/mL) could exhibit ~ 37% reduction in total biofilm protein recovery in contrast to the control set (Fig. 7D). To gain more confidence, fluorescence microscopic study was conducted between the piperine treated and untreated conditions. The result stated that the test organism could develop substantial biofilm in the absence of piperine (Fig. 7E). However, the test compound was found to reduce such clusters indicating the biofilm-disintegrating property of it against P. aeruginosa (Fig. 7E). Hence, the results indicated that piperine could disintegrate the pre-formed biofilm considerably. In literature, it was revealed that eDNA plays an important role in holding the biofilm integrity [3]. To understand the role of eDNA in biofilm integrity of P. aeruginosa, the developed biofilm was incubated with either DNase I or left untreated with the same. The result showed that the pre-existing biofilm got disintegrated under the influence of DNase I (Fig. 8A). The result suggested that eDNA could hold the biofilm structure efficiently. Then, we measured the amount of eDNA associated with biofilm under the absence and presence of piperine. The result indicated that the amount of eDNA was found to be the highest when the developed biofilm was not challenged with piperine (Fig. 8B). However, the amount of eDNA got reduced significantly when the pre-existing biofilm was incubated with piperine (Fig. 8B). To gain further confidence, the eDNA collected from both piperine treated and untreated biofilm was run through an agarose gel electrophoresis. The result suggested that the eDNA collected from the piperine untreated cells showed prominent band intensity (Fig. 8C). The band intensity got reduced noticeably for the eDNA which was collected from piperine treated cells (Fig. 8C). Further, the densitometry analysis was undertaken in which we noticed that the intensity of eDNA band of piperine untreated cells was found to be ~ 3.8 times higher than the eDNA band of piperine treated cells (Fig. 8D). Thus, the results demonstrated the biofilm-disintegrating property of piperine against P. aeruginosa.
P. aeruginosa efficiently developed biofilm with the advancement of time. A similar number of cells were allowed to grow at 37 °C for various time periods. In every set, cover slips were added aseptically. After the desired time period, planktonic cells were removed and the degree of biofilm formed by the test organism was analyzed. A CV assay profile. CV assay was followed to analyze the degree of biofilm formed by the test organism at different time intervals. Each experiment was repeated thrice. Error bars represented the standard error of the mean. The p values < 0.05 were marked with (*), p values < 0.01 were marked with (**), and p values < 0.001 were marked with (***) to show the statistical difference among the observations. B Fluorescence microscopic image analysis. Cover slips were recovered from the growth media at different time intervals followed by staining them with acridine orange and observed under a fluorescence microscope. The figure is representative image of 20 different fields of three independent experiments
Piperine showed efficient disintegration of the pre-formed biofilm of P. aeruginosa. An equal numbers of test organism were allowed to grow in fresh LB media at 37 °C for 24 h to develop biofilm. Then, the developed biofilm was either treated or untreated with piperine for 4 h at 37 °C. After that, planktonic cells were discarded, followed by measuring the disintegrating property of piperine against the test organism. A Biofilm profile. CV staining profile of both piperine treated and untreated biofilm cells. B Evaluation of biofilm disintegration profile. CV assay was taken into consideration to estimate the degree of biofilm disintegration under the influence of piperine. C Total biofilm protein profile. Total biofilm protein profile of both piperine treated and untreated cells were addressed by Lowry assay. D Estimation of total biofilm protein. Total biofilm protein was recovered, subjected to Lowry assay, and finally OD was recorded for both piperine treated and untreated cells. Each experiment was repeated thrice. Error bars represented standard error of the mean. The p values < 0.05 were marked with (*), p values < 0.01 were marked with (**), and p values < 0.001 were marked with (***) to show the statistical difference among the observations. E. Fluorescence microscopic image analysis. The degree of the change of the pre-formed microbial biofilm on the glass cover slips under the influence of piperine was observed under a fluorescence microscope. The figure is representative image of 20 different fields and from three independent experiments
Piperine exhibited considerable reduction in the production of eDNA during biofilm disintegration. A Role of eDNA in biofilm disintegration. An equal number of cells were allowed to grow for 24 h at 37 °C in glass tubes containing sterile LB to form biofilm over the tubes. Then, the pre-formed biofilms were either treated or untreated with DNase I. After that, the degree of biofilm remaining over the tubes under different conditions was determined by following CV assay. B Estimation of eDNA concentration. The eDNA was separately extracted from the piperine treated and untreated biofilm followed by measuring the concentration of the eDNA by recording the absorbance at 260 nm. Each experiment was repeated thrice. Error bars represented standard error of the mean. The p values < 0.001 were marked with (***) to show the statistical difference among the observations. C Gel electrophoresis of the extracted eDNA. The eDNA extracted from the piperine treated and untreated biofilm was run through an agarose gel electrophoresis. D Densitometry analysis. The density of each band of eDNA was measured by densitometry analysis. Each experiment was repeated thrice. Error bars represented standard error of the mean. P values < 0.05 were marked with (*) to show the statistical difference among the observations
The Tested Concentrations of Piperine Did not Exhibit any Antimicrobial Property Against P. aeruginosa
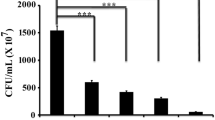
The current report so far stated that piperine at the concentrations of 8 and 16 µg/mL showed promising antibiofilm activity. However, the antimicrobial effect of the same concentrations of piperine is yet to be addressed against the test organism. To understand the effect of the tested concentrations (8 and 16 µg/mL) of piperine on the growth profile of the test organism, the pattern of the microbial growth curve was compared between the piperine treated and untreated cells. The result demonstrated that the bacterial growth curve remained almost similar for both piperine treated and untreated cells (Fig. 9A). This result indicated that the tested concentrations of piperine were found to show no antimicrobial action. Furthermore, the microbial viability was determined and compared between the piperine treated and untreated conditions. In this connection, we observed no significant difference in the viable microbial counts between the piperine treated and untreated cells (Fig. 9B). Additionally, clear zone assay and spot assays were also performed to affirm that piperine did not exhibit any antimicrobial activity against the test bacteria. In case of clear zone assay, bacterial lawn was prepared by pour plate method on LB agar plates. Thereafter, three holes were prepared in the plates which were loaded with different concentrations of piperine (0, 8, and 16 µg/mL). After the desired period of incubation, we did not notice any clear zone around any of the created holes (Fig. 9C). In the case of spot assay, bacterial cells (in equal number) were collected from both piperine treated and untreated set, serially diluted and further incubated for 24 h at 37 °C. The result did not show any effective variation in the growth of the bacterial culture in either of the treated or untreated samples (Fig. 9D). Thus, the results collectively confirmed that the tested doses (8 and 16 µg/mL) of piperine did not exhibit any antimicrobial property against the test organism.
Selected concentrations (8 and 16 µg/mL) of piperine did not exhibit any microbial growth inhibitory activity against P. aeruginosa. A Microbial growth curve analysis. A similar number of the test organism was inoculated in the sterile LB media under the presence and absence of piperine. Microbial cultures were taken out from all the growth media at regular time intervals followed by measuring the OD of the same at 600 nm. B Viable microbial count analysis. An equal volume of microbial culture was individually collected from both piperine-treated and untreated growth media. After that, viable microbial count was determined following CFU method. Each of the experiments was performed thrice. Error bars represented standard error of the mean. The p values above 0.05 were marked as N. S (no significant difference). C Clear zone analysis. A microbial lawn of the test organism was prepared by spreading the culture of the organism over the sterile LB agar plate. Different concentrations of piperine were loaded into the wells and incubated for 24 h at 37 °C to observe the development of the clear zone (if any) around the wells. D Microbial spot assay. An equal volume of microbial culture was separately collected from both piperine-treated and untreated growth media followed by performing the serial dilution of the same. After that, serially diluted samples were spotted on a sterile LB agar plate and incubated overnight at 37 °C to observe the change in microbial growth pattern (if any)
Discussion
P. aeruginosa, a Gram-negative human commensal, has been reported to cause chronic bacterial biofilm mediated infections at an alarming rate [1, 35]. The inefficacy of the conventional drugs on the biofilm matrix has become a major area of concern in the healthcare units. Thus, the current priority is to explore certain natural molecule/s that can effectively control the chronic infections by compromising the biofilm network. In this regard, piperine, a natural component of black pepper, has been selected for the present study as this molecule has been reported to show promising antibiofilm activity against Staphylococcus aureus [8]. However, the effect of the same on the microbial biofilm formation of P. aeruginosa is still unexplored. Thus, in the present study, efforts were targeted to understand the effect of piperine on the microbial biofilm profile of P. aeruginosa. Towards this direction, we started with the lower concentrations (8 and 16 µg/mL) of piperine for the biofilm management studies as it’s concentrations beyond 20 µg/mL started showing cytotoxicity [8]. Hence, several experiments such as crystal violet assay, estimation of total biofilm protein, EPS extraction assay and fluorescence microscopic observations were taken into consideration to confirm the biofilm inhibitory potential of the selected concentrations (8 and 16 µg/mL) of piperine against the test organism. It has also been reported in the literature that microorganisms associated with biofilm often release various virulence factors that resulted in the promotion of microbial pathogenicity [3]. Hence, in the present report, we examined whether the tested concentrations of piperine could affect the production of virulence factors (protease enzyme, rhamnolipid and pyocyanin production) from P. aeruginosa. Microorganisms associated with biofilm have been found to promote the production of protease enzyme that could help the causative organisms to get through the host tissue [30]. In this regard, we noticed that piperine was found to attenuate the secretion of protease enzyme from the test organism. Apart from the protease enzyme, we also looked into the secretion of rhamnolipid and pyocyanin from the test organism under the presence and absence of piperine. Rhamnolipid and pyocyanin happen to alter the cell membrane integrity and electron transport chain, respectively thereby affecting the host cell viability considerably [36, 37]. Hence, in the present study, we measured the production of rhamnolipid and pyocyanin from the test organism under the presence and absence of piperine wherein we noticed that piperine could inhibit the production of the mentioned virulence factors (rhamnolipid and pyocyanin) considerably. Thus, the results so far revealed that piperine could show efficient antibiofilm activity against the test organism. However, the underlying mechanism of microbial biofilm inhibition under the influence of piperine remains unexplored. In this connection, the literature survey suggested that cell surface hydrophobicity could be an important target for the management of biofilm as it has been reported to influence the biofilm formation of S. aureus considerably [3]. Therefore, by performing BATH assay, it was found that piperine could inhibit the cell surface hydrophobicity of P. aeruginosa. To understand the effect of the cell surface hydrophobicity on biofilm formation under the influence of piperine, a contour plot was constructed. Contour plot happens to be a 2-dimensional view of three variables. The contour plot suggested that with the rise of piperine concentrations, the cell surface hydrophobicity got decreased that resulted in the inhibition of microbial biofilm formation. Further studies revealed that microorganism showing effective motility could enhance biofilm formation considerably [12]. In this report, piperine was examined whether the compound could interfere with the swarming motility of the test organism. The result suggested that piperine was found to interfere with the microbial swarming motility that could reduce the microbial colonization leading to the formation of a compromised biofilm. Besides, existing literature revealed that accumulation of reactive oxygen species (ROS) could affect the microbial biofilm formation substantially [15]. Hence, in the present report, we measured the accumulation of ROS in the test organism under the presence and absence of piperine. We observed that piperine was found to accumulate ROS in the test organism that could attenuate the biofilm formation considerably. Further literature suggested that quorum sensing could be a sensitive target for the management of biofilm challenges as it happens to regulate various activities including microbial motility and biofilm formation [38, 39]. Quorum sensing happens to be a communication process in which microorganisms respond to external signal through a density dependent fashion [40]. After reaching a threshold density, microorganisms start secreting auto-inducers that lead to control the expression of quorum sensing genes [12]. P. aeruginosa has been reported to use LasI and LasR protein to practice quorum sensing [41]. LasI protein has been found to synthesize auto-inducer which interacts with the LasR protein and thereby activates LasR protein [42]. Activated LasR (when bound with auto-inducer) protein then promotes the expression of quorum sensing genes including lasI [38]. Towards exploring the underlying mechanism, we investigated whether piperine could affect the quorum sensing property of the test organism. To understand the same, we performed docking studies wherein we noticed that piperine could interact with the LasR protein as several interactions were noticed between them. To gain further confidence, molecular dynamics study was carried out in which we observed that a stable interaction was displayed between piperine and LasR protein. Then, to observe the impact of this interaction, we hypothesized that the expression of lasI gene could be affected under the influence of piperine. The result revealed that the extent of lasI gene expression got reduced under the increasing concentrations of piperine. Therefore, the result suggested that piperine could interfere with the quorum sensing property of the test organism. Thus, the results demonstrated that piperine was found to inhibit the microbial biofilm formation by targeting cell surface hydrophobicity, accumulating ROS, attenuating microbial motility, and affecting quorum sensing property of the test organism. Besides, efforts were put through to understand whether piperine could disintegrate the pre-existing biofilm of P. aeruginosa as the developed biofilm often exhibits serious threats to public healthcare [43]. Thus, in the present study, cells were grown first to develop biofilm followed by challenging the same with piperine. In this regard, we noticed that the organism could develop substantial biofilm after 24 h of growth as fluorescence microscopic image clearly stated that the highest degree of microbial clusters was spotted after 24 h of growth. After that, the developed biofilm was incubated with piperine to understand the effect of piperine on biofilm disintegration. Experiments like CV assay, estimation of total biofilm protein and fluorescence microscopic observations revealed that piperine showed efficient biofilm disintegrating potential against the test organism. We noticed that the extent of biofilm disintegration got increased with the rising concentrations of piperine. Existing literature documented that extracellular polymeric substances (EPS) could play an important role in holding the integrity of biofilm [44]. Further investigation indicated that among the several bio molecules spotted in EPS, eDNA happens to play a pivotal role as the degradation of the eDNA could promote the disintegration of the pre-existing biofilm [3]. To understand the effect of eDNA on the pre-existing biofilm stability of the test organism, the developed biofilm was separately incubated with the presence and absence of DNase I. In this regard, we observed that the degradation of eDNA by DNase I was found to exhibit biofilm disintegration considerably. Thus, the result stated that the availability of eDNA could increase the stability of biofilm considerably. Since piperine showed microbial biofilm disintegration, we hypothesized that the amount of eDNA might be reduced with the increasing concentrations of piperine. To understand the same, we recovered the eDNA from the test microorganisms growing under the presence and absence of piperine. In this connection, several experiments like eDNA concentration determination through spectrophotometer, eDNA band profile investigation through agarose gel electrophoresis and eDNA band intensity analysis through densitometry study revealed that the amount of eDNA got reduced considerably with the rising concentrations of piperine. All these experiments demonstrated that piperine could show efficient biofilm inhibition and disintegration against the test organism. To understand the antimicrobial effect (if any) of the tested concentrations (8 and 16 µg/mL) of piperine against the test organism, we performed a series of experiments (growth curve analysis, microbial viability estimation through colony counting, clear zone assay and spot assay) in which we observed that the selected concentrations (8 and 16 µg/mL) of piperine did not show any antimicrobial activity while showing antibiofilm action against P. aeruginosa.
Conclusion
Beneficial effects of plant derived natural molecules are found to contribute towards the emergence and spread of biofilm-associated health concern. Towards this direction, in this study, piperine, a plant derived alkaloids demonstrated evidence to become an excellent antibiofilm compound against a Gram-negative opportunistic human pathogen, P. aeruginosa by offering both biofilm inhibiting as well as disintegrating property.
Data Availability
The datasets produced during the present study are available from the corresponding author based on the reasonable request.
Code Availability
Minitab 19 (30 days’ trial version) was used for the present study to analyze the statistical observations.
References
Thi, M. T. T., Wibowo, D., & Rehm, B. H. (2020). Pseudomonas aeruginosa biofilms. International Journal of Molecular Sciences, 21(22), 8671.
Gupta, P., Sarkar, S., Das, B., Bhattacharjee, S., & Tribedi, P. (2016). Biofilm, pathogenesis and prevention-a journey to break the wall: A review. Archives of Microbiology, 198(1), 1–15.
Paul, P., Das, S., Chatterjee, S., Shukla, A., Chakraborty, P., Sarkar, S., Maiti, D., Das, A., & Tribedi, P. (2021). 1, 4-Naphthoquinone disintegrates the pre-existing biofilm of Staphylococcus aureus by accumulating reactive oxygen species. Archives of Microbiology, 203(8), 4981–4992.
Singh, S., Datta, S., Narayanan, K. B., & Rajnish, K. N. (2021). Bacterial exo-polysaccharides in biofilms: Role in antimicrobial resistance and treatments. Journal of Genetic Engineering and Biotechnology, 19(1), 1–19.
Bang, J. S., Oh, D. H., Choi, H. M., Sur, B. J., Lim, S. J., Kim, J. Y., Yang, H. I., Yoo, M. C., Hahm, D. H., & Kim, K. S. (2009). Anti-inflammatory and anti-arthritic effects of piperine in human interleukin 1β-stimulated fibroblast-like synoviocytes and in rat arthritis models. Arthritis Research & Therapy, 11(2), 1–9.
Quijia, C. R., & Chorilli, M. (2020). Characteristics, biological properties and analytical methods of piperine: A review. Critical Reviews in Analytical Chemistry, 50(1), 62–77.
Janakiraman, K., & Manavalan, R. (2008). Studies on effect of piperine on oral bioavailability of ampicillin and norfloxacin. African Journal of Traditional, Complementary and Alternative Medicines, 5(3), 257–262.
Das, S., Paul, P., Chatterjee, S., Chakraborty, P., Sarker, R. K., Das, A., Maiti, D., & Tribedi, P. (2022). Piperine exhibits promising antibiofilm activity against Staphylococcus aureus by accumulating reactive oxygen species (ROS). Archives of Microbiology, 204(1), 1–11.
BissoNdezo, B., TokamKuaté, C. R., & Dzoyem, J. P. (2021). Synergistic antibiofilm efficacy of thymol and piperine in combination with three aminoglycoside antibiotics against Klebsiella pneumoniae biofilms. The Canadian Journal of Infectious Diseases & Medical Microbiology, 2021(7029944), 1–8.
Tokam Kuate, C. R., BissoNdezo, B., & Dzoyem, J. P. (2021). Synergistic antibiofilm effect of thymol and piperine in combination with aminoglycosides antibiotics against four Salmonella enterica Serovars. Evidence-based Complementary and Alternative Medicine, 2021(1567017), 1–9.
Chung, P. Y., & Toh, Y. S. (2014). Anti-biofilm agents: Recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog Dis, 70(3), 231–239.
Chakraborty, P., Daware, A. V., Kumari, M., Chatterjee, A., Bhattacharyya, D., Mitra, G., Akhter, Y., Bhattacharjee, S., & Tribedi, P. (2018). Free tryptophan residues inhibit quorum sensing of Pseudomonas aeruginosa: A potential approach to inhibit the development of microbial bioflm. Archives of Microbiology, 200, 1419–1425.
Ghosh, S., Qureshi, A., & Purohit, H. J. (2019). D-Tryptophan governs biofilm formation rates and bacterial interaction in P. mendocina and S. aureus. J Biosci, 44(1), 1–10.
Zhang, J., Liang, X., Zhang, S., Song, Z., Wang, C., & Xu, Y. (2021). Maipomycin A, a novel natural compound with promising anti-biofilm activity against Gram-negative pathogenic bacteria. Frontiers in Microbiology, 11, 598024.
Mukherjee, K., Tribedi, P., Mukhopadhyay, B., & Sil, A. K. (2013). Antibacterial activity of long-chain fatty alcohols against Mycobacteria. FEMS Microbiology Letters, 338(2), 177–183.
Chakraborty, P., Paul, P., Kumari, M., Bhattacharjee, S., Singh, M., Maiti, D., Dastidar, D. G., Akhter, Y., Kundu, T., Das, A., & Tribedi, P. (2021). Attenuation of Pseudomonas aeruginosa biofilm by thymoquinone: An individual and combinatorial study with tetrazine-capped silver nanoparticles and tryptophan. Folia Microbiologica, 66(2), 255–271.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the Folin-phenol reagent. Journal of Biological Chemistry, 193, 265–275.
Decho, A. W., & Gutierrez, T. (2017). Microbial extracellular polymeric substances (EPSs) in ocean systems. Frontiers in Microbiology, 8, 922.
Paul, P., Chakraborty, P., Chatterjee, A., Sarker, R. K., Dastidar, D. G., Kundu, T., Sarkar, N., Das, A., & Tribedi, P. (2021). 1,4-Naphthoquinone accumulates reactive oxygen species in Staphylococcus aureus: A promising approach towards effective management of biofilm threat. Arch Microbial, 203(3), 1183–1193.
Cheng, T., Liang, J., He, J., Hu, X., Ge, Z., & Liu, J. (2017). A novel rhamnolipid-producing Pseudomonas aeruginosa ZS1 isolate derived from petroleum sludge suitable for bioremediation. AMB Express, 7(1), 1–14.
Chatterjee, P., Sass, G., Swietnicki, W., & Stevens, D. A. (2020). Review of potential Pseudomonas weaponry, relevant to the Pseudomonas-Aspergillus interplay, for the mycology community. Journal of Fungi, 6(2), 81.
Rosenberg, M., Perry, A., Bayer, E. A., Gutnick, D. L., Rosenberg, E., & Ofek, I. (1981). Adherence of Acinetobacter calcoaceticus RAG-1 to human epithelial cells and to hexadecane. Infection and Immunity, 33, 29–33.
Dwivedi, S., Wahab, R., Khan, F., Mishra, Y. K., Musarrat, J., & Al-Khedhairy, A. A. (2014). Reactive oxygen species mediated bacterial biofilm inhibition via zinc oxide nanoparticles and their statistical determination. PLoS ONE, 9, e111289.
Gronbech-Jensen, N., & Farago, O. (2014). Constant pressure and temperature discrete-time Langevin molecular dynamics. The Journal of Chemical Physics, 141(19), 194108.
Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD-visual molecular dynamics. J Molec Graph, 14, 33–38.
Pedretti, A., Villa, L., & Vistoli, G. (2002). VEGA: A versatile program to convert, handle and visualize molecular structure on windows-based PCs. Journal of Molecular Graphics and Modelling, 21(1), 47–49.
Paul, P., Chakraborty, P., Sarker, R. K., Chatterjee, A., Maiti, D., Das, A., Mandal, S., Bhattacharjee, S., Dastidar, D. G., & Tribedi, P. (2021). Tryptophan interferes with the quorum sensing and cell surface hydrophobicity of Staphylococcus aureus: a promising approach to inhibit the biofilm development. 3 Biotech, 11(8), 376.
Sarkar, S. (2020). Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA. Applied Microbiology and Biotechnology, 104(15), 6549–6564.
Sahu, P. K., Iyer, P. S., Oak, A. M., Pardesi, K. R., & Chopade, B. A. (2012). Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. Scientific World Journal, 2012, 1–10.
Rollefson, J. B., Stephen, C. S., Tien, M., & Bond, D. R. (2011). Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. Journal of Bacteriology, 193(5), 1023–1033.
Granato, E. T., Ziegenhain, C., Marvig, R. L., & Kümmerli, R. (2018). Low spatial structure and selection against secreted virulence factors attenuates pathogenicity in Pseudomonas aeruginosa. ISME Journal, 12(12), 2907–2918.
Krasowska, A., & Sigler, K. (2014). How microorganisms use hydrophobicity and what does this mean for human needs? Frontiers in Cellular and Infection Microbiology, 4, 112.
Bharadwaj, K. K., Sarkar, T., Ghosh, A., Baishya, D., Rabha, B., Panda, M. K., Nelson, B. R., John, A. B., Sheikh, H. I., Dash, B. P., Edinur, H. A., & Pati, S. (2021). Macrolactin A as a novel inhibitory agent for SARS-CoV-2 Mpro: Bioinformatics approach. Applied Biochemistry and Biotechnology, 193(10), 3371–3394.
Bharadwaj, K. K., Ahmad, I., Pati, S., Ghosh, A., Sarkar, T., Rabha, B., Patel, H., Baishya, D., Edinur, H. A., Kari, Z. A., Zain, M. R. A. M., & Rosli, W. I. W. (2022). Potent bioactive compounds from seaweed waste to combat cancer through bioinformatics investigation. Frontiers in Nutrition, 9(889276), 1–16.
Maurice, N. M., Bedi, B., & Sadikot, R. T. (2018). Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. American Journal of Respiratory Cell and Molecular Biology, 58(4), 428–439.
Shao, B., Liu, Z., Zhong, H., Zeng, G., Liu, G., Yu, M., Liu, Y., Yang, X., Li, X., Fang, Z., Zhang, J., & Zhao, C. (2017). Effects of rhamnolipids on microorganism characteristics and applications in composting: A review. Microbiological Research, 200, 33–44.
Glasser, N. R., Wang, B. X., Hoy, J. A., & Newman, D. K. (2017). The pyruvate and α-ketoglutarate dehydrogenase complexes of Pseudomonas aeruginosa catalyze pyocyanin and phenazine-1-carboxylic acid reduction via the subunit dihydrolipoamide dehydrogenase. Journal of Biological Chemistry, 292(13), 5593–5607.
Mishra, R., Panda, A. K., De Mandal, S., Shakeel, M., Bisht, S. S., & Khan, J. (2020). Natural antibiofilm agents: Strategies to control biofilm-forming pathogens. Frontiers in Microbiology, 11(566325), 1–23.
Urvoy, M., Labry, C., L’Helguen, S., & Lami, R. (2022). Quorum sensing regulates bacterial processes that play a major role in marine biogeochemical cycles. Frontiers in Marine Science, 9(834337), 1–25.
Rutherford, S. T., & Bassler, B. L. (2012). Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspectives in Medicine, 2(11), a012427.
Steindler, L., Bertani, I., De Sordi, L., Schwager, S., Eberl, L., & Venturi, V. (2009). LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Applied and Environment Microbiology, 75(15), 5131–5140.
Chakraborty, P., Dastidar, D. G., Paul, P., Dutta, S., Basu, D., Sharma, S. R., Basu, S., Sarker, R. K., Sen, A., Sarkar, A., & Tribedi, P. (2020). Inhibition of biofilm formation of Pseudomonas aeruginosa by caffeine: A potential approach for sustainable management of biofilm. Archives of Microbiology, 202(3), 623–635.
Galie, S., García-Gutiérrez, C., Miguélez, E. M., Villar, C. J., & Lombo, F. (2018). Biofilms in the foodindustry: Health aspects and control methods. Frontiers in Microbiology, 9(898), 1–18.
Di Martino, P. (2018). Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol, 4(2), 274.
Acknowledgements
Authors would like to express their deepest regards to Dr. Amlan Das (Research and Development Project Manager at National Institute of Biomedical Genomics, Kalyani, WB, India) for sharing the molecule (piperine) with us to carry out the desired research work.
Author information
Authors and Affiliations
Contributions
SD, PP, DGD, PC, SC, SS, and DM performed the experiments and analyzed the results. PT conceived the idea, designed the experiments, analyzed the results, and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Das, S., Paul, P., Dastidar, D.G. et al. Piperine Exhibits Potential Antibiofilm Activity Against Pseudomonas aeruginosa by Accumulating Reactive Oxygen Species, Affecting Cell Surface Hydrophobicity and Quorum Sensing. Appl Biochem Biotechnol 195, 3229–3256 (2023). https://doi.org/10.1007/s12010-022-04280-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04280-1