Abstract
Food-item authentication and traceability is an issue of primary concern, due to both socio-economical and health implications. DNA-based methods are increasingly being recognised as powerful tools to assess the reliability of supplier labels for any type of food. This is especially true for products characterised by a short shelf life and high-processing supply chain, such as seafood. In this work, a DNA barcoding approach was applied to assess the accuracy of species labelling in 150 cephalopod seafood products sold in the Italian market. Overall, high levels of mislabelling in squid, cuttlefish, and octopus items were identified, and in some cases, even species not included in the current food Regulations. Additionally, an application of the recently developed naked-eye detection tool ‘NanoTracer’, consisting in the combination of DNA barcoding with gold nanoparticle-based, was demonstrated to authenticate common cuttlefish (Sepia officinalis) seafood. The primer pairs used to set the fast detection system for S. officinalis were designed based on the most comprehensive DNA barcoding (COI and 16s rRNA) datasets ever assembled for cephalopods, assuring the specificity of the method. ‘NanoTracer’ allowed a simple, rapid, accurate and cost-effective authentication, revealing its potential adaptability to any type of seafood and other food categories.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seafood products are among the most requested food commodities in the world and their trade has strongly increased during the last 2 decades, mainly due to the population growth and to the higher consumers’ awareness toward healthy food items of high nutritional value (FAO 2016; European Commission 2016). World seafood consumption per capita exceeds 20 kg (FAO 2016) with European countries reaching an average of 25.5 kg (European Market Observatory for Fisheries and Aquaculture Products 2017). Overall, seafood supply chain is extremely various in terms of species and processing. At least 50% of the traded seafood products are transformed (i.e., sold as fillets, slices, minced) to reduce their perishability or increase their palatability. Such treatments lead to the partial or complete loss of morphological diagnostic traits, thus posing great risk to fraud events, which are intended as the practice of misleading consumers about their food products for financial gain (Barbuto et al. 2010). The literature indicates that the seafood supply chain is vulnerable to three main categories of problems dealing with adulteration (i.e., species substitution, adulteration and undeclared product extension), provenance (i.e., fishery substitution and chain of custody abuse) and ethical issues (e.g., adoption of illegal catch methods and lacking of measures to preserve animal welfare) (Fox et al. 2018). Besides socio-economical aspects, food fraud events are elements of great concern in the context of food allergies and other health issues. Possible allergic reactions due to the ingestion of mislabelled or adulterated seafood products put the consumers at increased risk and could also be life threatening (Sicherer et al. 2004). International regulations are progressively being introduced to force producers and distributors to comply with precise labelling requirements. For example, at the European level, the Regulation (EU) No. 1379/2013 establishes the obligation to provide the consumer with the commercial and scientific name of the product, together with other details on provenance and quality parameters of production (D’Amico et al. 2016; Tinacci et al. 2018a). Therefore, it is often compulsory to verify species identity at each step of the seafood supply chain using reliable tools able to work also on highly processed products, in which the raw species are no longer recognizable by simple ‘morphology-based’ inspection.
To date, several DNA-based methods have been developed for the authentication of food products and the detection of food substitution, adulteration, or dilution. These methods are based on a variety of techniques, mostly represented by conventional PCRs and sequencing, quantitative real-time PCRs (qRT-PCRs), microarrays, and High-Resolution Melting (HRM) analyses (Galimberti et al. 2013, 2015; Druml and Cichna-Markl 2014; Madesis et al. 2014; Applewhite et al. 2016; Jilberto et al. 2017). Each approach and its variants show variable advantages and disadvantages regarding, the discriminating power, cost, rapidity, and laboratory requirements (Madesis et al. 2014). For example, those methods based on Sanger sequencing or High-Throughput Sequencing (HTS) are probably the most accurate in terms of information power but are expensive in terms of time of analysis and could require dedicated resources and facilities. In the case of HTS tools, bioinformatic skills and high-quality reference databases are necessary to permit an efficient identification of food ingredients. Moreover, these approaches usually require moderate–high amounts of genetic material and are strictly dependent on DNA quality of extracts and fragmentation that could be strongly affected by processing of the food raw material (see Galimberti et al. 2013, 2015).
On the other hand, sequencing-free approaches such as qRT or HRM PCRs are more rapid but still require expensive instruments and specialized personnel to be adopted as routinely tools at the industrial scale. Overall, there is general consensus in using DNA barcoding-based methods to trace seafoods (Hebert et al. 2003; Barbuto et al. 2010). For example, the U.S. Food and Drug Administration recognized the utility of DNA barcoding, based on the sequence variability analysis at a 658 bp mtDNA COI region, for seafood identification (Yancy et al. 2008; Handy et al. 2011; Deeds et al. 2014). As a result, the approach is progressively being applied in several market surveys worldwide to verify the labelling compliance of seafood items (Barendse et al. 2019). However, due to the reduced shelf life of seafood products and different degrees of personnel specialization and equipment of the possible quality checkpoints, there is a general demand for the development and adoption of fast, simple and economic identification tools based on the DNA barcoding framework, also able to work in case of admixed and/or highly processed food items (Leal et al. 2015).
In the latest years, DNA-based and nano-biotechnologies were combined to further facilitate and speed up the detection of food product substitution. For example, a rapid system was recently developed (Valentini et al. 2017) to allow the detection of both substitution and adulteration in high-value fish and spice products. This method, called ‘NanoTracer’, combines DNA barcoding and nanoparticle-based naked-eye detection to allow for a rapid, cost-effective (less than 1€ per test) and analytically simple molecular traceability of food (Valentini et al. 2017).
In general, molecular-based methods have been typically tested on fish products (Pardo et al. 2016), mainly due to the largest abundance of reference genetic data. However, other seafood classes, among which the mollusks, could be a potential target for the introduction of novel validation approaches, given their high value on the market and large incidence of mislabelling events (see Arkhipkin et al. 2015; Guardone et al. 2017). For instance, cephalopods are an important seafood source for human consumption, and their exploitation has shown a positive trend in the last decades (FAO 2016), due to their palatability and nutritional quality (e.g. Zlatanos et al. 2006; Ozogul et al. 2008). Moreover, a recent study showed that cephalopod populations have increased in the last 60 years, possibly benefiting from the changing ocean environment (Doubleday et al. 2016). Commercially exploited cephalopods are conventionally classified as octopuses, squids, and cuttlefishes (Arkhipkin et al. 2015) and reached an annual global production of more than 3.5 million tons in 2016 (https://www.fao.org/fishery/topic/16140/en), with the main importers and consumers being Italy, Spain and Japan (FAO 2016). The correct identification and labelling of traded cephalopods is of imperative importance for food safety, due to the possible presence of toxic species (Wu et al. 2014), and the variable bioaccumulation capability of toxic metals (Pierce et al. 2008; Penicaud et al. 2017; Sangiuliano et al. 2017) and harmful algal toxins (Lopes et al. 2013) in different species.
In this work, we performed a large-scale survey on the authenticity of cephalopod seafood commercialised in Italy and mostly fished in the Atlantic Ocean and Mediterranean Sea. We collected squid, cuttlefish, and octopus products from several suppliers and checked their labelling through sequencing a portion of their DNA barcoding region cytochrome c oxidase subunit I. Additionally, we developed an application of the novel ‘NanoTracer’ technology to rapidly detect mislabelling in the cuttlefish Sepia officinalis, which is a cephalopod species commonly sold and highly appreciated in Italy and Europe.
Materials and methods
Sampling and DNA barcoding analysis
The sampling was carried out during 2017, and a total of 150 cephalopod samples were collected from commercialised seafood products, including octopuses, cuttlefishes, and squids. Specifically, samples were obtained from 13 among the main Italian seafood distributors and included a label with the species identification provided by the supplier and the geographic provenience indicated as FAO fishing area. To investigate the accuracy of these identifications, a DNA barcoding approach was employed.
DNA extracts were obtained from muscle tissues using Qiagen DNeasy Blood and Tissue Kit, following manufacturer's recommendations. A ~ 600 bp long portion of the cytochrome c oxidase subunit I (COI) was amplified using the primers and protocol described by Folmer et al. (1994) and PCR products were purified and directly sequenced using ABI technology. The obtained chromatograms were visually checked using Sequencher 4.1.4 (Gene Codes) and then deposited with the EMBL (GenBank accession numbers: MH292970-MH293112, MH473336-MH473342), as listed in Table S1. Sequences were aligned using MAFFT 7.110 (Katoh and Standley 2013) and the iterative refinement method E-INS-i. Intra- and inter-specific genetic distances were calculated as % p distance using MEGA X (Kumar et al. 2018), and variance was assessed with 1000 bootstrap replicates. To assess the accuracy of the species identifications provided by the seafood suppliers, each sequence was searched in: (i) the Animal Identification System in BOLD (Ratnasingham and Hebert 2007) and, in particular, in the Species Level Barcode Records Database, and in (ii) the NCBI BLASTn Database. Only species matches with a minimum value of 99% similarity were considered and compared to the species name labelled by the suppliers to obtain a percentage of the mislabelled products. 95% confidence intervals (α = 5) were also calculated for each proportion using Wilson's method (www.openepi.com).
NanoTracer detection system
Rapid DNA extractions from ~ 20 mg of tissue were performed using the Phire Animal Tissue Direct PCR Kit (ThermoFisher Scientific, Bremen, Germany). A set of primers was designed to specifically target a short sequence of the barcode region (46 bp) of Sepia officinalis (Table S2), following the method described by Valentini et al. (2017). The design of primers was performed on the most comprehensive dataset of Sepia COI sequences ever assembled to date, generated downloading all available sequences from GenBank and BOLD (501 sequences, 27 species). In addition, a primer pair specific for a region of the 16S ribosomal RNA conserved in cuttlefishes and relatives were generated based on the alignment of all available sequences retrieved from GenBank (n = 723) and was used for a positive control test (Table S2).
Asymmetric PCR (as-PCR) was performed on three species of Sepia, including the target S. officinalis, and two genetically similar species, Sepia hierredda and Sepia pharaonis. Specifically, 1 µL of genomic DNA was used as a template and added to 49 µL of reaction mix, containing 500 nM of excess primer (Integrated DNA Technologies), 33 nM of limiting primer (cTAG-primer), 2 mM MgCl2, 1X Flexi buffer, 1.25U GoTaq Hot Start Polymerase (Promega), 200 µM of each dNTP (Promega). The reaction was performed in a Bio-Rad T100 Thermal Cycler. As-PCR products were electrophoresed on a 18% polyacrylamide gel to assess the amplification reactions.
The colorimetric test was performed using two sets of 35 nm gold nanoparticles (AuNPs) functionalized with two single-stranded probes, each half-complementary to the tagged amplicon (Fig. S1), produced by as-PCR. DNA-functionalized AuNPs were prepared as previously reported (Valentini and Pompa 2016). Briefly, 15 nm AuNPs were synthetised by citrate reduction. Then, 35 nm AuNPs were prepared by seeded growth method of 15 nm AuNPs. After their characterization with UV–Vis, DLS and TEM (Fig. S2), they were functionalized with two different thiolated single-stranded probes. Their concentration was determined by UV–Vis. The colorimetric test was performed mixing 2 µL of as-PCR product with 650 pM of AuNPs mixture and reaching 0.8 M of NaCl. After 10 min at ca. 20 °C, a clear colour change was observed.
Results
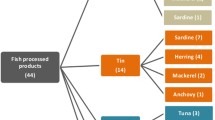
According to the suppliers’ labels, the 150 collected samples belonged to 7 genera and 11 species of commercially important coleoid cephalopods and were fished in the Mediterranean Sea (52%), Atlantic Ocean (47%), and Indian Ocean (1%). Samples were collected as whole animals or processed items (e.g., frozen and filleted items). From each sample, a COI sequence 626 bp long was generated. BOLD and BLASTn searches gave identical results, and sequences were assigned to 6 genera and 12 species (Table 1). The DNA barcoding approach revealed that 50 out of the 150 samples (33%) were mislabelled by the suppliers. Specifically, 27, 13, and 58% of cuttlefishes, octopuses, and squids, respectively, were wrongly identified (Table 1). The mislabelling regarded Sepia officinalis (29% identified as Sepia hierredda), Eledone moschata (71% identified as Eledone cirrhosa), Octopus vulgaris (5% identified as Octopus maya), Loligo forbesii (100% identified as Loligo vulgaris), Loligo vulgaris (5% identified as Loligo forbesii and 41% identified as Loligo reynaudii), and Todarodes sagittatus (100% identified as Illex coindetii). Overall, the total mean genetic distance was 15.7% and inter-specific genetic distances were high, ranging from 5 to 23.6% (Table S3). With the only exception of Sepia pharaonis, which showed an intra-specific distance of 9.3% and is likely a species complex (Anderson et al. 2007, 2010), all other intra-specific comparisons showed low values, spanning from 0 to 1.3% (Table S3). This pattern of genetic diversity further supports the reliability of DNA barcoding-based authentications.
The NanoTracer system was then exploited to easily discriminate Sepia officinalis from the two genetically similar species, S. hierredda and S. pharaonis (Fig. 1a). This AuNP-DNA-based tool was applied to 13 DNA samples, rapidly extracted from tissues of 5 S. officinalis species, 5 S. hierredda species, and 3 S. pharaonis species. The target as-PCR was performed on each sample and controlled by gel electrophoresis (Fig. S3). Amplification occurred specifically for S. officinalis (1–5), producing a double- and a single-stranded DNA amplicons, while S. hierredda (6–10) and S. pharaonis-containing samples (11–13) were not amplified (samples 9 and 13 presented some aspecific amplification, producing only the double stranded amplicon). The AuNP-based test was then performed on the as-PCR products. Sepia officinalis-containing samples turned purple, while all the other sample solutions kept the starting red colour. These results are reported in Fig. 1b, focusing on samples 9 (S. hieredda) and 13 (S. pharaonis) compared to the positive sample 1 (S. officinalis). As shown in the picture, despite some aspecific amplification, sample 9 and 13 remained red, demonstrating the high sensitivity and specificity of the colorimetric detection assisted by the AuNPs. Simultaneously, the positive control as-PCR and the colorimetric detection were performed on each sample, under the same conditions. Figure 1c depicts the results obtained on samples 1 (S. officinalis), 9 (S. hieredda) and 13 (S. pharaonis). All samples turned purple (Fig. 1c, above), as a result of the amplification (Fig. 1c, below), demonstrating the good performance of the reaction, as well as the efficiency of the rapid DNA extraction step.
NanoTracer application to authenticate common cuttlefish seafood. a NanoTracer strategy: rapid DNA extractions followed by as-PCRs and colorimetric tests. b Discrimination test on samples: 1. Sepia officinalis, 9. Sepia hierredda, 13. Sepia pharaonis, NC. MilliQ water. Above: colorimetric detection with AuNPs; Below: 18% polyacrylamide gel electrophoresis. c Positive control test on samples: 1. Sepia officinalis, 2. Sepia hierredda, 3. Sepia pharaonis, NC. MilliQ water. Above: Colorimetric detection with AuNPs; below: 18% polyacrylamide gel electrophoresis
Discussion
In the last years, DNA barcoding has been used in a number of studies to answer different questions in cephalopod diversity (e.g. Dai et al. 2012; Williams et al. 2012; Katugin et al. 2017) and the establishment of a cephalopod barcode of life database has also been proposed (Strugnell and Lindgren 2007). Currently, thousands of species-level DNA barcode records are available in the Barcode of Life Data System (https://www.barcodinglife.org), covering most of the described species and underlying a reliable DNA-based identification of cephalopod taxa. Moreover, DNA barcoding and DNA taxonomy in general are playing an important role in resolving taxonomic problems in different cephalopod taxa, allowing for instance the discovery of cryptic species (Anderson et al. 2010; Dai et al. 2012; Gebhardt and Knebelsberger 2015). Indeed, cephalopod species are not always easily discriminated using morphology alone, especially when dealing with damaged specimens, immature forms, and species with few diagnostic characters (Strugnell and Lindgren 2007). Moreover, being cephalopods a valuable seafood (FAO 2016), the DNA barcoding approach is a powerful tool to assess the reliability of species labelling in both whole organisms and highly processed items (Guardone et al. 2017; Wen et al. 2017). In addition, the current climate change may cause the biogeographic range expansion or shift of species (Parmesan and Yohe 2003), which can eventually overlap with other morphologically similar species, making the correct identification of organisms based on morphology and fishing area alone even more difficult.
To date, only a few studies used a molecular approach to assess the accuracy of species labelling in cephalopod seafood items, and in most cases, only on a small scale (Table 2), mainly because the principal objective of such studies was the development of a technique (Santaclara et al. 2007; Espiñeira et al. 2010), a survey on a specific group (Debenedetti et al. 2015) or on seafood in general (Armani et al. 2015; Tinacci et al. 2018a). However, previous works generally found high levels of mislabelling in cephalopod products, ranging from ~ 20 up to 60% (Table 2). For instance, Guardone et al. (2017) analysed 13 species of commercially available squids, cuttlefishes and octopuses (Table S4), finding that 40, 75, and 71% of the products were mislabelled, respectively, with a total percentage of wrong labelling of about 50% (n = 66) (Table 2). In this work, we analysed 150 cephalopod samples belonging to 11 species (according to the suppliers), finding that more than 30% of the products were wrongly labelled, with the squids being the most mislabelled items. In particular, Sepia officinalis, Eledone moschata, Loligo forbesii, Loligo vulgaris, and Todarodes sagittatus were the most subjected to errors. Therefore, our results are generally consistent with previous works in recovering moderate-to-high levels of mislabelling in an extremely valuable seafood resource such as cephalopods. Notably, the species Sepia hierredda and Loligo reynaudii are not included in the latest Italian Regulation (G.U. N° 266, November, 14 2017). Moreover, multiple samples labelled as L. vulgaris from FAO fishing zone 37 (Mediterranean Sea) corresponded to L. reynaudii, which has a known distribution limited to Southern Africa (Arkhipkin et al. 2015), raising doubts about the fortuity of the substitution. DNA-based seafood authentication is, therefore, important to determine the exact species and to trace the geographic origin of the products, to support sustainable fisheries and avoid public health implications. Indeed, mislabelling is generally believed to occur with not only the substitution of a higher value item with a lower value one (Naaum et al. 2016), but also ‘reverse substitutions’ have been documented, in which the product sold is of higher value than that labelled, but it likely comes from illegal fishing (Gordoa et al. 2017). Cephalopod species substitution may also cause health problems in the consumers due to the physiological and biological differences among species, and in one case a substitution with a potentially toxic pufferfish was also documented in the Italian market (Armani et al. 2015). All these issues, together with the relatively short shelf life of seafood products, make necessary the development of fast, precise, and sensitive authentication tools that could be used also by personnel without particular technical skills. Different methods have been proposed to verify the labelling of cephalopod items, including PCR and sequencing or gel visualization, PCR-Restriction Fragment Length Polymorphism, real-time PCR, and isothermal amplification combined with fluorescence detection (e.g. Santaclara et al. 2007; Ye et al. 2016, 2017; Wang et al. 2018). In this work, we explored applications of the recently developed ‘NanoTracer’ method (Valentini et al. 2017), that couples the amplification of the genomic DNA target by as-PCR to a naked-eye readout. This latter mechanism relies on the plasmonic shift, elicited by the target-induced aggregation of gold nanoprobes, resulting in a clear red-to-violet colour change. In particular, the ‘Nanotracer’ method employs 35 nm gold nanoparticles, since their high extinction coefficient allows an enhanced visual detection of the assay (Liu et al 2007). This robust technique was developed and applied to authenticate cephalopod products, and, in particular, Sepia officinalis (common cuttlefish). Possible impurities or PCR inhibitors due to the use of rapid DNA extraction methods did not interfere with the assay, as well as no effects on NanoTracer authentication performance were observed in case of processed cephalopods items. Given the short genomic regions selected as target for the positive controls and the species authentication assays, the system is also able to cope with the risk of high DNA fragmentation due to extreme processing along the whole food supply chain. Another strength of the NanoTracer assay derives from the accurate development towards the detection of its target species. This feature is pivotal when a certain food item could contain more ingredients over the declared one. We assessed the specificity of our developed assay testing it against two S. officinalis congeneric species, one of which was found to be a common substitution in our survey (Sepia hierredda). Interestingly, the test permitted to unequivocally authenticate S. officinalis products through naked-eye detection, also allowing the distinction between closely related species. Therefore, also in the case of cephalopods, this method represents a fast, cheap (i.e., less than 1 € per sample), and analytically simple solution for molecular traceability (Valentini et al. 2017) and can potentially be adapted to any food item of interest. Although the results of NanoTracer are purely qualitative, this test can be easily performed in situ and on a large industrial scale, further reducing the time required to obtain results, a characteristic that is crucial in short shelf-life products, such as cephalopods. Additional molecular-based tools are available to determine the quantitative traits of food adulteration or purity (e.g., HTS metabarcoding, ELISA and many others, see Galimberti et al. 2019), but the system developed in this study offers an efficient and universally customizable screening solution for authentication which is the first step required by international regulations for food control.
References
Anderson FE, Valinassab T, Ho CW, Mohamed KS, Asokan PK, Rao GS, Nootmorn P, Chotiyaputta C, Dunning M, Lu CC (2007) Phylogeography of the pharaoh cuttle Sepia pharaonis based on partial mitochondrial 16S sequence data. Rev Fish Biol Fish 17:345–352. https://doi.org/10.1007/s11160-007-9042-1
Anderson FE, Engelke R, Jarrett K, Valinassab T, Mohamed KS, Asokan PK, Zacharia PU, Nootmorn P, Chotiyaputta C, Dunning M (2010) Phylogeny of the Sepia pharaonis species complex (Cephalopoda: Sepiida) based on analyses of mitochondrial and nuclear DNA sequence data. J Molluscan Stud 77:65–75. https://doi.org/10.1093/mollus/eyq034
Applewhite L, Larkin P, Naaum AM (2016) Species identification using other tools. In: Naaum A and Hanner R (eds) Seafood authenticity and traceability. Academic Press, Cambridge, pp 133–148. https://doi.org/10.1016/B978-0-12-801592-6.00007-3
Arkhipkin AI, Rodhouse PG, Pierce GJ et al (2015) World squid fisheries. Rev Fish Sci Aquac 23:92–252. https://doi.org/10.1080/23308249.2015.1026226
Armani A, Guardone L, La Castellana R, Gianfaldoni D, Guidi A, Castigliego L (2015) DNA barcoding reveals commercial and health issues in ethnic seafood sold on the Italian market. Food Control 55:206–214. https://doi.org/10.1016/j.foodcont.2015.02.030
Barbuto M, Galimberti A, Ferri E, Labra M, Malandra R, Galli P, Casiraghi M (2010) DNA barcoding reveals fraudulent substitutions in shark seafood products: the Italian case of “palombo” (Mustelus spp.). Food Res Int 43:376–381. https://doi.org/10.1016/j.foodres.2009.10.009
Barendse J, Roel A, Longo C, Andriessen L, Webster LMI, Ogden R, Neat F (2019) DNA barcoding validates species labelling of certified seafood. Curr Biol 29:R198–R199. https://doi.org/10.1016/j.cub.2019.02.014
D’Amico P, Armani A, Gianfaldoni D, Guidi A (2016) New provisions for the labelling of fishery and aquaculture products: difficulties in the implementation of regulation (EU) n. 1379/2013. Mar Policy 71:147–156. https://doi.org/10.1016/j.marpol.2016.05.026
Dai L, Zheng X, Kong L, Li QI (2012) DNA barcoding analysis of Coleoidea (Mollusca: Cephalopoda) from Chinese waters. Mol Ecol Resour 12:437–447. https://doi.org/10.1111/j.1755-0998.2012.03118.x
Debenedetti F, Dalmasso A, Bottero MT, Gilli M, Gili S, Tepedino V, Civera T (2015) Application of DNA barcoding for controlling of the species from Octopus genus. Ital J Food Saf 3:196–199. https://doi.org/10.4081/ijfs.2014.4521
Deeds JR, Handy SM, Fry F Jr, Granade H, Williams J, Powers M, Shipp R, Weigt LA (2014) Protocol for building a reference standard sequence library for DNA-based seafood identification. J AOAC Int 97:1626–1633. https://doi.org/10.5740/jaoacint.14-111
Doubleday ZA, Prowse TA, Arkhipkin A et al (2016) Global proliferation of cephalopods. Curr Biol 26:R406–R407. https://doi.org/10.1016/j.cub.2016.04.002
Druml B, Cichna-Markl M (2014) High resolution melting (HRM) analysis of DNA—its role and potential in food analysis. Food Chem 158:245–254. https://doi.org/10.1016/j.foodchem.2014.02.111
Espiñeira M, Vieites JM, Santaclara FJ (2010) Species authentication of octopus, cuttlefish, bobtail and bottle squids (families Octopodidae, Sepiidae and Sepiolidae) by FINS methodology in seafoods. Food Chem 121:527–532. https://doi.org/10.1016/j.foodchem.2009.12.042
European Commission (2016) Facts and figures on the common fisheries policy. https://ec.europa.eu/fisheries/facts_figures_en. Accessed 13 May 2019
European Market Observatory for Fisheries and Aquaculture Products (2017) The EU fish Market, Edition 2017. https://www.eumofa.eu/documents/20178/108446/The+EU+fish+market+2017.pdf. Accessed 13 May 2019
FAO (2016) The state of world fisheries and aquaculture 2016: contributing to food security and nutrition for all. https://www.fao.org/3/a-i5555e.pdf. Accessed 13 May 2019
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fox M, Mitchell M, Dean M, Elliott C, Campbell K (2018) The seafood supply chain from a fraudulent perspective. Food Secur 10:939–963. https://doi.org/10.1007/s12571-018-0826-z
Galimberti A, De Mattia F, Losa A, Bruni I, Federici S, Casiraghi M, Martellos S, Labra M (2013) DNA barcoding as a new tool for food traceability. Food Res Int 50:55–63. https://doi.org/10.1016/j.foodres.2012.09.036
Galimberti A, Bruno A, Mezzasalma V, De Mattia F, Bruni I, Labra M (2015) Emerging DNA-based technologies to characterize food ecosystems. Food Res Int 69:424–433. https://doi.org/10.1016/j.foodres.2015.01.017
Galimberti A, Casiraghi M, Bruni I, Guzzetti L, Cortis P, Berterame NM, Labra M (2019) From DNA barcoding to personalized nutrition: the evolution of food traceability. Curr Opin Food Sci 29:41–48. https://doi.org/10.1016/j.cofs.2019.07.008
Gebhardt K, Knebelsberger T (2015) Identification of cephalopod species from the North and Baltic Seas using morphology, COI and 18S rDNA sequences. Helgoland Mar Res 69:259–271. https://doi.org/10.1007/s10152-015-0434-7
Gordoa A, Carreras G, Sanz N, Viñas J (2017) Tuna species substitution in the Spanish commercial chain: a knock-on effect. PLoS ONE 12:e0170809. https://doi.org/10.1371/journal.pone.0170809
Guardone L, Tinacci L, Costanzo F, Azzarelli D, D’Amico P, Tasselli G, Magni A, Guidi A, Nucera D, Armani A (2017) DNA barcoding as a tool for detecting mislabeling of fishery products imported from third countries: an official survey conducted at the Border Inspection Post of Livorno-Pisa (Italy). Food Control 80:204–216. https://doi.org/10.1016/j.foodcont.2017.03.056
Handy SM, Deeds JR, Ivanova NV, Hebert PDN, Hanner RH, Ormnos A, Weigt LA, Moore MM, Yancy HF (2011) A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J AOAC Int 94:201–210
Hebert PD, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond Ser B Biol Sci 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Italian Ministerial Decree n. 19105 from Ministero delle Politiche Agricole Alimentari e Forestali (G.U. No. 266, November 14, 2017)
Jilberto F, Araneda C, Larraín MA (2017) High resolution melting analysis for identification of commercially-important Mytilus species. Food Chem 229:716–720. https://doi.org/10.1016/j.foodchem.2017.02.109
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Katugin ON, Chichvarkhina OV, Zolotova AO, Chichvarkhin AY (2017) DNA barcoding for squids of the family Gonatidae (Cephalopoda: Teuthida) from the boreal North Pacific. Mitochondr DNA Part A 28:41–49. https://doi.org/10.3109/19401736.2015.1110792
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Leal MC, Pimentel T, Ricardo F, Rosa R, Calado R (2015) Seafood traceability: current needs, available tools, and biotechnological challenges for origin certification. Trends Biotechnol 33:331–336. https://doi.org/10.1016/j.tibtech.2015.03.003
Liu X, Atwater M, Wang J, Huo Q (2007) Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf B Biointerfaces 58:3–7. https://doi.org/10.1016/j.colsurfb.2006.08.005
Lopes VM, Lopes AR, Costa P, Rosa R (2013) Cephalopods as vectors of harmful algal bloom toxins in marine food webs. Mar Drugs 11:3381–3409. https://doi.org/10.3390/md11093381
Madesis P, Ganopoulos I, Sakaridis I, Argiriou A, Tsaftaris A (2014) Advances of DNA-based methods for tracing the botanical origin of food products. Food Res Int 60:163–172. https://doi.org/10.1016/j.foodres.2013.10.042
Naaum AM, Warner K, Mariani S, Hanner RH, Carolin CD (2016) Seafood mislabeling incidence and impacts. In: Naaum A and Hanner R (eds) Seafood authenticity and traceability. Academic Press, Cambridge, pp 3–26. https://doi.org/10.1016/C2014-0-00758-7
Ozogul Y, Duysak O, Ozogul F, Özkütük AS, Türeli C (2008) Seasonal effects in the nutritional quality of the body structural tissue of cephalopods. Food Chem 108:847–852. https://doi.org/10.1016/j.foodchem.2007.11.048
Pardo MÁ, Jiménez E, Pérez-Villarreal B (2016) Misdescription incidents in seafood sector. Food Control 62:277–283. https://doi.org/10.1016/j.foodcont.2015.10.048
Parmesan C, Yohe G (2003) A globally coherent fingerprint of climate change impacts across natural systems. Nature 421:37. https://doi.org/10.1038/nature01286
Penicaud V, Lacoue-Labarthe T, Bustamante P (2017) Metal bioaccumulation and detoxification processes in cephalopods: a review. Environ Res 155:123–133. https://doi.org/10.1016/j.envres.2017.02.003
Pierce GJ, Stowasser G, Hastie LC, Bustamante P (2008) Geographic, seasonal and ontogenetic variation in cadmium and mercury concentrations in squid (Cephalopoda: Teuthoidea) from UK waters. Ecotoxicol Environ Saf 70:422–432. https://doi.org/10.1016/j.ecoenv.2007.07.007
Ratnasingham S, Hebert PD (2007) BOLD: the barcode of life data system (https://www.barcodinglife.org). Mol Ecol Resour 7:355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
Sangiuliano D, Rubio C, Gutiérrez AJ, González-Weller D, Revert C, Hardisson A, Zanardi E, Paz S (2017) Metal concentrations in samples of frozen Cephalopods (cuttlefish, octopus, squid, and shortfin squid): an evaluation of dietary intake. J Food Prot 80:1867–1871. https://doi.org/10.4315/0362-028X.JFP-17-184
Santaclara FJ, Espiñeira M, Vieites JM (2007) Genetic identification of squids (families Ommastrephidae and Loliginidae) by PCR-RFLP and FINS methodologies. J Agric Food Chem 55:9913–9920. https://doi.org/10.1021/jf0707177
Sicherer SH, Muñoz-Furlong A, Sampson HA (2004) Prevalence of seafood allergy in the United States determined by a random telephone survey. J Allergy Clin Immunol 114:159–165. https://doi.org/10.1016/j.jaci.2004.04.018
Strugnell JM, Lindgren AR (2007) A barcode of life database for the Cephalopoda? Considerations and concerns. Rev Fish Biol Fish 17:337–344. https://doi.org/10.1007/s11160-007-9043-0
Tinacci L, Stratev D, Vashin I, Chiavaccini I, Susini F, Guidi A, Armani A (2018a) Seafood labelling compliance with European legislation and species identification by DNA barcoding: a first survey on the Bulgarian market. Food Control 90:180–188. https://doi.org/10.1016/j.foodcont.2018.03.007
Tinacci L, Guidi A, Toto A, Guardone L, Giusti A, D’Amico P, Armani A (2018b) DNA barcoding for the verification of supplier’s compliance in the seafood chain: how the lab can support companies in ensuring traceability. Ital J Food Saf 7:83–88. https://doi.org/10.4081/ijfs.2018.6894
Valentini P, Pompa PP (2016) A universal polymerase chain reaction developer. Angew Chem 55:2157–2160. https://doi.org/10.1002/anie.201511010
Valentini P, Galimberti A, Mezzasalma V, De Mattia F, Casiraghi M, Labra M, Pompa PP (2017) DNA barcoding meets nanotechnology: development of a smart universal tool for food authentication. Angew Chem 56:8094–8098. https://doi.org/10.1002/ange.201702120
Wang H, Tian H, Hao N, Wang X (2018) Development of a species-specific PCR assay for correct labeling of eight squid species from Loliginidae and Ommastrephidae Families (Mollusca: Cephalopoda). Food Anal Method 11:3071–3077. https://doi.org/10.1007/s12161-018-1287-x
Wen J, Tinacci L, Acutis PL et al (2017) An insight into the Chinese traditional seafood market: species characterization of cephalopod products by DNA barcoding and phylogenetic analysis using COI and 16S rRNA genes. Food Control 82:333–342. https://doi.org/10.1016/j.foodcont.2017.07.011
Williams RC, Newman SJ, Sinclair W (2012) DNA barcoding in Nautilus pompilius (Mollusca: Cephalopoda): evolutionary divergence of an ancient species in modern times. Invertebr Syst 26:548–560. https://doi.org/10.1071/IS12023
Wu YJ, Lin CL, Chen CH, Hsieh CH, Jen HC, Jian SJ, Hwang DF (2014) Toxin and species identification of toxic octopus implicated into food poisoning in Taiwan. Toxicon 91:96–102. https://doi.org/10.1016/j.toxicon.2014.09.009
Yancy HF, Zemlak TS, Mason JA et al (2008) Potential use of DNA barcodes in regulatory science: applications of the Regulatory Fish Encyclopedia. J Food Prot 71:210–217. https://doi.org/10.4315/0362-028X-71.1.210
Ye J, Feng J, Liu S, Zhang Y, Jiang X, Dai Z (2016) Identification of four squid species by quantitative real-time polymerase chain reaction. Mol Cell Probes 30:22–29. https://doi.org/10.1016/j.mcp.2016.01.001
Ye J, Feng J, Dai Z, Meng L, Zhang Y, Jiang X (2017) Application of Loop-Mediated Isothermal Amplification (LAMP) for rapid detection of jumbo flying squid Dosidicus gigas (D’Orbigny, 1835). Food Anal Method 10:1452–1459. https://doi.org/10.1007/s12161-016-0700-6
Zlatanos S, Laskaridis K, Feist C, Sagredos A (2006) Proximate composition, fatty acid analysis and protein digestibility-corrected amino acid score of three Mediterranean cephalopods. Mol Nutr Food Res 50:967–970. https://doi.org/10.1002/mnfr.200600003
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
On behalf of all authors, I declare that no conflict of interest exists regarding the submitted manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Maggioni, D., Tatulli, G., Montalbetti, E. et al. From DNA barcoding to nanoparticle-based colorimetric testing: a new frontier in cephalopod authentication . Appl Nanosci 10, 1053–1060 (2020). https://doi.org/10.1007/s13204-020-01249-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13204-020-01249-6