Abstract
Superoxide dismutase (SOD) proteins are crucial antioxidant enzymes that play critical roles in plant growth, development, and response to various abiotic stresses. The SOD gene family has been characterized in various plant species, but not in Medicago truncatula yet. Here, a total of 7 MtSOD genes were first identified from the whole genome of M. truncatula, including 1 MnSOD, 2 FeSODs, and 4 Cu/ZnSODs, which are unevenly distributed in five out of the eight chromosomes. Phylogenetic analysis showed that SOD proteins from M. truncatula and other plant species could be classified into two main categories (Cu/ZnSODs and Fe-MnSODs), which could be further divided into eight subgroups, and members within the same subgroup tended to share the same subcellular localization. In addition, MtSOD genes together with AtSODs and OsSODs within the same subgroup also displayed similar motif compositions and exon–intron structures. Most MtSOD genes were ubiquitously expressed in various tissues, particularly in leaves, seeds and root nodules at different developmental stages. Moreover, microarray analysis and high-throughput sequencing showed that most MtSOD genes were differentially expressed under salt, drought, and cold treatments, indicating their pivotal roles in stress response of M. truncatula. These findings provide useful information for the functional characterization of SOD family genes for growth, development, and stress response of M. truncatula.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As one of the most vital antioxidant enzymes in plant defense system, superoxide dismutase (SOD, EC 1.15.1.1) controls the generation of reactive oxygen species (ROS) and catalyzes the dismutation of superoxide anion radicals (O2−) to H2O2 (Gill et al. 2015; Zhou et al. 2017). According to the types of prosthetic metals interacting with their active sites, plant SODs can be categorized into Cu/ZnSOD, FeSOD, and MnSOD (Feng et al. 2016a; Mittler et al. 2004; Yan et al. 2016; Zhou et al. 2017). In plants, Cu/ZnSOD is frequently observed in cytoplasm, peroxisomes, chloroplasts, and/or extracellular space. Besides, FeSODs are mostly located in the cytoplasm and chloroplasts, while MnSOD is located in the matrix of mitochondria (Corpas et al. 2006; Kliebenstein et al. 1998; Pilon et al. 2011).
Overexpression of genes that encode different isoforms of SOD can confer resistance to various abiotic stresses in plants, including heat, cold, drought and salinity. For instance, overexpression of a Cu/ZnSOD from Puccinellia tenuiflora (PutCu/Zn-SOD) conferred tolerance to a number of abiotic stresses in transgenic Arabidopsis (Wu et al. 2016). In durum wheat, the transcription of an MnSOD gene (TdMnSOD) was induced by different abiotic stresses, and recombinant yeast cells and transgenic Arabidopsis plants of TdMnSOD exhibited enhanced tolerance to multiple abiotic stresses (Kaouthar et al. 2016). These findings indicate that SODs may play essential roles in regulating stress tolerance of plants. In addition, the stress tolerance of plants is usually improved by enhancing the antioxidative defense capacity and decreasing the levels of ROS. For example, overexpressing AhCuZnSOD gene from Arachis hypogaea (Negi et al. 2015), KcCSD gene from Kandelia candel (Jing et al. 2015), and TaSOD2 gene from wheat (Wang et al. 2016a) could improve the tolerance to salt stress with a higher antioxidative defense capacity. In Sedum alfredii, overexpression of SaCu/ZnSOD in Arabidopsis could confer tolerance to oxidative stress via increasing antioxidative defense capacities, such as SOD and peroxidase activities (Li et al. 2017). These reports demonstrate the crucial roles of SODs in the adaptation of plants to abiotic stresses possibly through the elimination of ROS generated due to abiotic stresses.
In recent years, numerous SOD family members have been systematically investigated in some plant species at the genome level, including Arabidopsis thaliana (Kliebenstein et al. 1998), Dimocarpus longan (Lin and Lai 2013), Populus trichocarpa (Molina-Rueda et al. 2013), Oryza sativa (Nath et al. 2014), Sorghum bicolor (Filiz and Tombuloğlu 2015), Musa acuminata (Feng et al. 2015), Solanum lycopersicum (Feng et al. 2016a), Volvariella volvacea (Yan et al. 2016), Cucumis sativus (Zhou et al. 2017), Gossypium hirsutum, G. arboreum, and G. raimondii (Wang et al. 2016b, 2017; Zhang et al. 2016). However, there is no report about the identification and characterization of SOD gene family in M. truncatula. In the present study, 7 SOD genes, including 1 MSD gene, 2 FSD genes and 4 CSD genes, were identified and characterized in M. truncatula. In addition, their basic characteristics and expression profiles were systematically investigated.
Materials and methods
Identification of M. truncatula SOD genes
The accession numbers and corresponding amino acid sequences of SOD proteins from Arabidopsis and rice were obtained from previous research (Kliebenstein et al. 1998; Nath et al. 2014), and were used as queries to perform a BLASTp search against the NCBI database and M. truncatula genome website (https://phytozome.jgi.doe.gov/Mtruncatula). The redundant sequences were discarded based on alignment to each other, and each resulting candidate sequence was put into Pfam database (http://pfam.xfam.org/) and the SMART protein domain annotation resource (http://smart.embl-heidelberg.de/) for checking the existence of the SOD domain.
The online ProtParam website (http://ca.expasy.org/tools/protparam.html) was employed to compute the physicochemical characteristics of each MtSOD protein, such as isoelectric point (pI), grand average of hydropathy index (GRAVY) and molecular weight (MW). The subcellular location of each MtSOD protein was predicted with the help of the online server of WoLF PSORT (https://www.genscript.com/tools/wolf-psort).
Protein alignment and phylogenetic analysis
We downloaded the SOD protein sequences of Arabidopsis thaliana (8 members), Oryza sativa (8 members), and Cucumis sativus (9 members) according to the previous research (Kliebenstein et al. 1998; Nath et al. 2014; Zhou et al. 2017). In addition, 9 S. lycopersicum, 8 S. bicolor, 6 Brachypodium distachyon, 10 G. raimondii, and 9 Setaria italica SOD protein sequences were obtained from the Phytozome website (https://phytozome.jgi.doe.gov/). The names and accession numbers of all the SOD proteins are provided in Table S1. The full-length protein sequences of these SOD proteins were submitted to Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) for alignment with the default parameter settings, and the resulting alignments were visualized and edited with the Genedoc software. The MEGA 5.0 software (http://www.megasoftware.net/) was employed to build a phylogenetic tree based on the neighbor-joining (NJ) method (Tamura et al. 2011), and the bootstrap parameter was set to 1000.
Conserved domain and gene structure analysis
The conserved motifs among SOD proteins of M. truncatula, Arabidopsis, and rice were detected with the online tool of Multiple Expectation Maximization for motif Elicitation (MEME, http://meme-suite.org) (Bailey et al. 2009), using the following parameters: the maximum number of motifs were 10; and the motif widths were restricted between 6 and 50. For gene structure analysis, the coding sequences (CDS) and genomic DNA (gDNA) sequences of the SOD genes of M. truncatula, Arabidopsis, and rice were downloaded from the M. truncatula genome database (https://phytozome.jgi.doe.gov/Mtruncatula), the TAIR website (http://www.arabidopsis.org/), and the rice genome resource (http://rice.plantbiology.msu.edu/), respectively. Then, these sequences were submitted to GSDS (Gene structure Display Server, http://gsds.cbi.pku.edu.cn) to determine the gene structures of these SOD genes.
Chromosomal distribution and gene duplication
The physical locations of the MtSOD genes were obtained from the M. truncatula genome database (https://phytozome.jgi.doe.gov/Mtruncatula), and drafted to the chromosomes of M. truncatula using the GenomePixelizer software (http://www.atgc.org/GenomePixelizer) (Kozik et al. 2002). The segmental gene duplication event was marked based on the previously described criteria: the sequence alignment length covered more than 80% of the longer gene, and the identity of the aligned regions was over 70% (Jia et al. 2017; Song et al. 2017). The tandem duplicated genes were defined as two or more homologous genes located on one single chromosome with a distance of 100 kb without any intervening gene (Kayum et al. 2017).
In silico expression patterns of MtSOD genes
To investigate the expression levels of the MtSOD genes in different tissues and under salt stress, the raw microarray data were retrieved in MtGEA (https://mtgea.noble.org/v3/). The expression of each MtSOD gene was normalized and calculated with the quantile algorithm using the Expression Console software supplied by Affymetrix, and visualized as previously described (Xuan et al. 2016; Zhou et al. 2018b).
Transcriptome sequencing (RNA-seq) was performed to investigate the expression patterns of MtSOD genes under various abiotic stresses. Four-week-old M. truncatula seedlings were transferred to various stress conditions including salt, drought, and cold as described previously (Song et al. 2017), and the seedlings were collected at different time points (0, 2, 6, and 12 h) and then frozen in liquid nitrogen for later total RNA extraction. The construction and deep sequencing of cDNA libraries were conducted using the Illumina HiSeq 3000 platform (Genergy Biotechnology, Shanghai, China). Data preprocessing and analyses were performed based on the methods described in the previous study (Song et al. 2017). The transcriptions of MtSOD genes were computed as FPKM and visualized as heatmap with hierarchical clustering following previous descriptions (Yang et al. 2017; Zhou et al. 2018a).
Results
Genome-wide identification of SOD family genes in M. truncatula
To identify MtSOD family members from M. truncatula, BLASTP was employed to search the M. truncatula genome with the protein sequences of AtSODs and OsSODs as queries. As a result, seven putative members were identified, and their amino acid sequences were checked by searching with SMART and Pfam tools for the existence of a SOD domain, which is the typical characteristic of SOD gene family proteins. The results showed that the M. truncatula genome harbored 1 MnSOD, 2 FeSODs and 4 Cu/ZnSODs (Table 1). All the 4 Cu/ZnSODs (MtCSD1–MtCSD4) contained a conserved Sod_Cu domain (Pfam: PF00080), which is the typical characteristic of Cu/ZnSOD proteins. Amongst them, MtCSD2 contained an additional heavy metal-associated domain (Pfam: PF00403), which was also found in some Cu/ZnSOD proteins from other plant species, such as SbSOD3 (Filiz and Tombuloğlu 2015), SlSOD4 (Feng et al. 2016a), and CsCSD3 (Zhou et al. 2017). The 2 FeSODs (MtFSD1 and MtFSD2) and 1 MnSOD (MtMSD) each contained an N-terminal SOD alpha-hairpin domain (Pfam: PF00081) and a C-terminal SOD domain (Pfam: PF02777), suggesting that they belong to Fe/MnSODs.
The gDNA length of MtSOD genes ranged substantially from 1617 bp (MtCSD4) to 5363 bp (MtMSD), which encoded proteins with lengths from 152 (MtCSD4) to 312 (MtCSD2) aa. The predictions of physical and chemical characteristics by the ProtParam server showed that the MW of the MtSOD proteins varied from 15.23 to 35.33 kDa, with calculated pI ranging from 5.45 to 7.90, and GRAVY values ranging from − 0.690 (MtFSD1) to − 0.012 (MtCSD1) (Table 1). In addition, pairwise identity analysis showed that MtSOD proteins shared 8.46–69.08% identity at the amino acid level (Table S2). Amongst them, the identity of MtMSD and MtFSD proteins was 32.55–44.92%, while that of MtCSD proteins was 26.97–69.08% (Table S2). Additionally, WoLF PSORT analysis showed that MtCSDs and MtFSDs were located in the chloroplast or cytoplasm, whereas MtMSD was localized in the mitochondria (Table 1).
Phylogenetic relationships of SOD gene family members between M. truncatula and other plant species
To study the phylogenetic history of SOD gene family members between M. truncatula and other plant species, 74 full-length SOD protein sequences from M. truncatula (7 MtSODs), C. sativus (9 CsSODs), S. lycopersicum (9 SlSODs), O. sativa (8 OsSODs), S. bicolor (8 SbSODs), A. thaliana (8 AtSODs), B. distachyon (6 BdSODs), G. raimondii (10 GrSODs), and S. italica (9 SiSODs) were aligned using Clustal Omega to generate a phylogenetic tree. As a result, these SOD proteins fell into two major groups (Fig. 1). All of the Cu/ZnSODs constituted Group I, which could be further classified into four subgroups (a–d). MnSODs were clustered with FeSODs into Group II, which could also be subdivided into four subgroups (e–h), implying that MnSODs and FeSODs might originate from a common ancestor of plants. MtSODs were distributed in each subgroup, except for subgroup f, which included only monocotyledonous FeSODs (Fig. 1). In contrast, subgroup h harbored dicotyledonous FeSODs, while subgroup g had both monocotyledonous and dicotyledonous FeSODs. Similarly, subgroups a–d and subgroup e also included Cu/ZnSODs and MtSODs from monocots and dicots, and these SODs could be clearly separated into monocot-specific and dicot-specific groups, suggesting their independent evolution after the divergence of monocots and dicots. In addition, all MtSODs were clustered with dicotyledonous SODs, implying a closer relationship of MtSODs with dicot SOD proteins than with monocot SOD proteins (Fig. 1). It is noteworthy that MtSOD2 was clustered with CsCSD3 and chloroplastic Cu/ZnSODs from other plants in subgroup d, suggesting that it is a copper chaperone for superoxide dismutase (CCS) protein.
Phylogenetic analysis of SOD proteins from M. truncatula and other plant species. Multiple sequence alignments of the full-length SOD protein sequences from M. truncatula, C. sativus, S. lycopersicum, O. sativa, S. bicolor, A. thaliana, B. distachyon, G. raimondii, and S. italica were performed with Clustal Omega, and the phylogenetic tree was constructed with MEGA 5.0 software by the neighbor-joining method using 1000 bootstrap replicates. The numbers indicated for each clade represent the bootstrap support values given as percentages
Conserved domain analysis of MtSOD proteins
To study the conservation among the seven MtSOD proteins, we carried out multiple sequence alignments of the sequences from SOD proteins of M. truncatula and other plant species based on their different types. The two conserved Cu/ZnSOD signatures (GFH[VLI]H[EA][YL]GDTT and GNAG[GA]R[VL]ACG) were present in MtCSD1, MtCSD3 and MtCSD4 (Fig. 2a), but absent in MtCSD2, which contained two conserved metal-binding motifs (MXCXXC and CXC) (Fig. 2b). In addition, MtCSD1, MtCSD3 and MtCSD4 harbored the metal-binding sites for Cu2+ and Zn2+. The FeSOD signature ([AE][QEL][VA]WNH[DEH]FFWES) and conserved metal-binding domain (D[VL]WEHAYY) were found in FeSODs and Fe/MnSODs, respectively (Fig. 2c, d).
Comparison of the amino acid sequences of MtSOD proteins. a Multiple sequence alignment of the deduced amino acid sequences of Cu/ZnSOD proteins. Two conserved Cu/ZnSOD signatures (GFH[VLI]H[EA][YL]GDTT and GNAG[GA]R[VL]ACG) are underlined. The metal-binding sites for Cu2+ and Zn2+ are marked with regular and inverted triangles, respectively. b Multiple sequence alignment of the deduced amino acid sequences of CCS proteins. The conserved metal-binding motifs (MXCXXC and CXC) are underlined. c Multiple sequence alignment of the deduced amino acid sequences of FeSOD proteins. The FeSOD signature ([AE][QEL][VA]WNH[DEH]FFWES) and conserved metal-binding domain (D[VL]WEHAYY) are underlined. Two conserved metal-binding His residues are marked with regular triangles. d Multiple sequence alignment of the deduced amino acid sequences of MnSOD proteins. The conserved metal-binding domain (DVWEHAYY) is underlined. Six conserved residues (His, Gln, and Asp) are marked with regular triangles
For clarification of the conserved domains of MtSODs, we employed MEME program to identify the conserved motifs in the SOD proteins of M. truncatula, Arabidopsis and rice, and ten conserved motifs were identified (Fig. 3). Amongst them, motifs 7, 1, 10 and 2 were related to the Sod_Cu domain. Motifs 4, 8 and 6 were specific to the N-terminal SOD domain, while motifs 5, 9, 3 and 10 were associated with the C-terminal SOD domain. It is noteworthy that SOD proteins gathered in the same group tended to have similar motif distribution patterns, which further supported the group classification. All of the Cu/ZnSODs had motifs 7, 1, 10 and 2, except for two CCS members, MtCSD2 and CuZn-SOD-CCh, which contained only two conserved domains, motif 1 and motif 7 (Fig. 3). Interestingly, motif 8 was absent in MnSODs but present in FeSODs, suggesting their divergence in motif composition. In addition, two FeSODs, AtFSD2 and MtFSD1 had double motif 9, implying their particular functions (Fig. 3).
Phylogenetic relationship and conserved motif analysis of SOD proteins from M. truncatula, Arabidopsis, and rice. Left: phylogenetic analysis for the SOD proteins from Arabidopsis, rice, and M. truncatula through neighbor-joining method with 1000 bootstrap replicates. The proteins can be further divided into eight subgroups as marked by different colors. Right: conserved motif analysis of the SOD proteins from Arabidopsis, rice, and M. truncatula using the online tool MEME. The ten motifs are assigned to differently colored boxes
Structural features and chromosomal location of MtSOD genes
The structural features of MtSODs, AtSODs and OsSODs were examined by GSDS program. Generally, these SOD genes had 5–9 introns, and genes gathered in the same group tended to have similar structural features (Fig. 4). For example, in Group I, most members in subgroups a and c harbored 6 introns, with the exception of cCuZn-SOD1, which had the largest number of introns (9), while the members of subgroups b and d shared a strictly conserved intron number (7 and 5, respectively). In Group II, the members (MnSODs) in subgroup e shared the same number of introns (5), while the intron number of FeSODs (subgroups f–h) varied from 6 to 8.
Gene structure analysis of SOD genes from M. truncatula, Arabidopsis, and rice based on phylogenetic relationship. Left: phylogenetic analysis for the SOD proteins from Arabidopsis, rice, and M. truncatula. Right: structural features of the SOD genes from Arabidopsis, rice, and M. truncatula were examined using the online tool GSDS. Lengths of introns, exons, and upstream/downstream of each SOD gene are proportionally presented
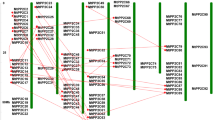
The MtSOD genes were distributed in 5 of the 8 chromosomes of M. truncatula (Fig. 5). Chromosomes 3 and 4 each possessed two MtSOD genes, while chromosomes 1, 6 and 7 each included only one gene.
Tissue-specific expression profiles of MtSOD genes
To study the expression patterns of MtSOD genes, the expression data were downloaded from MtGEA (https://mtgea.noble.org/v3/). The results indicated that 9 MtSOD genes were expressed in all the tested tissues (Fig. 6). However, MtCSD2, MtCSD3, and MtFSD2 exhibited low transcript abundance, and their highest expression was detected in seeds at 36 days after pollination (DAP), leaves, and petioles, respectively. In addition, the expression of three genes (MtCSD1, MtCSD4 and MtMSD) was relatively higher in seeds, suggesting their functions in early seed development (Fig. 6a). The expression of MtSOD genes during nodule development was also examined. MtCSD4 and MtMSD were more highly expressed in root nodules, while the expression of MtFSD2 in root nodules was much lower and even undetectable (Fig. 6b).
Expression profiles of MtSOD genes in different developmental stages including panicle development (a) and root development (b). The average log signal values of MtSOD genes were downloaded from the Medicago truncatula gene expression Atlas (MtGEA) Project database (https://mtgea.noble.org/v3/). The color scale (representing log signal values) is shown at the bottom. dap, days after pollination
Expression profiles of MtSOD genes under abiotic stress conditions
To assess the roles of MtSOD genes in response to abiotic stress, the expression profiles of MtSOD genes under salt stress were analyzed using a publicly available microarray of M. truncatula (Li et al. 2009). As a result, compared with in the untreated control sample, nearly all MtSOD genes displayed strongly down-regulated expression levels at all time points, with the exception of MtCSD4, whose expression significantly increased after treatment of 6 h, and subsequently declined at 24 and 48 h (Fig. 7a).
Expression patterns of MtSOD genes in response to various abiotic stresses based on data of microarray (a) and transcriptome (b–d). a Two-week-old M. truncatula seedlings were grown in hydroponics media with 180 mM NaCl for 0, 6, 24, and 48 h (Li et al. 2009). The microarray data were retrieved from the Medicago truncatula gene expression Atlas (MtGEA) Project database (https://mtgea.noble.org/v3/), and represented as the relative signal intensity values. Transcriptome sequencing (RNA-seq) was performed to investigate expression profiles of MtSOD genes in response to salt (b), drought (c), and cold (d) as described in our previous study (Song et al. 2017). The FPKM-normalized values were represented by a color gradient from low (blue) to high expression (red)
To further evaluate the expression profiles of MtSOD genes under abiotic stress, we performed high-throughput sequencing to determine the expression profiles of MtSOD genes under various stresses including salt, drought, and cold according to our previous study (Song et al. 2017). Under salt stress, the expression levels of MtCSD1, MtCSD2 and MtCSD4 started to rise at the early stage of stress (2 h), and subsequently declined at 6 and 12 h, while the expression of the rest MtSOD genes decreased significantly at some time points (Fig. 7b). Similar expression pattern of MtSOD genes was observed under drought stress. Most of MtSOD genes showed significant decreases in expression under drought stress, except for MtCSD2 and MtCSD4, whose expression increased sharply and reached the highest level at 2 h, and declined at 6 and 12 h (Fig. 7c). Cold stress significantly decreased the expression of all MtSOD genes (Fig. 7d).
Discussion
In this study, we identified and characterized 7 SOD genes in the M. truncatula genome by genome-wide method, including 1 MnSOD, 2 FeSODs, and 4 Cu/ZnSODs. Previous studies have revealed that different plants harbor various numbers of SOD genes (Zhou et al. 2017), and M. truncatula has a smaller number of SOD genes than several other plant species, including Arabidopsis (Kliebenstein et al. 1998), rice (Nath et al. 2014), tomato (Feng et al. 2016a), sorghum (Filiz and Tombuloğlu 2015), and cucumber (Zhou et al. 2017). Segmental and tandem duplication events were found in SOD family genes of a number of plant species (Feng et al. 2015, 2016a; Wang et al. 2016b, 2017; Zhou et al. 2017), but neither segmental nor tandem duplication events have been identified among MtSOD genes, suggesting that segmental and tandem duplications are not involved in SOD gene family expansion in M. truncatula, and the smaller number of MtSOD genes may be due to the whole-genome duplication, which was also discovered in other plants (Feng et al. 2015; Wang et al. 2017).
A phylogenetic tree generated among 74 SOD proteins from M. truncatula and 8 other plant species showed that these SOD proteins fell into two major categories termed as Cu/ZnSODs and Fe-MnSODs. Among Fe-MnSODs, FeSODs and MnSODs from these plants were clustered together and separated by a high bootstrap value, indicating that they might originate from common ancestral genes (Miller 2012; Wang et al. 2017; Zhou et al. 2017). The two major categories can be further divided into eight subgroups, and the members within the same subgroup tended to share the same subcellular localization (Fig. 1). For example, MtCSD4 was clustered together with SlSOD1, CsCSD1, cCuZn-SOD1, cCuZn-SOD2 and other cytoplasmic Cu/ZnSODs into subgroup a, while MtCSD1 together with CsCSD5, SlSOD3, pCuZn-SOD and other chloroplastic Cu/ZnSODs were clustered into subgroup b. The MnSODs of different plant species were clustered in subgroup e, all of which were predicted to be located in the mitochondria (Fig. 1; Table 1). MtCSD2 was clustered with SlSOD4, CsCSD3, and CuZn-SOD-CCh in subgroup d, which exhibited independent evolutionary trajectories from other Cu/ZnSODs (Fig. 1). Two specific conserved metal-binding motifs, MXCXXC and CXC, were found to be present in MtCSD2 as well as in CsCSD3, MaCCS, and CuZn-SOD-CCh (Fig. 2b), suggesting that these SODs are homologous to copper chaperones for SOD, which is in agreement with previous reports (Feng et al. 2016b; Nath et al. 2014; Zhou et al. 2017). In addition, the protein motif distribution and gene structures also reveal that the SOD proteins have strong evolutionary conservation. For example, similar motif distributions were found in FeSODs, with one motif (motif 9) doubled in AtFSD2 and MtFSD1. The same motif patterns were found in MnSODs and Cu/ZnSODs, with the exception of MtCSD2 and CuZn-SOD-CCh, which contained only two conserved domains (Fig. 3). In addition, SOD genes from M. truncatula, Arabidopsis and rice showed similar distributions in the phylogenetic subgroups and intron–exon organizations, suggesting a high conservation of these genes in the evolutionary process. For example, several gene pairs exhibited similar numbers of exons/introns and exon length, such as MtCSD4/AtCSD1/cCuZn-SOD2, MtCSD1/AtCSD2/pCuZn-SOD, MtCSD3/CuZn-SOD-L, MtCSD2/CuZn-SOD-CCh, MtMSD/Mn-SOD1/AtMSD1/AtMSD2, MtFSD2/AtFSD3/Fe-SOD3, and MtFSD1/AtFSD2 (Fig. 4). Thus, we speculate that they might have common biological functions.
Previous studies have reported that SOD genes may exhibit two distinctly different expression patterns, namely constitutive expression and tissue-specific expression (Feng et al. 2016a; Zhou et al. 2017). In this study, microarray analyses of MtSOD genes at different developmental stages revealed that most of the MtSOD genes were ubiquitously expressed (Fig. 6a), which is in accordance with the results of previous reports. In addition, compared with in other tissues, MtCSD3, MtFSD1, and MtFSD2 displayed relatively higher expression levels in the leaves (Fig. 6a), suggesting that SOD is a crucial enzyme in scavenging ROS generated from the photosynthesis of plants (Wang et al. 2017; Zhang et al. 2016). Similar findings have been reported in some other plants, including cucumber (Zhou et al. 2017), cotton (Wang et al. 2017; Zhang et al. 2016), and tomato (Feng et al. 2016a). Additionally, MtCSD4 and MtMSD showed high transcriptions in root nodules during different developmental stages, while MtCSD1 displayed relatively higher expression levels at the early stage of root nodule development (Fig. 6b), suggesting that they may be involved in the nodulation of M. truncatula. Moreover, genes belonging to the same functional clade have similar tissue expression patterns.
The transcripts of SOD genes were found to be altered under different abiotic stresses (Wang et al. 2017; Zhang et al. 2017; Zhou et al. 2017). To explore whether the MtSOD genes are related to abiotic stress responses or not, we analyzed the microarray and high-throughput sequencing data from M. truncatula. Transcript abundance analysis demonstrated that the transcriptions of several MtSOD genes increased significantly under stresses. For example, the RNA-seq data showed that MtCSD2 and MtCSD4 were induced at the early time point under salt and drought conditions (Fig. 7b, c), suggesting their crucial roles in scavenging ROS caused by various stresses in M. truncatula. However, most MtSOD genes showed down-regulated expression at certain time points under various abiotic stresses, especially under cold stress (Fig. 7). Similar findings were also obtained in several plant species, including banana (Feng et al. 2015), tomato (Feng et al. 2016a), cucumber (Zhou et al. 2017) and cotton (Wang et al. 2017; Zhang et al. 2016). For instance, both SlDOD3 and MtCSD1 were clustered in subgroup c (Fig. 1), and were down-regulated under salt and drought stress (Fig. 7b, c) (Feng et al. 2016a). GrFSD2 is the homologous gene of MtFSD2 in G. raimondii (Fig. 1), and their expression levels were found to be dramatically decreased by salt and cold stress conditions (Fig. 7b, d) (Zhang et al. 2016). These results imply that MtSOD genes possibly have diverse functions in stress response of M. truncatula.
Conclusion
In this study, genome-wide characterization and in silico analysis of the SOD family genes in M. truncatula were performed. As a result, 7 MtSOD genes (1 MnSOD, 2 FeSODs, and 4 Cu/ZnSODs) were identified. We also carried out a detailed analysis of their chromosomal locations, phylogenetic relationships, conserved domains, gene structures, tissue-specific expression profiles as well as expression profiles in response to certain abiotic stresses. The findings described in this study may help future investigation of the possible biological functions of SOD genes to regulate the growth, development and abiotic stress response of M. truncatula.
References
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Corpas FJ, Fernandez-Ocana A, Carreras A, Valderrama R, Luque F, Esteban FJ, Rodriguez-Serrano M, Chaki M, Pedrajas JR, Sandalio LM, del Rio LA, Barroso JB (2006) The expression of different superoxide dismutase forms is cell-type dependent in olive (Olea europaea L.) leaves. Plant Cell Physiol 47:984–994
Feng X, Lai Z, Lin Y, Lai G, Lian C (2015) Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom 16:823
Feng K, Yu J, Cheng Y, Ruan M, Wang R, Ye Q, Zhou G, Li Z, Yao Z, Yang Y, Zheng Q, Wan H (2016a) The SOD gene family in tomato: identification, phylogenetic relationships, and expression patterns. Front Plant Sci 7:1279
Feng X, Chen F, Liu W, Thu MK, Zhang Z, Chen Y, Cheng C, Lin Y, Wang T, Lai Z (2016b) Molecular characterization of MaCCS, a novel copper chaperone gene involved in abiotic and hormonal stress responses in Musa acuminata cv. Tianbaojiao. Int J Mol Sci 17:441
Filiz E, Tombuloğlu H (2015) Genome-wide distribution of superoxide dismutase (SOD) gene families in Sorghum bicolor. Turkish J Biol 39:49–59
Gill SS, Anjum NA, Gill R, Yadav S, Hasanuzzaman M, Fujita M, Mishra P, Sabat SC, Tuteja N (2015) Superoxide dismutase—mentor of abiotic stress tolerance in crop plants. Environ Sci Pollut Res Int 22:10375–10394
Jia Q, Xiao ZX, Wong FL, Sun S, Liang KJ, Lam HM (2017) Genome-wide analyses of the soybean F-box gene family in response to salt stress. Int J Mol Sci 18
Jing X, Hou P, Lu Y, Deng S, Li N, Zhao R, Sun J, Wang Y, Han Y, Lang T, Ding M, Shen X, Chen S (2015) Overexpression of copper/zinc superoxide dismutase from mangrove Kandelia candel in tobacco enhances salinity tolerance by the reduction of reactive oxygen species in chloroplast. Front Plant Sci 6:23
Kaouthar F, Ameny FK, Yosra K, Walid S, Ali G, Faical B (2016) Responses of transgenic Arabidopsis plants and recombinant yeast cells expressing a novel durum wheat manganese superoxide dismutase TdMnSOD to various abiotic stresses. J Plant Physiol 198:56–68
Kayum MA, Park JI, Nath UK, Saha G, Biswas MK, Kim HT, Nou IS (2017) Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom 18:885
Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118:637–650
Kozik A, Kochetkova E, Michelmore R (2002) GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics 18:335–336
Li D, Su Z, Dong J, Wang T (2009) An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genom 10:517
Li Z, Han X, Song X, Zhang Y, Jiang J, Han Q, Liu M, Qiao G, Zhuo R (2017) Overexpressing the Sedum alfredii Cu/Zn superoxide dismutase increased resistance to oxidative stress in transgenic Arabidopsis. Front Plant Sci 8:1010
Lin YL, Lai ZX (2013) Superoxide dismutase multigene family in longan somatic embryos: a comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol Breed 32:595–615
Miller AF (2012) Superoxide dismutases: ancient enzymes and new insights. FEBS Lett 586:585–595
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Molina-Rueda JJ, Tsai CJ, Kirby EG (2013) The Populus superoxide dismutase gene family and its responses to drought stress in transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a). PLoS One 8:e56421
Nath K, Kumar S, Poudyal RS, Yang YN, Timilsina R, Park YS, Nath J, Chauhan PS, Pant B, Lee C-H (2014) Developmental stage-dependent differential gene expression of superoxide dismutase isoenzymes and their localization and physical interaction network in rice (Oryza sativa L.). Genes Genom 36:45–55
Negi NP, Shrivastava DC, Sharma V, Sarin NB (2015) Overexpression of CuZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep 34:1109–1126
Pilon M, Ravet K, Tapken W (2011) The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim Biophys Acta 1807:989–998
Song J, Mo X, Yang H, Yue L, Mo B (2017) The U-box family genes in Medicago truncatula: key elements in response to salt, cold, and drought stresses. PLoS One 12:e0182402
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang M, Zhao X, Xiao Z, Yin X, Xing T, Xia G (2016a) A wheat superoxide dismutase gene TaSOD2 enhances salt resistance through modulating redox homeostasis by promoting NADPH oxidase activity. Plant Mol Biol 91:115–130
Wang W, Xia M, Chen J, Deng F, Yuan R, Zhang X, Shen F (2016b) Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene 6:18–29
Wang W, Zhang X, Deng F, Yuan R, Shen F (2017) Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom 18:376
Wu J, Zhang J, Li X, Xu J, Wang L (2016) Identification and characterization of a PutCu/Zn-SOD gene from Puccinellia tenuiflora (Turcz.) Scribn. et Merr. Plant Growth Regul 79:55–64
Xuan Y, Zhou ZS, Li HB, Yang ZM (2016) Identification of a group of XTHs genes responding to heavy metal mercury, salinity and drought stresses in Medicago truncatula. Ecotoxicol Environ Saf 132:153–163
Yan JJ, Zhang L, Wang RQ, Xie B, Li X, Chen RL, Guo LX, Xie BG (2016) The sequence characteristics and expression models reveal superoxide dismutase involved in cold response and fruiting body development in Volvariella volvacea. Int J Mol Sci 17:34–46
Yang Z, Gong Q, Qin W, Cheng Y, Lu L, Ge X, Zhang C, Wu Z, Li F (2017) Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol 17:113
Zhang J, Li B, Yang Y, Hu W, Chen F, Xie L, Fan L (2016) Genome-wide characterization and expression profiles of the superoxide dismutase gene family in Gossypium. Int J Genom 2016:8740901
Zhang L, Sun L, Zhang L, Qiu H, Liu C, Wang A, Deng F, Zhu J (2017) A Cu/Zn superoxide dismutase gene from Saussurea involucrata Kar. et Kir., SiCSD, enhances drought, cold and oxidative stress in transgenic tobacco. Can J Plant Sci 97:816–826
Zhou Y, Hu L, Wu H, Jiang L, Liu S (2017) Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Int J Genom 2017:7243973
Zhou Y, Hu L, Ye S, Jiang L, Liu S (2018a) Genome-wide identification of glutathione peroxidase (GPX) gene family and their response to abiotic stress in cucumber. 3 Biotech 8:159
Zhou Y, Zeng L, Chen R, Wang Y, Song J (2018b) Genome-wide identification and characterization of the stress associated protein (SAP) gene family encoding A20/AN1 zinc finger proteins in Medicago truncatula. Arch Biol Sci 70:87–98
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31560076 and 31760074).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, J., Zeng, L., Chen, R. et al. In silico identification and expression analysis of superoxide dismutase (SOD) gene family in Medicago truncatula. 3 Biotech 8, 348 (2018). https://doi.org/10.1007/s13205-018-1373-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1373-1