Abstract
Sugarcane (Saccharum spp.) is a major sugar crop grown in tropical and sub-tropical areas throughout the world which is vulnerable to high temperature stress due to climate change. In this present study, we have generated a transcriptome profile of sugarcane variety Co 99004 exposed to high-temperature stress (47 °C). The Illumina Nexseq2500 platform yielded a total of 39.28 and 13.44 million reads, corresponding to 3.9 and 1.3 gigabase pair (Gb) of the processed reads for control and high-temperature-stressed samples, respectively. Initially, the reads were de novo assembled into 118,017 unigenes with an average length of 780 bp. The longest sequence in the assembly was 21 kb. Further, these transcripts were BLASTed against GO, KEGG and COG databases to identify the novel genes/transcripts expressed due to elevated temperature conditions. The different expression analysis showed 1137 transcripts which were up-regulated during heat temperature stress when compared to control conditions. Analysis of relative gene expression showed phytepsin, ferredoxin-dependent glutamate synthase, and stress protein DDR-48 threefold increased expression during heat stress. These findings reveal novel targets for subsequent research on the genomics genetic manipulation and molecular mechanism of elevated stress tolerance in sugarcane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane is one of the world’s major C4 crops that are mainly grown in tropical and sub-tropical regions for sugar and bioenergy. Extreme weather conditions and the changing environment with increased atmospheric (CO2), temperature, and irrational precipitation are the key factors affecting sugarcane production worldwide, especially in many developing countries (Zhao and Li 2015). Abiotic stresses such as drought, salinity, high temperatures and oxidative stress are often interconnected, through the abscisic acid signaling pathway and may cause membrane damage or macromolecule degradation by the formation of ROS (Wang et al. 2003). High temperature is often a primary factor affecting the rate of plant growth, development, and reproduction (Hatfield and Prueger 2015). According to the earlier reports, the harmful effects of high temperature on plants lead to significant yield loss of 10–12.5% (Kaushal et al. 2016). Increased temperature causes loss of cell water content which results in reduction of the cell size and ultimately growth (Ashraf et al. 2004). During high-temperature stress, reduction in net assimilation rate (NAR) causes reduced relative growth rate (RGR) which was confirmed in maize and millet (Wahid et al. 2007) and sugarcane (Srivastava and Ashok 2012).
Heat stress results from increased temperature adequate to damage plant tissues. Diferent tropical plant species vary in their responses to temperature stress (ranging from 35–45 °C) throught their life cycle (Hall 1992, 2001; Mahmood et al. 2010; Hasanuzzaman et al. 2013). Heat stress has a pronounced effect on germination percentage and early seedling growth in a number of plant species including sugarcane (Wahid and Shabbir 2005; Wahid et al. 2010). However, plant age and the duration of exposure to heat stress are important in sugarcane (Wahid et al. 2007). Heat stress applied to sugarcane reduced the Hill reaction, chlorophyll fluorescence and electron transport at PSII (Ebrahim et al. 1998). Despite ample water supply to roots, water potential and its components were severely affected in sugarcane leaves under heat stress (Wahid and Close 2007). As a heat tolerance strategy, sugarcane showed the synthesis of primary metabolites, viz., free proline, glycine betaine and soluble sugars, and secondary metabolites, viz., carotenoids, soluble phenolics and anthocyanins were closely associated with the heat resistance mechanism. Enzymatic antioxidants also combat heat stress-induced oxidative damage in sugarcane (Jain et al. 2007).
Sugarcane shows heat sensitivity on the far side, 36 °C, as evident from its diminished growth and water relations (Wahid et al. 2010). Despite this, heat tolerance mechanisms are relatively less understood in sugarcane. Identifying novel genes and studying their expression patterns in response to heat stress will provide a molecular basis for improving heat tolerance in crops (Yang et al. 2006; Qin et al. 2008). The available studies showed that canopy temperature is an important factor in the growth and production of new leaves in sugarcane (Robertson et al. 1998; Hatfiled et al. 2011). Despite sugarcane’s economic importance, its highly complex polyploid genome has not been sequenced so far. Because probably over 5000 sugarcane genes stay undiscovered (Cardoso-Silva et al. 2014), there is a desire for brand spanking new sequencing efforts of sugarcane transcriptomics derived from many tissues and treatments, so as to extend the fundamental set of sequence info and assist biotechnological ways. Emerging genomic technologies, such as next-generation sequencing, will allow more efficient transcriptomics and will help in laying a platform for sugarcane genomics (Manners 2011; Jain 2011).
Recently, high throughput of deep-sequencing technology has become a powerful tool for identifying specific genes and analyzing transcriptome profiles. Digital gene-RNA-Seq is a new approach for expression analysis and has identified a large number of genes involved in stress response and plant development (Hao et al. 2011; Wang et al. 2012). RNA-Seq, a next-generation sequencing (NGS) technology, is used for profiling gene expression and plant breeding programs in many plants (Li et al. 2016). To our knowledge, heat stress in sugarcane has received much less attention than the other abiotic stresses. Sugarcane varietal evolution in the future requires yield stability even under harsh climates; understanding of the metabolic and molecular signal transcription processes and the interaction to high temperatures are absolutely necessary. Therefore, our research will provide a valuable resource for understanding the mechanism of heat stress tolerance in sugarcane and will contribute to the researchers in precise genome editing for improved stress tolerance of sugarcane.
Materials and methods
Experimental design and library preparation
Seven sugarcane genotypes were selected based on their performance from the previous studies conducted at the Plant Physiology Section, Division of Crop Production, ICAR-Sugarcane Breeding Institute, Coimbatore. It consists of five commercial canes (Co 06022, Co 0315, Co 8021, Co 86032 and Co 99004) and two wild spp. (Spontaneum spp. Viz., Taiwan-96 and SES-150) genotypes. All the seven genotypes were planted in 6 inches pots filled with a mixture of red soil, sand and farmyard manure, which was previously homogenized with enough water to bring the soil to field capacity. The experiment was laid out in a completely randomized block design (CBD) with three replications. Normal agronomic practices were followed for all of these plants. To impose heat stress, three pots/genotype was chosen for the study, experimental sugarcane genotypes received heat stress, a temperature of 4–5 °C above its optimum temperature range, an increase which corresponds tightly to climate change model predictions. Control plants were grown under optimal conditions at 37/28 ± 2 °C day/night with a 12 h photoperiod. Heat-stressed plants were grown at 45/32 ± 2 °C during the day/night with a 12 h photoperiod and for a total of 15 days, with 60–70% relative humidity and light intensity 395–410 μmol m−2 s−1.
Based on physiological and biochemical assays, among the varieties, Co 99004 was selected as the heat stress-tolerant sugarcane variety. Meristematic tissues were collected from three pots (one pot/replicate) from both control and treated samples at formative phase of the crop (150 days). These samples were considered as one biological replicate, immediately frozen in liquid nitrogen and then stored at − 80 °C for total RNA isolation. The total RNA was isolated from the meristematic tissues of sugarcane genotype Co 99004 (using TRIzol reagent (Ambion, Life technologies, North America.) according to the manufacturer’s instructions. The integrity and the concentration of the total RNA was determined for each sample by Nanodrop 2000C spectrophotometer (Thermo Scientific, USA). The A260/A280 ratio was calculated to confirm the purity of RNA. A part of it was outsourced for transcriptome sequencing and another part was equalized to a concentration of 0.1 µgµl−1 and stored for RT-PCR reactions.
An equal amount of total RNA (4 µg) from both control and treated (high temperature) samples was used for library construction and transcriptome sequencing. Poly-A containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. For purification, the mRNA was fragmented into small pieces using divalent cations at elevated temperature. The cleaved RNA fragments were used for first-strand cDNA (reverse transcriptase and random primer were used), followed by second-strand cDNA synthesis (DNA polymerase I and RNase H were used). Those cDNA fragments were processed by the end repair process, addition of a single ‘A’ base and ligation of the adapters. The product was purified and enriched with PCR to create the final cDNA library. Bio-analyzer plots were used at every step to assess the mRNA quality, enrichment success, fragmentation sizes and the final library sizes. Qbit was used for measuring the quantity of the library before sequencing.
Trancriptome sequencing, pre-processing and de novo transcriptome assembly
These libraries were sequenced on Illumina HiSeq 2500 instrument. After sequencing, raw reads were processed to remove adapter sequences and trimming of bases and reads of low quality. High-quality reads were processed by Cutadapt software (version 1.8) (https://github.com/marcelm/cutadapt). Sickle 1.33 (https://github.com/najoshi/sickle) software was used for checking the quality of transcriptome sequences which described the base quality score distribution, base composition distribution and GC distribution. Bowtie2 (version 2.2.6) (https://Bowtie2-bio.sourceforge.net/index.shtml) software removed contaminations in RNA-seq very fast and sensitively. Finally, clean reads were produced from raw reads. Clean reads were normalized and assembled into expressed transcripts as full-length sequences. The duplicated genes were found using Trinity (version 2.3.2) software (https://trinityrnaseq.sourceforge.net). CD-HIT (version 4.6) software tool ((https://cd-hit.org) was utilized to perform the sequence into a non-redundant transcripts set of gene-oriented clusters and comparing protein or nucleotide sequences. Transcript clustering was done using the CD-HIT-v4.5.4 at 95% identity and 95% query coverage. The assembled transcripts were blasted against all Viridiplantae kingdom protein sequences from Uniprot, GO (Gene Ontology) terms, KEGG (Kyoto Encyclopedia of Genes and Genomes) and KAAS (https://www.genome.jp/tools/kaas/). The unigenes were aligned to the COG (Clusters of Orthologous Group) database to predict and classify possible functions.
Differential gene expression analysis, functional annotation of transcripts and validation of DGE
In-house script tool was used for a sequence of RNA pool again, filtered the contigs with FPKM ≥ 1 for every fragment was sequenced and captured the unique contigs from multiple samples. The transcripts with different level fold changes of FPKM in response to high- temperature stress were compared to FPKM value of the control sample. More than twofold changes and adjusted p value p < 0.05 were identified as differentially expressed genes by DeSeq (version1.16.0) database (https://www.bioconductor.org/packages/release/bioc/html/DeSeq.html). The plant metabolic pathway analysis was performed based on the PMN database (https://www.plantcyc.org/). The assembled transcripts were annotated using CANoPI (Contig annotator Pipeline). The transcripts were blasted against the NRNCBI database and then functionally annotated through gene ontology annotation, pathway annotation, gene and protein annotation, and organism annotation. The assembled mRNA were queried to non-redundant protein database from NCBI and checked for similarity (under the parameters of E-value ≤ 1e−5, similarity ≥ 40%) to existing known protein from various plant genomes. Organisms’ similarity distribution and top hits for organism were found using BLASTX. By using CANoPI, significant BLASTX hit and UniProt hit provided the protein status, subcellular location of protein and EC numbers for sequences. The transcripts were compared with the GO database to classify unigenes based on biological processes, molecular function and cellular components. A cluster of orthologous genes: eggNOG database was used to annotate any sequences present and its corresponding orthologous groups.
To validate the differential expression, nine genes were chosen and quantitative -time polymerase chain reaction (qRT-PCR) was performed with a Step-One plus Real-Time PCR system (Applied Biosystems, Canada). Each reaction was carried out in triplicate; the primers used for qRT-PCR were collected from the NCBI database. For qRT-PCR experiments, dissociation curve analysis was also performed to check the primer specificity. The cocktail for qRT-PCR reaction contained 50 ng cDNA, 2.5 pmol primers, 12.5 µl of 2X SYBRGREEN Master Mix (Qiagen) and with nuclease-free water and the total reaction mixture was made up to 25 µl. The reaction was performed for 40 cycles (denaturation for 10 min at 95 ○C followed by annealing and extension at 1 min for 60 ○C).The cycle threshold (Ct) values of the candidate genes were used to evaluate their expression stability by using NormFinder applet for MS Excel. Norm Finder allows the user to determine variance as well as the stable value of each candidate gene. Using a reference gene (actin), the fold change of target gene expression levels comparing the meristematic tissues of both control and treated samples were determined using the 2Δ−ΔΔCT method (Livak and Schmittgen 2001). Statistical significance was determined using Student’s one tailed t test with significant differences for p < 0.05.
Results and discussion
Seven sugarcane clones were exposed to high-temperature stress and screened for their performance through growth parameters such as stay green nature and tillering ability. Based on these basic data analysis one sugarcane clone Co 99004 was identified to be relatively tolerant to high-temperature conditions with significant differences in the growth parameters. Hence, the clone Co 99004 was identified for the transcriptome analysis.
De novo assembly and functional annotation of transcripts
To identify the molecular mechanisms involved in sugarcane in response to high temperature stress, we constructed a transcriptome profile with the help of Illumina Sequencing-Paired End libraries from control (C) and treated (T) meristematic tissues of Co 99004. The HiSeq 2500 platform yielded 5.5 and 3.2 million raw reads for C and T, respectively. These raw reads were checked for quality using Fast QC and the low-quality sequences (phred score < 20 bp) of the paired end reads and vague raw reads were trimmed using SILVA database (https://www.arb-silva.de/) from raw reads. During pre-processing the raw reads of 28.9 and 57.6% for control and treated were removed, respectively, which resulted in 39,280,628 (39 million) and 13,437,294 (13.4 million) clean paired-end reads, corresponding to 3.8 and 1.3 Gb clean bases, for C and T, respectively, and used for further assembly. The Trinity software was used to generate the de novo transcriptome assembly of sugarcane meristematic tissue for high-temperature stress response. We generated an transcriptome assembly of 1,42,859 contigs with a mean GC of 49.64%. The length distribution of the assembly is shown in Fig. 1. Approximately, 59.6% of the assembled transcripts were ~ 200 to > 5000 bp in length. The redundancies in the assembly were removed by CD-HIT-EST, after which the assembled transcripts were 118,017 sequences with a mean GC content of 49.44%. The longest sequence in the assembled transcriptome was 21,435 bp which had a similarity of (99%) with a protein of Maturase K (Intron maturase) from Saccharum officinarum; the same has been submitted in NCBI and acquired an accession number (NCBI Acc. No. 49659490). Transcript of length ≥ 200 bp was considered for transcript expression estimation and annotations (Table 1).
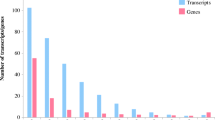
In this study, we have constructed the gene expression profile of Co99004 for heat stress tolerance with the help of Illumina Sequencing-Paired End libraries. RNA-Seq is one of many applications that can be performed using deep transcriptome sequencing (Metzker 2010). The Trinity assembler was used for De novo transcriptome assembly analysis. The longest sequence in the assembled transcriptome was 21,435 bp, which is a similar to putative chloroplast-encoded group II intron maturase and involved in the regulation mechanisms relating to plant development and, indirectly, to photosynthesis which was in accordance with Barthet and Hilu (2007). Transcriptome annotation provides insight into the function and biological process of transcripts and the proteins they encode. It consists of inter-linkages in between biological information of biochemical and biological functions, involved in regulations, interactions and expression to the genomic elements to improve genomic annotations. To understand the features and functions of the assembled transcripts, we used BLASTX algorithm for annotation against different protein databases, viz., NCBI plant non-redundant (nr), UniProt, GO, KEGG pathway, KOG, COG and eggNOG. The parameters set were hits with an e-value cutoff of 10–5. A total 85,176 (72.1% of clean reads) transcripts were annotated, of which we found that 50,882 (59.7%) of assembled transcripts had at least one significant hit, mapped with 268 organisms to the NR database under the e-value of 1 × 10–5. During the BLASTX search, 44% of transcripts were distributed between 1e−5 and 1e−50 and the similarity score was 74% that was distributed 80–99 (Fig. S1). As expected in the NR mapping, we found that 36,364 (71.5%), 5495 (10.8%), 3752 (7.37%) and 1192 (2.34%) assembled transcripts were aligned to Sorghum bicolor, Zea mays, Setaria italica and Oryza sativa subsp. japonica respectively. The top hit organism distribution of the transcriptome showed that 35,000, 25,000, and 20,000 transcripts were similar to Sorghum, Zea mays and Setaria italica, respectively. Only few transcripts were mapped to Saccharum species (Fig. 2); this might be due to the unavailability of sugarcane genome and gene sequences for open access, which has been reported by many researchers (Li et al. 2016; Prathima et al. 2017; Kasirajan et al. 2018). Among the 50,882 transcripts, 45,171 (38%) transcripts were annotated using UniPort database to determine their GO functional annotations. These transcripts were categorized into “biological process,” 30,070 (66.5%), molecular function,” 43,815 (96.9%) and cellular components 28,367 (62.7%). Out of the transcripts assigned to biological processes, 1.67, 1.24, 1.22, 1.15 and 0.76% of transcripts were annotated into regulation of transcription, DNA template, “DNA integration” and “carbohydrate metabolic process”, respectively. Similarly in the “molecular function” group, the transcripts were assigned to “ATP binding” (4.17%), “zinc ion binding” (2.68%), “nucleus” (2.44%), “nucleic acid binding” (2.37%) and “protein kinase activity” (1.23% transcripts), ranked the highest, and in the cellular component, 11.4, 3.14, 3.07 and 1.57% transcripts were annotated as a subcategory of “integral component of membrane”, “nucleus”, “DNA binding” and “cytoplasm”, respectively (Fig. S2a, b, c).
Clustering and KEGG orthology classification
We annotated the transcripts subsequently with the Clusters of Orthologous Groups (COG) protein database and classified into functional categories. A total of 6901 (5.84%) transcripts were annotated and classified into 50 functional categories according to their putative functions. Most of the transcripts 4692 (67.9%) grouped under hypothetical/unknown function or with no similarities to previously reported sequences/genes, denoated as “S”, which indicates the presence of putative novel genes that are reported here for the first time and may be specific to sugarcane. The second largest category (5.46%) comprised of (K) “transcription”-related transcripts, followed by 4.26, 3.29, 3.19 and 2.52% transcripts of (O) “-translational modification, protein turnover, chaperones,” (U) indicating active lipid, calcium signaling and metabolic networks have been initiated in the meristematic tissues of Co 99004 to manage the heat stress and “intracellular trafficking, secretion, and vesicular transport,” (J) which included the proteins for endomembrane trafficking, “translation, ribosomal structure and biogenesis,” and (G) “carbohydrate transport and metabolism”, respectively (Fig. S3).
The KEGG database provides systemic functional information of biochemical pathways and functions of gene products in addition to annotations of sequences. Major KEGG biochemical pathways groups were “genetic information processing” (1603 transcripts), “metabolism” (1248 transcripts), “cellular processes” (689 transcripts) “environmental information processing” (599 transcripts), and “unclassified” (108 transcripts). Among the categories, 37.7% transcripts were grouped in the genetic information processing which included folding, sorting and degradation, translation, transcription and replication and repair. The second largest percentages of transcripts were distributed in “metabolism” (29.38% transcripts) and it was grouped into various subcategories such as carbohydrate metabolism, amino acid metabolism, energy and lipid metabolism and so on. “Cellular processes” is one of the major KEGG biochemical pathways, 16.22% transcripts were classified under this category (Fig. S4). It includes transport and catabolism, cell growth and death, cellular community and cell mortality. Signal transduction, signaling molecules interactions and membrane transport were as a subcategory of the “environmental information processing”, and 14.10% transcripts were classified under this group.
Differential gene expression analysis and GO ontology for the DE transcripts
To identify the differentially expressed genes we used FPKM (The fragments per kb per million fragments) (Trapnell et al. 2009). We considered the transcripts with FPKM values > = 1.0 and with p-value < 0.05, log2fold change (FC) > 2.0 for further analysis (Ma et al.2012). The numbers of transcripts were 60,733 and 72,763 for control and treated samples respectively; which is referred to as the high temperature response transcripts (HTRTs) in the present study in the later sections. Totally, 2308 transcripts were differentially expressed in the meristematic tissues of sugarcane in response to heat stress, which is shown in the volcano plot (up-regulated in red; down-regulated as blue) Fig. S5. Among the heat-responsive DE transcripts, 1137 transcripts were up-regulated and 1171 transcripts were down-regulated in heat stress-tolerant sugarcane cultivar Co 99004 in comparison to control. These up- and down-regulated transcripts were distributed in different ranges of fold change. Among the upregulated transcripts the highest number of the transcripts (640) fell in the four times fold change, while 227,145 transcripts grouped under three- and twofold increase in expression respectively. There were also six- and one transcript/s with six- and sevenfold change, respectively (Fig S6). Differentially expressed transcripts of heat stress-tolerant sugarcane variety were annotated using UniPort database to draw the GO tree to classify the up- and down-regulated transcripts into putative functional groups. On the basis of Gene Ontology (GO) analysis, we analyzed 1137 up-regulated transcripts and categorized into 82 GO functional subcategories, which were summarized into three main categories “biological process” “metabolic function” and “cellular process” (Fig. 3): In the “biological process” ontology transcripts were mainly distributed into “biological process” (250 transcripts, 20.50%), “cellular process” (212 transcripts, 17.39%), “metabolic process” (210 transcripts, 17.22%) and “biosynthetic process” (113 transcripts, 9.2%). GO analysis also showed 242 transcripts (25.34%) had “binding” function, “nucleotide binding” 128 transcripts (13.40%) and 106 (11.09%) with “catalytic activity” function in “molecular function” ontology. The subgroup of “cellular component” (155, 22.59%), “membrane” (124, 18.07%), “intracellular” (72, 10.49%) and “cell” (70, 10.20% transcripts) ranked the highest in the category of “cellular component”.
GO term distribution of the down-regulated differentially expressed transcripts 1171 were functionally categorized into 76 subcategories, which were summarized into three main categories: “biological process” “metabolic process” and “cellular process”. GO analysis showed 249 (19.69%) transcripts had “biological process, “cellular process” 194 (15.34%) and 175 (13.84%) with “metabolic process” function in “Biological process” ontology. The subgroup of “binding” (216, 25.68%), “hydrolase activity” (90, 10.70%) and “catalytic activity” (82, 9.75% transcripts) ranked the highest in the category of “molecular function”. In the “cellular component” ontology transcripts were mainly distributed in “cellular component” (161, 22.39%), “membrane” (128, 17.80%) and “cell” (80, 11.12%). In addition, some transcripts showed no similarity to any known protein database; there were probabilities that they were putative long noncoding RNAs or novel genes in sugarcane variety (Cardoso-Silva et al. 2014). More experiments are required to characterize them (Metzker 2010).
KEGG pathway classification of the differentially expressed transcripts
The orientations of the differentially expressed upregulated transcripts in metabolic pathways were analyzed by querying the KEGG database. The total upregulated transcripts (1137) were assigned to five specific pathways, including metabolism, cellular processes, environmental information processing, genetic information processing and hypothetical/unknown. The largest category contained 69.8% of up-regulated DE transcripts with hypothetical/uncharacterized function. The remaining 30.19% of upregulated transcripts sequences were found to be involved in 30 predicted KEGG metabolic pathways. The pathways with the most representation by the upregulated transcripts were “metabolic” pathways (61% transcripts) which included “carbohydrate metabolism” (26%), “amino acid metabolism” (26%), “energy metabolism” (20%), “enzyme families” (13%) and so on. The second largest percentage of up-regulated transcripts were distributed in “genetic information processing” (17% transcripts) and it grouped into various subcategories. They were, “Folding, sorting and degradation” (64%), “translation” (27%) and “transcription” (9%). in the “cellular processes” categories, the most significant enriched KEGG pathways were transport and catabolism (43%), followed by cell growth and death (43%), and cellular community-eukaryotes (14%). Additionally, the most abundant subcategories in KEGG “environmental-information processing” (11%) category was “signal transduction” (71%) and “membrane transport” (29%). The above functional annotations indicated that the clustered up-regulated transcripts represented an extensive catalog encompassing a large proportion of the genes expressed in response to heat stress (Fig. 4a).
The down-regulated transcripts (1171) were assigned into five specific pathways; the largest category contained 54.99% of down-regulated transcripts with hypothetical//uncharacterized function (Fig. 4b). The remaining 45% of down-regulated transcripts were found to be involved in 18 predicted KEGG metabolic pathways. The pathways with the most representation by the down-regulated transcripts were “metabolic” pathways (40% transcripts) which included “energy metabolism” (36%), “enzyme families” (17%), “carbohydrate metabolism” (8%) and so on. The second the largest percentage of down-regulated transcripts were distributed in “genetic information processing” (30% transcripts) and it was grouped into various subcategories such as “folding, sorting and degradation” (44%), “translation” (26%) and “transcription” (26%). In the “environmental-information processing” categories, the most significant enriched KEGG pathways were signal transduction (47%), followed by signaling molecules and interaction (40%) and membrane transport (13%). Additionally, the most abundant subcategories in KEGG “cellular processes” (13%) category were “transport and catabolism” (67%) and “cell growth and death” (33%). The most represented KEGG biochemical pathways classification were “metabolic pathways” (23%), “photosynthesis—antenna proteins” (17%), “protein processing in endoplasmic reticulum” (13%), “biosynthesis of secondary metabolites” (8%) and so on.
There were common differential expressed transcripts detected between up- and down-regulated transcripts (Fig. 5). Totally, 458 transcripts and 305 transcripts were up- and down-regulated, respectively, in response to heat stress in sugarcane. Among the 458 up-regulated transcripts, 328 transcripts were found to be commonly up-regulated in both control and heat stress. Among the down-regulated transcripts, 292 transcripts were down-regulated in both treated and control. 130 transcripts uniquely up-regulated during heat stress, while no transcripts were uniquely down-regulated. The most represented classification was metabolic pathways, biosynthesis of secondary metabolites, carbon metabolism, glyoxylate and dicarboxylate metabolism, glycine, serine and threonine metabolism, alanine, aspartate and glutamate metabolism, glycolysis/gluconeogenesis, carbon fixation in photosynthetic organisms, pyruvate metabolism and so on. Mangrauthia et al. (2016) reported earlier that the high temperature altered the expression of genes involved in fatty acid, amino acid and energy metabolism. At a cellular level, heat stress triggers multiple and often interconnected signal transduction pathways that lead from initial perception to a finally enhanced thermotolerance by Boyko et al. (2010). However, as evident from the various induced response mechanisms, the adaptation of plants to heat stress requires energy and, thus, occurs at the expense of metabolic processes such as cellular growth (Barnabas et al. 2008).
Transcripts associated with heat stress and real- time validation
The molecular mechanism regulating the heat tolerance in sugarcane is still unclear; however, genes associated with other abiotic stresses have been studied extensively. We analyzed the commonly up-regulated transcripts in treated and found that theses genes listed are releated to heat stress tolerance (putative l-ascorbate peroxidase 6 (APX6)—response to oxidative stress [GO:0006979], hypothetical protein (SORBI_001G418600)heat shock protein 70 family, PREDICTED: stress protein DDR-48-like (LOC100822915), PREDICTED: thioredoxin F, chloroplastic (LOC100834089 BRADI_2g58440)—cellular response to oxidative stress [GO:0034599], hypothetical protein (SORBI_3002G188500)—trehalose metabolism in response to stress [GO:0070413], hypothetical protein (SORBI_001G363100)—response to stress [GO:0006950], heat shock protein 26.8—small heat shock protein (HSP20) family, hypothetical protein (SORBI_007G126100)—trehalose metabolism in response to stress [GO:0070413], catalase-2-like—response to oxidative stress [GO:0006979], hypothetical protein F775_06747—cellular response to oxidative stress [GO:0034599], glutamate dehydrogenase 2 (F775_29860)—response to salt stress [GO:0009651], 70 kDa heat shock protein—heat shock protein 70 family, hypothetical protein (SORBI_3010G161600)—response to oxidative stress [GO:0006979], hypothetical protein (ZEAMMB73_Zm00001d002898)—response to oxidative stress [GO:0006979] and endoplasmin (TRIUR3_31474)— response to heat [GO:0009408]).
In this study, heat shock protein families 20, and 70 with other oxidative stress tolerance proteins such as trehalose, glutamate dehydrogenase were only expressed during stress, which are involved in the regulation of transcription and post-translational modifications. Expression of HSP protein families along with other genes and transcription factors was predominant, which might be due to cross talk between abiotic stresses and their regulatory network. Our DGE analysis revealed that several genes code for dehydration-responsive element binding 1A such as grain softness protein, malate synthase1, lipoxygenase, avenin-like protein, bidirectional sugar y = transporters SWEET-13-like and a hypothetical protein of sorghum (Fig. 6). Park et al. (2009) showed that the expression of HSP and thioredoxin reportedly conferred tolerance to heat stress. However, Kotak et al. during (2007) said, “Recent reports suggest that reactive oxygen species also contribute to heat stress signaling”. The pathway analysis of differentially expressed genes at the high temperature suggested the altered expression of transcripts associated with heat shock proteins (HSP 20 and HSP 70 family) in rice (Mangrauthia et al. 2016). This evidence shows us that transcripts encoding proteins in response to heat stress can act in the pathway of stress response in sugarcane.
Differentially expressed transcripts (DETs) in heat-tolerant sugarcane variety of Co 99004 were identified by RNA-Seq and were confirmed by quantitative real-time PCR (qRT-PCR) experiment. We selected ten DETs as candidates from RNA-Seq. Integrated DNA technology software (IDT) was used to design for forward and reverse primers of these ten candidate genes and actin gene were used as the reference gene. qRT-PCR and DEGs were used to determine the gene expression pattern (Fig. 7), Among the ten chosen genes, phytepsin (involved in vacuole via trafficking through the Golgi apparatus), ferredoxin-dependent glutamate synthase (which participates in nitrogen metabolism) and stress protein DDR-48 (responsive for defense regulation) had threefold increased expression during heat stress. Among the differently expressed heat shock proteins, HSP of 70 gene family was the prominent player in sugarcane. These genes might be promising targets for manipulation for elevated temperature stress.
Conclusion
To conclude, we have employed RNA-Seq analysis to develop a transcriptome profile for a heat-tolerant clone Co 99004 and characterized candidate genes during formative stage of growth. A total of 1,42,859 transcripts were assembled and the longest sequence in the assembled transcripts was 21,435 bp. We used BLASTX algorithm for mapping of the assembled transcripts against different assorted protein databases, viz., NCBI plant non-redundant (nr), UniProt, GO, and KEGG pathway; KOG, COG and egg NOG. DGE analysis revealed that a total of 1137 and 1171 transcripts were significantly up-regulated and down-regulated, respectively. The functional annotation of these transcripts revealed that they were involved in the response oxidative stress, heat shock protein and glutamate pathway. Nine transcripts were validated for their biological significance through qRT-PCR and RT-PCR for their expression in leaf tissues. The results showed high consistency between qRT-PCR and RNA-Seq methods. The transcripts were produced by this study which will facilitate our understanding on the molecular basis for heat tolerance in sugarcane variety (Co 99004) and further improvement of heat tolerance mechanisms in sugarcane.
Data availability
All the clean reads have been submitted as sequence read archive (SRA) in NCBI with the Accession no SRR7252568, SRR7252569 for control and treatment.
References
Ashraf M (2004) Some important physiological selection criteria for salt tolerance in plants. Flora 199:361–376
Barnabas B, Jager K, Feher A (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31:11–38
Barthet MM, Hilu KW (2007) Expression of MATK: functional and evolutionary implications. Am J Bot 94:1402–1412
Boyko A (2010) Trans-generational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer like proteins. PLoS ONE 5:e9514
Cardoso-Silva CB, Costa EA, Mancini MC, Balsalobre TWA, Canesin LEC, Pinto LR (2014) De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE 9:e88462. https://doi.org/10.1371/journal.pone.0088462
Ebrahim MKH, Zingsheim O, El-Shourbagy MN, Moor PH, Komor E (1998) Growth and sugar storage in sugarcane grown at temperatures below and above optimum. Plant Physiol 153:593–602
Hall AE (1992) Breeding for heat tolerance. Plant Breed Rev 10:129–168
Hall AE (2001) Crop developmental responses to temperature, photoperiod, and light quality. In: Hall AE (ed) Crop response to environment. CRC Press, Boca Raton, pp 83–87
Hao QN, Shou XA, Sha AH, Wang C, Zhou R, Chen SL (2011) Identification of genes associated with nitrogen use efficiency by genome wide transcriptional analysis of two soybean genotypes. BMC Genomics 15:525
Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M (2013) Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci 14:9643–9684
Hatfield JL, Prueger JH (2015) Temperature extremes: effect on plant growth and development. Weather Clim Extremes 10:4–10
Hatfield JL, Boote KJ, Kimball BA, Ziska LH, Izaurralde RC, Ort D, Thomson A, Wolfe D (2011) Climate impacts on agriculture: Implications for crop production. Agron J 103:351–370
Jain M (2011) A next generation approach to the characterization of a non model plant transcriptome. Curr Sci 101:1435–1439
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143(4):1467–1483
Kasirajan L, Nam V, Hoang AF, Frederik Botha C, RobertHenry J (2018) Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci Rep 8:11612. https://doi.org/10.1038/s41598-018-30033-4
Kaushal N, Bhandari K, Siddique KHM, Nayyar H (2016) Food crops face rising temperatures: an overview of responses, adaptive mechanisms, and approaches to improve heat tolerance. Cogent Food Agric 2:1–42
Kotak S, Larkindale J, Lee U, Koskull-Doring PV, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10:310–316
Li M, Liang Z, Zeng Y, Jing Y, Wu K, Liang J, He S, Wang G, Mo Z, Tan F, Li S, Wang L (2016) De novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.). BMC Genomics 17:195
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] Method. Methods 25:402–408
Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12:161
Mahmood S, Wahid A, Javed F, Basra SMA (2010) Heat stress effects on forage quality characteristics of maize (Zea mays) cultivars. Int J Agric Biol 12:701–706
Mangrauthia SK, Agarwal S, Sailaja B, Sarla N, Voleti SR (2016) Transcriptome analysis of Oryza sativa (Rice) seed germination at high temperature shows dynamics of genome expression associated with hormones signaling and abiotic stress pathways. Tropical Plant Biol. https://doi.org/10.1007/s12042-016-9170-7
Manners JM (2011) Functional genomics of sugarcane. In: Kader JC, Delseny M (eds) Advances in Botanical Research, vol 60. Academic Press, Cambridge, pp 89–168
Metzker ML (2010) Sequencing technology-The next generation. Nat Rev Genet 11(1):31–46
Park SK, Jung YJ, Lee JR, Lee YM, Jang HH, Lee SS (2009) Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol 150:552–561
Prathima Thirugnanasambandam PP, Hoang NV, Furtado A, Botha FC, Henry RJ (2017) Association of variation in the sugarcane transcriptome with sugar content. BMC Genomics 18:909
Qin D, Wu H, Peng H, Yao Y, Ni Z, Li Z, Zhou C, Sun Q (2008) Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using wheat genome array. BMC Genomics 9:432
Robertson MJ, Bonnett GD, Hughes RM, Muchow RC, Campbell JA (1998) Temperature and leaf area expansion of sugarcane: integration of controlled-environment, field and model studies. Aust J Plant Physiol 25:819–828
Srivastava K, Ashok (2012) Sugarcane production: impact of climate change and its mitigation. Biodivers J Biol Divers 13:214–227. https://doi.org/10.13057/biodiv/d130408
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Wahid A, Close T (2007) Expression of dehydrins under heat stress and their relationship with water relations of sugarcane leaves. Biol Plant 51:104–109. https://doi.org/10.1007/s10535-007-0021-0
Wahid A, Shabbir A (2005) Induction of heat stress tolerance in barley seedlings by pre-sowing seed treatment with glycine-betaine. Plant Growth Regul 46:133–141. https://doi.org/10.1007/s10725-005-8379-5
Wahid A, Gelani S, Ashraf M, Foolad M (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223. https://doi.org/10.1016/j.envexpbot.2007.05.011
Wahid A, Farooq M, Rashead R, Gelani S, Rasul E (2010) Sugarcane under thermal stress: some biotechnological consideration. In: Ashwani K (ed) Plant genetic transformation and molecular marker. Pointer Publisher, Jaipur, pp 109–123
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang G, Zhu QG, Meng QW, Wu CA (2012) Transcript profiling during salt stress of young cotton seedlings via Solexa Sequencing. Act Physiol Plant 34:107–115
Yang KA, Lim GJ, Hong JK, Cheong YH, Chung WS, Lee KO, Lee SY, Cho MJ, Co L (2006) Identification of cell wall genes modified by a permissive high temperature in Chinese cabbage. Plant Sci 171(1):175–182
Zhao D, Li YR (2015) Climate change and sugarcane production: potential impact and mitigation strategies. Inter J Agron. https://doi.org/10.1155/2015/547386
Acknowledgements
The authors would like to thank the Science and Engineering Board, Department of Science and Technology for funding (Grant No.SB/YS/LS-165/2013) and ICAR—Sugarcane Breeding Institute for providing the necessary infrastructure. We thank Dr. C Palaniswami, Head, Division of Crop Production and Dr. A. Selvi, Principal Scientist, ICAR—SBI for their guidance and support throughout this research. We also thank Genotypic Technologies, Bangalore, for the transcriptome sequencing work.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by GR, LK and KS. The first draft of the manuscript was written by KS, and LK critically revised the manuscript. All authors read and approved the final manuscript. GR and LK contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.








Rights and permissions
About this article
Cite this article
Raju, G., Shanmugam, K. & Kasirajan, L. High-throughput sequencing reveals genes associated with high-temperature stress tolerance in sugarcane. 3 Biotech 10, 198 (2020). https://doi.org/10.1007/s13205-020-02170-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02170-z