Abstract
In this study, nine strains of Pseudomonas aurantiaca and P. chlororaphis, and two isolates of Pseudomonas sp.: At1RP4 and RS-1, were characterized for the in-vitro production of secondary metabolites in LB, DMB, and King’s B media, and of the genes responsible for the production of antagonistic metabolites. Based on 16S rRNA gene sequence, isolates At1RP4 and RS-1 were identified as strains of P. aeruginosa and P. fluorescens. Five phenazine derivatives comprising phenazine, phenazine-1-carboxylic acid (PCA), 2-hydroxyphenazine-1-carboxylic acid (2-OH-Phz-1-COOH), phenazine-1,6-dicarboxylic acid (Phz-1,6-di-COOH), and 2-hydroxyphenazine (2-OH-Phz) were produced by all strains in all three culture media including DMB, King’s B and LB. However, 2,8-dihydroxyphenazine, 6-methylphenazine-1-carboxylic acid, pyrrolnitrin, and the ortho-dialkyl-aromatic acids, were produced by the P. aurantiaca and P. chlororaphis strains. In addition, all strains produced 2-acetamidophenol, pyochelin, and diketopiperazine derivatives in variable amounts in all three culture media used. Highest levels of quorum-sensing signal molecules including PQS, 2-Octyl-3-hydroxy-4(1H)-quinolone, and hexahydro-quinoxaline-1,4-dioxide were recorded for P. aeruginosa At1RP4. Moreover, all strains produced volatile hydrogen cyanide (0.95–6.68 µg/L) and the phytohormone indole-3-acetic acid (0.42–13.9 µM). Production of extracellular lipase and protease was recorded in all pseudomonads, whereas, cellulase production and phosphate solubilization were variable. Genes for hydrogen cyanide and phenazine-1-carboxylic acid were detected in all eleven strains while the gene for pyrrolnitrin biosynthesis was amplified in P. aurantiaca and P. chlororaphis strains. Comparative metabolomic analysis provided detailed insights about the strain-specific metabolites in pseudomonads, and their pseudo-relative quantification in different bacterial growth media to be used as single-strain biofertilizer and biocontrol inoculums.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fluorescent pseudomonads are well documented for their biocontrol potential, and for their ability to successfully colonize surfaces and the internal tissues of plants at high densities. These bacteria can compete with soil microbes, particularly for the suppression of phytopathogens, and are known to produce a range of antagonistic secondary metabolites. A large body of literature describes the production of bioactive metabolites, including 2,4-diacetylphloroglucinol, pyrrolnitrin, pyoluteorin, phenazines, pyocyanin, rhizoxin, pyoverdines, hydrogen cyanide, and cyclic-lipopeptides (CLPs) by fluorescent pseudomonads, and their role in stimulating plant growth, and managing soil and plant health (Welbaum et al. 2004; Compant et al. 2005; Hayat et al. 2010; Saharan and Nehra 2011). Fluorescent pseudomonad species such as P. fluorescens, P. aeruginosa, P. aurantiaca, P. putida, P. pyrrocinia, and P. aureofaciens have been characterized and show antimicrobial activities with varying degree of antagonism (De-Weger et al. 1995; Shahid et al. 2017). The antimicrobial ability of this genus is mainly attributed to the production of phenazines. Phenazines constitute a large group of bright colored compounds containing nitrogen-heterocyclic rings, which are effective against diverse fungal and bacterial phytopathogens. Only a few strains of fluorescent pseudomonads which are actively involved in microbial competition in plant rhizosphere are known to simultaneously produce different phenazine derivatives (Kumar et al. 2005; Naik and Sakthivel 2006; Yu et al. 2018). Phenazine-1-carboxylic acid (PCA), phenazine-1-carboxamide (PCN), pyocyanin (toxin), and 2-hydroxyphenazine are the most abundant and commonly reported phenazines produced by florescent pseudomonads (Chin-A-Woeng et al. 1998; Pathma et al. 2010; Huang et al. 2011). Other important phenazines include phenazine-1,6-dicarboxylic acid, 2-hydroxyphenazine-1-carboxylic acid, di-hydroxyphenazines and phenazine-1-carboxylic acid derivatives. Thus far, research has been limited to the reporting of the production of these few dominant phenazine compounds, and less focus has been given to exploring other phenazine derivatives in the same bacterial strain.
Another widely used broad-spectrum agricultural biofungicide of Pseudomonas spp. is pyrrolnitrin. This pyrrole-based metabolite was first reported in P. aureofaciens and P. fluorescens (Kirner et al. 1998), and it was found active against wide range of fungi, including Deuteromycota, Ascomycota, and Basidiomycota. Many variants of pyrrolnitrin with low antifungal activity (such as isopyrrolnitrin, monodechloropyrrolnitrin, and oxypyrrolnitrin) have also been reported in Pseudomonas spp. (Ligon et al. 2000). Nevertheless, few reports of pyrrolnitrin production are available from P. aurantiaca and P. chlororaphis.
To meet its iron demands, Pseudomonas spp. are known to synthesize low molecular-weight iron-chelating agents called siderophores. Microbial siderophores sequester the available rhizosphere iron, making it unavailable for plant pathogens, thereby suppressing their growth. Many siderophores of fluorescent pseudomonads (such as pyoverdines, pyochelin, pseudobactins and quinolones) have been previously reported (Matthijs et al. 2008). Most, research studies have focused on the plate assay (i.e., petri-dish bioassays) detection of siderophores, which provides little information about the type of siderophore produced by an organism. Ortho-dialkyl-substituted aromatic acids, with moderate antibacterial activities, have been found to be a new class of compounds produced by this genus, and indicate higher biosynthetic capacity of pseudomonads than previously anticipated. Lahorenoic acid derivatives were reported in P. aurantiaca PB-St2 for the first time by our group (Mehnaz et al. 2013) and, to date, there are only two other reports of these metabolites from this genus (Yasmin et al. 2017). In addition to certain antimicrobial metabolites, biofilm formation and the production of certain virulence factors is regulated by two special quorum-sensing systems in pseudomonads. These systems contain two distinct classes of quorum-sensing signals: N-acyl homoserine lactones and 4-quinolones (Diggle et al. 2006; Bauer et al. 2016). The latter class has a significant role in bacterial survival under challenging environmental conditions, and is known to have a self-regulatory mechanism.
Although many of these antimicrobial compounds have long been reported in Pseudomonas spp., the metabolic profile of this genus, particularly of P. aurantiaca and P. chlororaphis, has not yet been explored in a single study under different culture conditions. In the present study, eleven isolates of fluorescent pseudomonads including P. fluorescens, P. aeruginosa, P. chlororaphis subsp. aurantiaca, and P. chlororaphis subsp. chlororaphis were metabolically characterized for the production of quorum-sensing signals, antimicrobial compounds, toxins, volatile organic compounds, siderophores, aromatic acids, and phenazine derivatives in LB, King’s B, and Davis minimal broth (DMB) media, and for the presence or absence of species-specific metabolites. We have previously characterized the P. chlororaphis subsp. aurantiaca and P. chlororaphis subsp. chlororaphis strains for their antifungal activities and plant growth promoting (PGP) potential, where they significantly increased wheat growth (Shahid et al. 2017). Genes that encoded secondary metabolites were also amplified, and the monoisotopic m/z values of these metabolites were recorded with high mass accuracy.
A detailed analysis of Pseudomonas spp. species-specific metabolites and the production of these metabolites under specific culture conditions enable the selection of the best strain for a particular field and in-vitro applications. Moreover, the use of single-strain inocula, with both broad-spectrum antagonistic and plant growth promoting traits, has the potential to be more efficient and user-friendly. Bacteria with multifaceted beneficial traits may be promising future candidates as biofertilizers and plant wardens against phytopathogens, particularly for South Asian crops.
Materials and methods
Bacterial strains and culture conditions
In this study, eight identified strains of P. chlororaphis subsp. aurantiaca: PB-St2, ARS-38, FS-2, GS-1, GS-3, GS-4, GS-6, GS-7, and one strain of P. chlororaphis subsp. chlororaphis: RP-4, were used. P. aurantiaca strains FS-2, GS-1, GS-3, GS-4, GS-6, and GS-7 were isolated from cactus, PB-St2 from sugarcane, ARS-38 from cotton, whereas, P. chlororaphis RP-4 was isolated from para grass. In addition to these, two unidentified bacterial isolates—RS-1 (isolated from rice rhizosphere) and At1RP4 (isolated from Atriplex rhizoplane), were also selected for this study, based on their antifungal abilities (Fig. S1). Bacterial strains were revived from glycerol stocks and streaked on King’s B agar plates (King et al. 1954). Plates were incubated at 28 °C for 48 h and observed for culture purity.
Biochemical and molecular characterization of isolates RS-1 and At1RP4
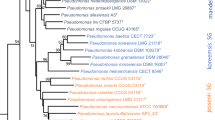
QTS-24 bacterial identification kits (DESTO Laboratories, Pakistan) were used for biochemical characterization of the RS-1 and AT1RP4 isolates, according to the manufacturer’s protocol. Bacterial DNA was isolated using a genomic-DNA purification kit (Thermo Fisher Scientific, Catalog number: K0512) from 24-h old bacterial cultures grown in King’s B broth. The 16S rRNA gene was amplified from the genomic DNA of isolates RS-1 and At1RP4 by PCR with the forward primer FGPS 1509–153 (5′-AAGGAGGTGATCCAGCCGCA-3′) and the reverse primer FGPS 4–281 (5′-AGAGTTTGATCCTGGCTCAG-3′, Normand 1995). PCR conditions are described in Table 1. A 50-μL reaction mixture was prepared from 5 μL of 10X Taq buffer, MgCl2 2 μL (25 mM), 2 μL of Taq polymerase (5 U), 2 μL of dNTPs (2.5 mM), 1 μL each of forward and reverse primer (20 pmol), 35 μL of dH2O, and 2 μL of the template DNA (> 50 ng/μL). All chemicals were purchased from Fermentas, USA. PCR products were purified using the GeneJET PCR purification kit (Thermo Fisher Scientific, Catalog number: K0701) and PCR products were sequenced with both forward and reverse primers from Eurofins, USA (Samples were shipped in SimpleSeq tubes, and Sanger sequencing was performed by using ABI 3730xl 96-capillary DNA Analyzers at the commercial facility of Eurofins, USA). Sequences were compared to the NCBI GenBank sequence database, using the BLAST search tool. Phylogenetic analysis was performed using Phylogeny.fr platform (Dereeper et al. 2008). Nucleotide sequences of isolates RS-1 and AT1RP4 were aligned with MUSCLE (ver. 3.8.31). Ambiguous regions containing gaps and poorly aligned sequences were removed using Gblocks (ver. 0.91b) with the following parameters: minimum length of the block after gap cleaning, 10; no gap positions were allowed in the final alignment; all segments with contiguous non-conserved positions bigger than 8 were rejected; minimum number of sequences for a flank position, 85%. Phylogenetic tree was constructed using the maximum likelihood method implemented in the PhyML program (v3.1/3.0). The HKY85 substitution model was selected, assuming an estimated proportion of invariant sites of 0.001, and 4 gamma-distributed rate categories to account for rate-heterogeneity across sites. The gamma shape parameter was estimated directly from the data (γ = 96.300). Reliability for internal branch was assessed using the aLRT test (SH-Like). Enterobacter cloacae (Accession No. MF574396) was used as an outgroup. The percentage of replicate trees in which associated taxa clustered together in the bootstrap data sets was 100. There were a total of 1,408 positions in the final dataset of RS-1 and 1371 positions in final dataset of At1RP4. These sequences were deposited in the GenBank database, and accession numbers were obtained (i.e., RS-1: MF574397; AT1RP4: MF574400).
Identification and pseudo-relative quantification of secondary metabolites by liquid chromatography–tandem mass spectrometry (LC–MS/MS)
Preparation of samples
The metabolites for individual strains were extracted from 100 mL of 96-h-old bacterial cultures of FS-2, ARS-38, RP-4, PB-St2, GS-1, GS-3, GS-4, GS-6, GS-7, RS-1, and At1RP4, grown in three media separately: King’s B (King et al. 1954), LB (Bertani 2004), and glycerol-DMB (Davis 1949). Cultures were centrifuged at 3900×g for 30 min (Allegra TM X-22R Centrifuge, Beckman Coulter) and the supernatants were acidified to pH 2 with 6 N HCl. The acidified supernatants were extracted twice with 100 mL of ethyl acetate, and the organic layers were collected, dehydrated with anhydrous sodium sulfate and evaporated to dryness in a nitrogen evaporator. Residues of the extracts were individually re-suspended in 4 mL of methanol-chloroform [1:1 (v/v)], and sonicated for 5 min in a water bath to completely dissolve them. A 200 µL aliquot of each resulting solution was mixed with 200 µL of methanol and 100 µL of H2O. A 10 µL sample was injected for each LC–MS analysis (Shahid et al. 2017).
Pellets of each bacterial strain were individually re-suspended in 5 mL of acetone, sonicated for 10 min, and centrifuged at 3900×g and 4 °C for 20 min. Supernatants were transferred to 15-mL falcon tubes and evaporated to dryness. Dried residues were re-suspended in 2 mL of methanol-chloroform [1:1 (v/v)] and sonicated for 2 min. A 200-µL aliquot of each of the resultant solution was mixed with 200 µL of methanol and 100 µL of H2O. All samples were stored at 4 °C for further analysis. Extracts of sterilized un-inoculated LB, King’s B, and DMB media were used as controls.
Liquid chromatography mass spectrometry (LC–MS/MS) and data analyses
Sample solutions were injected onto an Eclipse Plus C18 RRHD column (2.1 × 100 mm, 1.8 μm; Agilent, Santa Clara, CA, USA) and separated by reverse-phase liquid chromatography using a Waters Acquity UPLC system (Milford, MA, USA) at a flow rate of 0.4 mL/min. The UPLC system was coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with an EASY-Max NG electrospray ion source. The mobile phases were water/0.01% formic acid (A) and 100% acetonitrile/0.01% formic acid (B). Secondary metabolites were separated on the column with a 20-min binary solvent elution gradient (0 min, 5% B; 0–15 min, 5–100% B; 15–17 min, 100% B) followed by a 3-min column equilibration at 5% B between injections. The MS parameters were as follows: positive-ion electrospray spray voltage, 3.9 kV; negative-ion electrospray spray voltage, 2.5 kV; capillary temperature, 325 °C; S-lens Level, 60%; sheath gas, 50; sweep gas, 1; and auxiliary gas 10. The MS survey scan was carried out over the mass range m/z 80–1800, with the data recorded in the centroid mode and at a mass resolution of 120 K FWHM (m/z 200), AGC target 4E5, and one microscan with a maximum injection time of 50 ms in the Quadrupole isolation mode. A lock mass at m/z 391.28426 from di-(2-ethylhexyl) phthalate (a ubiquitous plasticizer) was used for real-time internal mass calibration during the LC–MS runs with the Fourier transform (FT) MS detection. In the LC–MS/MS runs, the top five most intense ions, with their charge state of 1 and ion counts of > 5000 in each survey scan were selected for MS/MS by collision-induced dissociation (CID) in the linear ion trap or by “higher-energy” collisional dissociation (HCD) in the collision cell in the upfront of the C-trap. Other parameters included: activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum injection time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 30% for the CID operations. Activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum inject time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 25% were used for the HCD operations.
Raw data files were recorded and processed using the XCalibur 4.1.31.9 (Thermo Scientific) software suite. For relative quantification of secondary metabolites produced by Pseudomonas strains, each sample solution was injected in triplicate and the peak areas were averaged. Bar graphs were plotted with error bars to show the analytical standard deviations.
Identification and quantification of indole-3-acetic acid (IAA)
Preparation of samples
Bacterial cultures were grown in 10 mL of King’s B medium (King et al. 1954) supplemented with L-tryptophan (100 mg/L). After 7 days of growth, bacterial cells were centrifuged at 3900×g for 30 min (Allegra TM X-22R Centrifuge, Beckman Coulter) and the pH of the supernatants was adjusted to pH 2.0 with 6 N HCl. The acidified supernatants were extracted twice with an equal volume (10 mL) of ethyl acetate. After centrifugation, the upper organic layers were collected, pooled, and evaporated to dryness (Goswami et al. 2015).
Preparation of indole-3-acetic acid standard curve and LC–MS quantification
The reference standard of indole-3-acaetic acid (IAA, C10H9NO2, monoisotopic neutral mass = 175.0633, Sigma-Aldrich) was dissolved in methanol:chloroform:H2O [50:25:25, v/v] at 0.25 mM to form stock standard solution S1. This solution was serially diluted at a dilution ratio of 1 to 4 with the same solvent to make solutions S2 to S10. Table S1 lists the concentrations of the serially diluted standards. Ten-μL aliquots of the standard solutions and the sample solutions were injected into the column for the LC-FTMS runs, as described above. The concentrations in the samples were determined from the peak areas of the extracted ion chromatograms of IAA detected at m/z 176.0706 [M + H]+ by interpolation of the linear-regression calibration curve for this compound (Fig. S2).
Detection of HCN production by Pseudomonas isolates RS-1 and At1RP4
Qualitative determination of hydrocyanic acid (HCN) production was done by streaking each bacterial isolate on King’s B agar plates. A filter paper saturated with an alkaline picrate solution (g/L: picric acid, 2.5; Na2CO3, 12.5; pH 13) was placed into the lid of the petri plate (Millar and Higgins 1970). After 2–4 days incubation at 28 ± 2 °C, an assessment of HCN production was made by the change in color of filter paper from yellow to brown/reddish brown. P. aurantiaca PB-St2 was used as a positive control. All other strains have previously been reported as positive for HCN production by plate assay (Shahid et al. 2017).
Quantitative determination of HCN production by Pseudomonas strains
Hydrogen cyanide produced by bacterial strains was quantified by the method of Lambert et al. (1975). Bacterial strains were cultured on modified King’s B agar plates, supplemented with 4.4 g/L of glycine. Filter paper discs (3 cm2) saturated with 1 M NaOH, were placed in the center of petri-plate lids and the plates were sealed with parafilm and incubated at 28 ± 2 °C for 48 h. After incubation, filter paper discs were removed, individually extracted with 5 mL of 1 M of NaOH and titrated with 4.25 mL of acetic acid. Hydrogen cyanide in the NaOH solution was quantified by allowing the NaOH solution to react with the barbituric acid-pyridine reagent, and the absorbance at 575 nm were spectrophotometrically determined. The amount of cyanide (µg/L KCN) was calculated using a standard curve of KCN in NaOH (Fig. S3).
Siderophore production by Pseudomonas isolates RS-1 and At1RP4
Siderophore production was detected by the O-CAS method (Schwyn and Neilands 1987). Bacterial culture-containing plates were incubated at 28 °C for 24 h. CAS medium was overlaid on incubated culture plates, and a change in color from blue to orange (hydroxamate-type siderophore) or purple (catechol-type siderophore) was considered positive result. P. aurantiaca PB-St2 was used as a positive control. Sterilized LB medium and water were used as the negative control. All other strains have previously been reported positive for siderophore production by plate assay (Shahid et al. 2017).
Identification of genes encoding antagonistic secondary metabolites in Pseudomonas strains
Detection of the genes encoding phenazine-1-carboxylic acid (PCA), pyrrolnitrin, and hydrogen cyanide (HCN) was done using gene-specific primers. Primers used in this study were prnA (F′-GTGTTCTTCGACTTCCTCGG and R′-TGCCGGTTCGCGAGCCAGA) for pyrrolnitrin, PCA2a- (5′-TTGCCAAGCCTCGCTCCAAC-3′), PCA3b- (5′-CCGCGTTGTTCCTCGTTCAT-3′) for phenazine-1-carboxylic acid (Zhang et al. 2006) and AC (a′5′-ACTGCCAGGGGCGGATGTGC-3′ and b′5′-ACGATGTGCTCGGCGTAC-3′) for hcnBC hydrogen cyanide (Kim et al. 2013). PCR cycles were performed as described in reference publications with little modification, and PCR conditions are shown in Table 1. PCR products were analyzed on 1% agarose gel. The respective bands were excised and purified using a gel extraction kit. PCR products were sequenced (Eurofins) and analyzed using the NCBI BLAST (blastn) and alignment tools.
Detection of hydrolytic enzymes produced by Pseudomonas strains
Lipase production was checked on 1% Tween-20 LB agar plates by the method of Sierra (1957). Pikovskaya agar medium was used for the detection of phosphatase production (Pikovskaya 1948). Protease production was checked using skim milk agar plates (Alnahdi 2012). Extracellular cellulase production was checked on 1% CMC-LB agar plates (Kasana et al. 2008). Bacterial cultures, grown overnight, were spot inoculated on separate media plates and incubated at 28 °C. For the detection of lipase and protease, plates were incubated for 48 h; for cellulase detection, plates were incubated for 4–5 days. Pikovskaya agar medium plates were incubated for 10 days. Precipitation zones around bacterial colonies for lipase and clear halo zones around bacterial growth for protease, cellulase (after staining), and phosphatase were considered to be positive test results.
Results
Biochemical and molecular characterization of isolates RS-1 and At1RP4
At1RP4 and RS-1 were biochemically characterized for carbon-source utilization and the presence/absence of specific enzymes with the help of QTS-24 identification kits (Table S2). For 16S rRNA gene, sequence data were searched through NCBI BLAST. Results confirmed the strain RS-1 as P. fluorescens and At1RP4 as P. aeruginosa. Phylogenetic tree based on 16S rRNA gene sequences showed the homology of these strains with closely related strains and are shown in Fig. S4. Sequence lengths, accession numbers, and percent homology of each strain are summarized in Table S3.
Identification and pseudo-relative quantification of secondary metabolites by liquid chromatography mass spectrometry (LC–MS/MS)
Secondary metabolites produced by Pseudomonas strains were first detected by LC/ESI-FTMS in the full-mass scan mode. The first set of peaks observed belong to the phenazine family, where strong signals corresponding m/z [M + H]+ were recorded for seven different phenazines derivatives and for other metabolites including pyrrolnitrin (PRN), pyochelin, Lahorenoic acids A-C, maculosin, cyclo-l-Pro-l-Val, and cyclo-l-Pro-l-Met. In addition, three quorum-sensing signal molecules were detected in these samples (Table 2). Chemical formulas, monoisotopic neutral masses, observed m/z values, and retention times of these compounds have been described in Table S4. All eleven strains produced five derivatives of phenazines–phenazine, phenazine-1-carboxylic acid (PCA), 2-hydroxyphenazine-1-carboxylic acid (2-OH-Phz-1-COOH), phenazine-1,6-dicarboxylic acid (Phz-1,6-di-COOH), and 2-hydroxyphenazine (2-OH-Phz)—in all three culture media (Fig. S5). High levels of all phenazine derivatives were observed in LB medium, except for 2-OH-Phz-1-COOH whose maximum levels were in King’s B and DMB, in varying amounts, by different strains (Fig. 1a–e). All strains, except for P. aeruginosa AT1RP4 and P. fluorescens RS-1, produced 2,8-dihydroxyphenazine and 6-methylphenazine-1-COOH (Fig. 1f, S7).
Pseudo-relative quantification of phenazines in King’s B, LB, and DMB culture media. Phenazine (a), Phz- 1,6-di-COOH (b), Phenazine-1-COOH (c), 2-OH-Phz (d), 2-OH-Phz-1-COOH (e), 6-methyl-Phz-1-COOH (f). Data is presented with error bars to show the analytical standard deviations for three replicates
All P. aurantiaca subsp. aurantiaca strains (PB-St2, ARS-38, FS-2, GS-1, GS-3, GS-4, GS-6, GS-7, and P. aurantiaca subsp. chlororaphis RP-4) produced pyrrolnitrin. Its maximum concentration was observed in King’s B medium, but it was not produced by P. aeruginosa At1RP4 and P. fluorescens RS-1. The highest amount of pyrrolnitrin was observed for P. chlororaphis RP-4, followed by P. aurantiaca FS-2. All strains were positive for 2-acetamidophenol and siderophore pyochelin, production. Variable amounts of 2-acetamidophenol were produced in three culture media used in this study, however, maximum levels of pyochelin were observed in P. aeruginosa At1RP4 grown in King’s B medium (Fig. S6, Fig. 2 a–c).
The ortho-dialkyl-substituted aromatic acid derivatives, Lahorenoic acid A, B and C, were produced by P. aurantiaca and P. chlororaphis strains (Fig. S6). The highest levels of these three derivatives were observed in DMB medium, while negligible amounts of these secondary metabolites were observed in LB and King’s B media (Fig. 3a–c). The maximum maculosin levels were in King’s B broth, while more cyclo-l-Pro-l-Val and cyclo-Met-Pro-diketopiperazine were detected in LB medium (Fig. 3d–f).
Although the quorum-sensing signal molecules (2-heptyl-3-hydroxy-4(1H)-quinolone (PQS), 2-octyl-3-hydroxy-4(1H)-quinolone, and hexahydro-quinoxaline-1,4-dioxide) were produced by all strains, but the maximum amounts of 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) and 2-octyl-3-hydroxy-4(1H)-quinolone were observed in P. aeruginosa AT1RP4 in DMB medium (Fig. 4a, b). All pseudomonads produced hexahydro-quinoxaline-1,4-dioxide in variable amounts in LB and King’s B media (Fig. 4c).
The identified molecules were structurally confirmed by LC–MS/MS. The MS/MS spectra from ion fragmentation with HCD and CID were searched against the available MS/MS databases including CFM-ID (Allen et al. 2014), METLIN (Smith et al. 2005) and GNPS (Wang et al. 2016). The MS/MS spectra were also compared with previously published spectra in the literature, especially for those compounds still not entered into any of the databases. The extracted MS and MS/MS spectra of detected compounds are shown in Figures S8-S17.
Indole-3-acetic acid (IAA) production by Pseudomonas strains
All Pseudomonas strains were positive for IAA production. The highest IAA production was observed in P. fluorescens RS-1 (13.914 µM) followed by P. aurantiaca ARS-38 (1.7714 µM) and GS-7 (1.6138 µM). Rest of the strains produced indole-3-acetic acid in the range of 0.41982–1.3882 µM (Fig. S18).
HCN and siderophore production by Pseudomonas strains
P. aeruginosa At1RP4 and P. fluorescens RS-1 also produced siderophores in plate bioassays. Both strains (At1RP4 and RS-1) produced hydrogen cyanide in the plate assay and turned indicator filter paper from yellow to reddish brown (Fig. 5). The highest HCN production (6.683 µg/L) was observed in P. chlororaphis subsp. aurantiaca ARS-38, followed by P. chlororaphis RP-4 (6.286 µg/L). All other strains produced HCN in the range of 0.946–4.681 µg/L (Fig. S18). P. aurantiaca and P. chlororaphis strains have previously been reported to be positive for siderophore and HCN production in plate assays (Shahid et al. 2017).
Identification of genes encoding antagonistic secondary metabolites
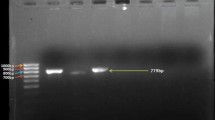
The presence of phenazine-1-carboxylic acid (pca) and 2-OH-Phz (phzO) genes have previously been reported in P. aurantiaca strains PB-St2, ARS-38, FS-2, GS-1, GS-3, GS-4, GS-6, GS-7, and P. chlororaphis RP-4 (Shahid et al. 2017). P. aurantiaca strain PB-St2 was used as the positive control as its genome is published and has the genetic clusters coding for PCA, 2-OH-PHZ, and HCN (Mehnaz et al. 2014). P. aeruginosa At1RP4 and P. fluorescens RS-1 were also positive for the pca gene and showed 1.2 kb band. In addition, all P. aurantiaca and P. chlororaphis strains were also positive for the prnA gene and indicated 1 kb band on agarose gel, while 586 bp products for hcn gene were also detected in all eleven strains (Fig. S19A–C). PCR products were sequenced (Eurofins, USA) and compared with PB-St2 genome (Accession no. AYUD00000000).
Detection of hydrolytic enzymes produced by Pseudomonas strains
All strains were positive for production of protease and lipase enzymes. Two strains – P. aeruginosa At1RP4 and P. fluorescens RS-1 solubilized insoluble phosphate on Pikovskaya agar plates. P. chlororaphis subsp. aurantiaca strains ARS-38, FS-2, GS-4, GS-6, P. chlororaphis subsp. chlororaphis RP-4, and P. fluorescens RS-1 were positive for cellulase production (Fig. 5).
Discussion
Pseudomonas spp. are ubiquitous in nature with a range of agricultural, environmental, and industrial applications. This genus is highly appreciated for its immense agricultural potential due to its plant growth promoting traits and range of anti-phytopathogenic antibiotics (Welbaum et al. 2004; Compant et al. 2005; Hayat et al. 2010; Saharan and Nehra 2011). Among pseudomonads, P. chlororaphis subsp. aurantiaca, and P. chlororaphis subsp. chlororaphis are not common inhabitants of soils, and only few strains have been characterized so far. These species recently received attention for their successful survival in diverse plant hosts, their adaptability to living in extreme environments, and they are established biocontrol agents for their antagonistic activities (Compant et al. 2005; Yasmin et al. 2017). To date, more than thirty compounds have been characterized in these species, but the list of unidentified compounds is long and there is only limited data describing the maximum metabolic potential of these species in a single study (Saharan and Nehra 2011). P. chlororaphis subsp. aurantiaca strains PB-St2, ARS-38, FS-2, GS-1, GS-3, GS-4, GS-6, GS-7, P. chlororaphis subsp. chlororaphis RP-4, P. aeruginosa At1RP4, and P. fluorescens RS-1, were isolated from diverse hosts and showed broad-spectrum antifungal activities. These were subjected to detailed polyphasic characterization under minimal (DMB), rich (LB), and specific (King’s B) culture media.
LC–MS/MS analyses revealed that the biosynthetic activities of these strains were higher than previously anticipated. Overall, all strains produced five phenazine derivatives including phenazine, phz-1,6-di-COOH, PCA, 2-OH-phz, and 2-OH-phz-1-COOH. In-vitro studies have reported the production of PCA by several species of fluorescent pseudomonads. P. fluorescens LBUM636 has been shown to produce PCA, where it significantly inhibited the growth of Phytophthora infestans, but production of other phenazine derivatives has not been reported (Morrison et al. 2017). Likewise, P. putida strains have been genetically engineered for heterologous gene expressions to analyze the synthesis of natural products, but few reports have described the inherent potential of wild-type strains for the range of phenazine derivatives produced (Loeschcke et al. 2015). P. aeruginosa strain PA14 has been evaluated for production of PCA, 1-hydroxyphenazine, and pyocyanin for its biocontrol of C. elegans, but production of 2-OH-phz-1-COOH and phz-1,6-di-COOH were not shown (Cezairliyan et al. 2013). P. aurantiaca PB-St2 and P. chlororaphis PA23, however, were reported earlier to produce 2-hydroxyphenazine, PCA, and 2-OH-phz-1-COOH. Nonetheless, these strains were not analyzed for production of phz-1,6-di-COOH (Selin et al. 2010; Mehnaz et al. 2013). 2,8-Di-OH-Phz and 6-methyl-Phz-1-COOH derivatives were not detected in P. aeruginosa At1RP4 or P. fluorescens RS-1, and were exclusively produced by the P. aurantiaca and P. chlororaphis strains. On the basis of 16S rRNA gene sequence, strain RS-1 was earlier reported as P. putida, but it solubilized gelatin which is the characteristic feature of P. fluorescens.
High production of these phenazine derivatives was observed in LB medium, except for 2-OH-phz-1-COOH where the maximum production was in DMB medium. Variable amounts of these phenazine derivatives were produced by all strains, and comparable production of phenazines was found in P. chlororaphis and P. aurantiaca. However, the highest amount of phz-1,6-di-COOH was produced by P. aeruginosa At1RP4. High production of phenazines in the presence of rich medium has been shown previously in a study where P. chlororaphis O6 was cultivated in glucose-supplemented-nutrient broth (NB) and NB without glucose, but production of these derivatives was not evaluated in minimal medium or in the presence or absence of glycerol (Park et al. 2011). In our current study, phenazine production was enhanced in LB medium that contains scant amounts of carbohydrate sources (Bertani 2004) as compared to King’s B and DMB media that are supplemented with glycerol. These results are consistent with the findings that phenazine biosynthesis requires the products of glucose catabolism using enzymes in the shikimic acid pathway (Park et al. 2011). Negligible production of 2-hydroxyphenazine was observed in P. aeruginosa At1RP4 and P. fluorescens RS-1, in all three types of culture media used. This suggests that 2-hydroxylated phenazines were synthesized in P. aeruginosa and P. fluorescens but in very low amounts. 2-acetamidophenol utilizes the same shikimic acid pathway for its production and is known to synergistically enhance antifungal ability of PCA (Park et al. 2011). High production of this compound was also observed in LB medium, where P. aurantiaca GS-6 was the highest producer of this metabolite.
Pyrrolnitrin production was enhanced with the addition of glycerol in King’s B medium suggesting that this simple carbon source reciprocally affects the production of secondary metabolites in fluorescent pseudomonads. The observation of pyrrolnitrin synthesis in P. aurantiaca and P. chlororaphis resembled the regulation of antimicrobial metabolites in P. chlororaphis O6 and P. fluorescens Pf-5 where the addition of glucose stimulated phenazine and 2,4-DAPG production, while decreasing pyrrolnitrin and pyoluteorin synthesis (Kraus and Loper 1995; Park et al. 2011). The siderophore, pyochelin production was highest in P. aeruginosa At1RP4, followed by P. chlororaphis RP-4 in King’s B medium. These results are consistent with previous literature reports where P. aeruginosa PUPa3 has been shown to produce pyochelin under iron-limited conditions (Kumar et al. 2005).
The highest yield of ortho-dialkyl-substituted aromatic acids (Lahorenoic acids A-C) was observed in DMB, while P. aurantiaca PB-St2 and GS-4 showed significant production of all three derivatives. Lahorenoic acid derivatives are a relatively new class of moderate antibacterial metabolites of pseudomonads, and there are only few reports of their production to date (Mehnaz et al. 2013; Shahid et al. 2017; Yasmin et al. 2017). This data is consistent with their first report in P. aurantiaca PB-St2, where these compounds were detected in DMB medium (Mehnaz et al. 2013). The biosynthetic pathway and the putative genes involved in the production and regulation of these compounds in pseudomonads is still not known.
Many research studies regard small cyclicpeptide, i.e., diketopiperazines as sterilization artifacts, but the literature supports their synthesis by dedicated non-ribosomal peptide synthetase (NRPS) and their antifungal, antibacterial, and antiviral activities. The P. stutzeri strain has been reported to produce several cyclicpeptide derivatives with anti-Pythium insidiosum activity (Thongsri et al. 2014). Cyclodipeptides from P. aeruginosa PAO1 have been analyzed for apoptosis in cancer cell lines where they successfully exhibited cytotoxicity (Vázquez et al. 2015). The diketopiperazine derivatives maculosin, cyclo-l-Pro-l-Val, and cyclo-Met-Pro-diketopiperazine were detected in our current study, and have been previously shown to have inhibitory effects against Saccharomyces cerevisae, rice pathogen Pyricularia oryzae, Aspergillus fumigatus and Penicillium roquefortii by inhibiting family 18 chitinases (Smaoui et al. 2011). Variable amounts of these cyclicpeptide derivatives were produced by all strains in DMB, LB, and King’s B media, suggesting that there are diverse biosynthesis genes involved in the production of these metabolites under different culture conditions. Although, 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) and 2-octyl-3-hydroxy-4(1H)-quinolone were produced by all strains, but their maximum yield was observed in P. aeruginosa At1RP4, suggesting these two are primary quorum-sensing signals of this specie (Tashiro et al. 2013). Nevertheless, hexahydro-quinoxaline-1,4-dioxide production was observed by all pseudomonads, exhibiting the differential regulation of these QS signal molecules in different Pseudomonas spp. (Minagawa et al. 2012).
PCR analyses of all Pseudomonas strains exhibited presence of gene clusters directly involved in the synthesis of hydrogen cyanide, PCA, and pyrrolnitrin. In addition, all strains were positive for phytohormone indole-3-acetic acid and volatile HCN production, which is a well-established phenomenon in fluorescent pseudomonads, and several Pseudomonas strains have been widely reported to stimulate plant growth through these traits (Welbaum et al. 2004; Compant et al. 2005; Hayat et al. 2010; Saharan and Nehra 2011). Moreover, pseudomonads which are extensively used as agricultural bacteria have been examined for production of extracellular proteases, cellulases, lipases, and phosphatase where these traits give them the additional quality of being biofertilizers (Oteino et al. 2015). Pseudomonas strains used in this study have also been explored for these characteristic enzymes in plate bioassays, where they exhibited variable production of these hydrolytic enzymes.
Comparative metabolomic analysis in rich, minimal, and specific culture media showed similarities and differences among the traits and secondary metabolites produced by the eleven strains of fluorescent pseudomonads that were used in this study. Detailed analysis of the secondary metabolites of these strains provided new insights about their adaptation to diverse environmental niches and their strain-specific metabolites. Whether the differential effects of DMB, LB, and King’s B media on metabolite regulation and synthesis are directly related to variation in metabolic precursors still remain to be determined. Our current study describes the effects of different culture media as nutritional and antibiotic regulators of the secondary-metabolite biosynthesis pathways. This will improve the use of these strains under field conditions, to promote the production of desired antibiotics for competitive suppression and biocontrol of phytopathogens.
References
Allen F, Pon A, Wilson M, Greiner R, Wishart D (2014) CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acid Res 42:W94-99
Alnahdi HS (2012) Isolation and screening of extracellular proteases produced by new isolated Bacillus sp. JAPS 2:71–74
Bauer JS, Hauck N, Christof L, Mehnaz S, Gust B, Gross H (2016) The systematic investigation of the quorum sensing system of the biocontrol strain Pseudomonas chlororaphis subsp. aurantiaca PB-St2 unveils aurI to be a biosynthetic origin for 3-oxo-homoserine lactones. PLoS ONE 11:0167002
Bertani G (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186:595–600
Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM (2013) Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1003101
Chin-A-Woeng TFC, Bloemberg GV, Van-der-Bij AJ, Van-der-Drift KMGM, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker PAHM, Tichy H, de Bruijn FJ, Thomas-Oates JE, Lugtenberg BJJ (1998) Biocontrol by phenazine-1-carboxamide producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. MPMI 11:1069–1077
Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA (2005) Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. Strain PsJN Appl Environ Microbiol 71:1685–1693
Davis BD (1949) The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci 35:1–10
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465-469
De-Weger LA, Van-der Bij AJ, Dekkers LC, Simons M, Wijffelman CA, Lugtenberg BJ (1995) Colonization of the rhizosphere of crop plants by plant-beneficial pseudomonads. FEMS Microbiol Ecol 17:221–227
Diggle SP, Cornelis P, Williams P, Camara M (2006) 4-Quinolone signaling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol 296:83–91
Goswami D, Thakker JN, Dhandhukia PC (2015) Simultaneous detection and quantification of indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA) produced by rhizobacteria from L-tryptophan (Trp) using HPTLC. J Microbiol Methods 110:7–14
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60:579–598
Huang L, Chen MM, Wang W, Hu HB, Peng HS, Xu YQ, Zhang XH (2011) Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Applied Microbiol Biotechnol 89:169–177
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Current Microbiol 57:503–507
Kim JS, Pauline MM, David EC (2013) Application of PCR primer sets for detection of Pseudomonas sp. functional genes in the plant rhizosphere. J Agric Chem Environ 2:8–15
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307
Kirner S, Hammer PE, Hill DS, Altmann A, Fischer I, Weislo LJ, Lanahan M, van Pee KH, Ligon JM (1998) Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J Bacteriol 180:1939–1943
Kraus J, Loper JR (1995) Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 61:849–854
Kumar RS, Ayyadurai N, Pandiaraja P, Reddy AV, Venkateswarlu Y, Prakash O, Sakthivel N (2005) Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J Appl Microbiol 98:145–154
Lambert JL, Ramasamy J, Paukstelis JV (1975) Stable reagents for the colorimetric determination of cyanide by modified Konig reactions. Anal Chem 47:916–918
Ligon JM, Hill DS, Hammer PE, Torkewitz NR, Hofmann D, Kempf HJ, van-Pee KH (2000) Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag Sci 56:688–695
Loeschcke A, Thies S (2015) Pseudomonas putida-a versatile host for the production of natural products. Appl Microbiol Biotechnol 99:6197–6214
Matthijs S, Budzikiewicz H, Schafer M, Wathelet B, Cornelis B (2008) Ornicorrugatin, a new siderophore from Pseudomonas fluorescens AF76. Z Naturforsch 63:8–12
Mehnaz S, Saleem RSZ, Yameen B, Pianet I, Schnakenburg G, Pietraszkiewicz H, Valeriote F, Josten M, Sahl HG, Franzblau SG, Gross H (2013) Lahorenoic acids A-C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J Nat Prod 76:135–141
Mehnaz S, Bauer JS, Gross H (2014) Complete genome sequence of the sugar cane endophyte Pseudomonas aurantiaca PB-St2, a disease-suppressive bacterium with antifungal activity toward the plant pathogen Colletotrichum falcatum. Genome Announc 2:e1108–e1113. https://doi.org/10.1128/genomeA.01108-13
Millar R, Higgins VJ (1970) Association of cyanide with infection of bird’s foot trefoil by Stemphylium loti. Phytopathology 60:104–110
Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N (2012) RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. https://doi.org/10.1186/1471-2180-12-70
Morrison CK, Arseneault T, Novinscak A, Filio M (2017) Phenazine-1-carboxylic acid production by Pseudomonas fluorescens LBUM636 alters Phytophthora infestans growth and late blight development. Phytopathology 107:273–279
Naik PR, Sakthivel N (2006) Functional characterization of a novel hydrocarbonoclastic Pseudomonas sp. strain PUP6 with plant-growth-promoting traits and antifungal potential. Res Microbiol 157:538–546
Normand P (1995) Utilisation des séquences 16S pour le positionnement phylétique d’un organisme inconnu. Oceanis 21:31–56
Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00745
Park JY, Oh SA, Anderson AJ, Neiswender J, Kim JC, Kim YC (2011) Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett Appl Microbiol 52:532–537
Pathma J, Ayyadurai N, Sakthivel N (2010) Assessment of genetic and functional relationship of antagonistic fluorescent pseudomonads of rice rhizosphere by repetitive sequence, protein coding sequence and functional gene analyses. J Microbiol 48:715–727
Pikovskaya R (1948) Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:e370
Saharan B, Nehra V (2011) Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 21. http://astonjournals.com/lsmr
Schwyn B, Neilands J (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Selin C, Habibian R, Poritsanos N, Athukorala SN, Fernando D, de Kievit TR (2010) Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol Ecol 71:73–83
Shahid I, Rizwan M, Baig DN, Saleem RS, Malik KA, Mehnaz S (2017) Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis subsp. aurantiaca strains isolated from cotton, cactus, and para grass. J Microbiol Biotechnol 27:480–491
Sierra G (1957) A simple method for the detection of lipolytic activity of micro-648 organisms and some observations on the influence of the contact between cells and fatty 649 acid substrates. Antonie Leeuwenhoek 23:15–22
Smaoui S, Mellouli L, Lebrihi A, Coppel Y, Fguira LFB, Mathieu F (2011) Purification and structure elucidation of three naturally bioactive molecules from the new terrestrial Streptomyces sp. TN17 strain. Nat Prod Res 25:806–814
Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27:747–751
Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N (2013) Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ 28:13–24
Thongsri Y, Aromdee C, Yenjai C, Kanokmedhakul S, Chaiprasert A, Prariyachatigul C (2014) Detection of diketopiperazine and pyrrolnitrin, compounds with anti-Pythium insidiosum activity, in a Pseudomonas stutzeri environmental strain. Biomed Pap 158:378–383
Vázquez RD, González O, Guzmán-Rodríguez J, Perez ALD, Zaarzosa AO, Bucio JL, Carmen VM, Garcia JC (2015) Cytotoxicity of cyclodipeptides from Pseudomonas aeruginosa PAO1 leads to apoptosis in human cancer cell lines. BioMed Res Intern. https://doi.org/10.1155/2015/197608
Wang M et al (2016) Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol 34:828–837
Welbaum GE, Sturz AV, Dong Z, Nowak J (2004) Managing soil microorganisms to improve productivity of agroecosystems. Crit Rev Plant Sci 23:175–193
Yasmin S, Hafeez FY, Mirza MS, Rasul M, Arshad HMI, Zubair M, Iqbal M (2017) Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front Microbiol 8:1895. https://doi.org/10.3389/fmicb.2017.01895
Yu JM, Wang D, Pierson LS III, Pierson EA (2018) Effect of producing different phenazines on bacterial fitness and biological control in Pseudomonas chlororaphis 30–84. Plant Pathol J 34:44
Zhang Y, Fernando WG, Kievit TRD, Berry C, Daayf F, Paulitz T (2006) Detection of antibiotic-related genes from bacterial biocontrol agents with polymerase chain reaction. Can J Microbiol 52:476–481
Acknowledgements
This work received financial support from Higher Education Commission (HEC) of Pakistan. The funding was awarded to Izzah Shahid for PhD research work. The metabolomics research at the UVic-Genome BC Proteomics Centre was supported by funding to “The Metabolomics Innovation Centre (TMIC)” through the Genome Innovations Network (GIN) from Genome Canada, Genome Alberta, and Genome British Columbia for operations (205MET and 7203) and for technology development (215MET and MC3T) in metabolomics. The authors would like to thank Dr. Carol E. Parker for assistance with English language editing.
Author information
Authors and Affiliations
Contributions
IS performed experiments and prepared manuscript initially. CHB and JH provided the conceptual framework for the experiments. DNB helped in genetic analysis. DH assisted in LC–MS and data analysis. SM supervised the research as well as feedback and guidance during manuscript development. KAM managed resources and critically read the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahid, I., Han, J., Hardie, D. et al. Profiling of antimicrobial metabolites of plant growth promoting Pseudomonas spp. isolated from different plant hosts. 3 Biotech 11, 48 (2021). https://doi.org/10.1007/s13205-020-02585-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02585-8