Abstract
The isolation of seed-endophytic bacteria (SEB) is a promising approach for the selection of maize plant growth-promoting bacteria (PGPB). With the hypothesis that maize seeds harbor SEB that occupy different niches and show plant-growth-promoting abilities, we aimed to isolate and characterize the potential PGPB from these seeds. The bacteria from commercial seeds (BRS Gorutuba) and axenically grown maize-seedlings were isolated, molecularly fingerprinted, and genetically characterized by amplified ribosomal DNA restriction analysis (ARDRA). All SEB were evaluated for their promotion of early root growth. The selected strains were identified by 16S rRNA sequencing and evaluated for their plant growth-promotion traits. A pot experiment was conducted to assess the ability of the SEB to promote maize-growth and nutrient accumulation. Fifty-one bacterial strains were retrieved, mostly isolated directly from the seeds. All the isolated bacteria represented different strains according to their molecular fingerprinting. ARDRA clustering revealed six clusters influenced by their plant tissue/organ of origin. Twenty-nine SEB were selected based on their influence on early root growth. The 16S rRNA sequences classified the SEB as Bacillus (22), Paenibacillus (2) and Acinetobacter (5). The inoculation of Bacillus ESA 674 improved the shoot dry mass in 57% and the Acinetobacter ESA 662 improved the root growth by 235%, both compared to the uninoculated control. At least 12 bacteria improved nutrient content in the shoots. The Bacillus spp. ESA 674 and ESA 652 outstood in improving maize nutrition by increasing the accumulation of several nutrients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Plant growth-promoting bacteria (PGPB) are biological agents that facilitate the establishment and development of plants through several biological mechanisms (García-Fraile et al. 2015; Aeron et al. 2020). In agriculture, PGPB can be used as inoculants and biofertilizers to increase the plant yield, reducing the production costs and environmental impacts of agriculture. The development of commercial inoculants is guided by extensive research efforts on the isolation and selection of efficient PGPB for various crops. For example, commercial inoculants with selected Azospirillum brasilense strains for maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum) and Brachiaria spp. (Hungria et al. 2010, 2016) are available in the Brazilian market.
In addition to those A. brasilense strains, several research groups are focusing on the isolation and evaluation of new bacteria from several nonlegume crops, such as sorghum (Sorghum bicolor) (Schlemper et al. 2018; da Silva et al. 2018; Antunes et al. 2019), melon (Cucumis melo) (Seido et al. 2019), and sunflower (Helianthus annuus) (Ambrosini et al. 2016), maize (Cavalcanti et al. 2020; Ikeda et al. 2020), among others. In these studies, the bacteria were obtained through the harvest of the plant in the field, and the use of their soft tissues for isolation procedures in the laboratory.
In addition to obtaining microbial isolates from field-grown plants, some studies have focused on evaluating the diversity of seed endophytic bacteria (SEB) (Verma and White Jr 2019). This approach has already revealed bacteria with biotechnological applications, such as the production of hydrolytic enzymes (Bodhankar et al. 2017) and plant growth promotion (Puente et al. 2009; Bodhankar et al. 2019). SEB can spread and colonize plant tissues (Mano et al. 2006; Hardoim et al. 2012; Bodhankar et al. 2017) and interact with other plant-associated bacteria. For example, Bacilio-Jiménez et al. (2001) reported that the SEB from rice inhibits the colonization of host plants by Azospirillum brasilense, a remarkable PGPB, indicating the important role of seed-borne bacteria in crop management.
SEB can be obtained vertically (from the mother plant) or horizontally (from the environment) (Truyens et al. 2015; Chowdhury et al. 2019). The management of SEB can result in seeds carrying beneficial bacteria that favor plant establishment and development. Mitter et al. (2017) enriched the composition of the SEB community with Paraburkholderia phytofirmans PsJNT in maize and wheat through inoculation of flowers. Wheat plants derived from these seeds showed higher development and grain yield. These results show the potential of the management of the SEB community as a strategy to increase plant growth and production. However, P. phytofirmans PsJNT was isolated from the roots of onion (Allium cepa) plants (Sessitsch et al. 2005). Onion root endophytic compartments are full of water (>90% water content) and form a mild environment, differing from the endophytic environment of cereal seeds, for example, those of maize, which provide a dry (10–13% water content) harsh environment. The selection of PGPB from seeds for SEB community management should be carried out by identifying plant growth-promoting SEB. Then, isolation and evaluation of the plant growth promotion abilities of the SEB is a crucial step.
Brazil is the third-largest maize producer in the world. In the 2018/2019 season, Brazilian maize production was over 96 thousand tons, and internal consumption reached 65 thousand tons (CONAB 2019). At the Northeast Region of Brazil (which encompasses the Brazilian semiarid belt), the average maize yield is approximately 2530 kg ha−1, while in the whole country, this value is approximately 5350 kg ha−1 (CONAB 2019). The lower yields of maize fields in northeastern Brazil are primarily due to the low technological inputs adopted by the growers and the harsh environmental conditions (low rainfall and high temperatures) in the region. For this reason, the development and dissemination of sustainable technologies are urgently needed to increase maize production in this region and similar regions worldwide. For these reasons, the identification of new PGPB for the production of inoculants and/or enriched seeds is one of the most promising strategies (Santos et al. 2019; Aeron et al. 2020).
Therefore, we hypothesized that commercial seeds of maize cv. BRS Gorutuba harbors a community of SEB that occupies several ecological niches, spreading throughout the maize seedlings, in axenic conditions, and shows different plant growth-promoting abilities. This work aimed to isolate and characterize the potential PGPB originating from commercial maize seeds.
2 Methods and materials
2.1 Maize seeds, experimental setup, and isolation procedure
Commercial seeds of maize cv. BRS Gorutuba were used. The seeds were produced in the seed-producing field of Embrapa Produtos e Mercados at Petrolina municipality (Pernambuco State, Brazil: lat -9.053611; long -40.275556). The seed batch was produced in the summer of 2017 (from January to April) by applying conventional maize management practices according to the recommendations for the region. The seeds were not coated with fungicide, and the isolation procedures and experimental implementation were conducted in July 2017.
Three samples of 100 g of seeds were separated and surface disinfected through submersion in ethanol 96°GL for 30 s and sodium hypochlorite 2% (v v−1) for 10 min followed by eight washes in distilled, autoclaved water (DAW) (Vincent 1970). After the disinfection procedure, the water used in the last seed wash was inoculated in nutrient-agar (NA) medium to assess the efficiency of the disinfection process. In the same way, 10 randomly selected seeds were streaked on the surface of the same medium to assess the presence of bacteria on the seed surface. The dishes were incubated at 28 °C for five days. No contamination was observed after this procedure.
An amount of 50 g from each seed sample was crushed in a blender with 450 mL of NaCl 0.85% (w/v). The solutions were serially diluted to 10−4 for inoculation in the solid media. The serial dilution was conducted in triplicate. At the same time, in 1 L borosilicate bottles containing 350 mL of agar-water 1.5% (w v-1), previously disinfected seeds were carefully laid on the surface of the medium (5 seeds per bottle). The bottles were incubated in a growth chamber at 26 °C with a photoperiod of 12 h for 15 days. At 4, 9, and 13 days after germination (DAG), 5 mL of DAW was added per bottle. At 5 DAG, three spare plants were gently removed, and two plants were left in each bottle.
At 15 DAG, the plants were harvested. The whole plants were carefully removed from the agar, and the roots were separated from the shoots. To obtain bacteria from the regions equivalent to the “rhizosphere”, seedling roots and shoots, three different parts of the maize plants were analyzed. To assess the bacteria colonizing the “rhizosphere” (the agar medium close to the roots, the equivalent to the rhizosphere in the root-soil interface), 20 g of roots were added to 500 mL flasks containing 180 mL of NaCl 0.85% (w/v) and shaken vigorously (300 rpm) in an orbital shaker for 10 min. The roots were subjected to surface disinfection, and the solutions were diluted serially until 10−4.
To the isolation of endophytic bacteria within the root and shoots of seedlings, aliquots with 20 g of each roots or shoots were surface disinfected as described above for the seeds, the water from the last rinse and the surfaces of roots, and shoots were used to streak NA Petri dishes to assess the efficiency of the surface disinfection. The 20 g aliquots of roots and shoots were crushed in a blender with 180 mL of NaCl 0.85% (w v-1). The solutions were serially diluted to 10−4 for plating. The serial dilutions were conducted in triplicate.
For plating, aliquots of 100 μL of all serial dilutions of seeds, “rhizosphere”, seedling roots, and seedling shoots were inoculated in Petri dishes containing nutrient-agar (NA), Dyg’s (Rodrigues Neto et al. 1986) and YMA (Vincent 1970) media and spread with the aid of a Drygalski loop. The dishes were incubated at 28 °C, and colony growth was observed daily for five days. The inoculated dilutions that reached approximately 10–30 colonies per plate were purified in the same medium. The cultures were characterized according to their morphological characteristics [colony size (mm), color, and shape (circular or irregular), border type (regular or irregular), and amount of mucus (low or high production)] and stored in 25% (v v-1) glycerol at -80 °C.
2.2 Molecular diversity of the culture collection
The DNA of all bacterial strains was extracted with the Brasílica 13-BR200 commercial kit (LGC Biotecnologia, São Paulo, Brazil) and stored at -20 °C until PCR. The bacteria were fingerprinted using the BOX-A1 (CTACGGCAAGGCGACGGCTGACG) (Versalovic et al. 1994), (GAC)5 (Gadanho and Sampaio 2002) and (GTG)5 (Švec et al. 2005) primers. The reactions were performed in a Veriti 96 well thermocycler (Applied Biosystems, USA) and submitted to horizontal electrophoresis (TBE 0.5X buffer) in agarose gel 1.2% (w v−1) at 120 V for 3 h to BOX-A1 and 2 h to the other two markers.
Additionally, we performed an amplified ribosomal DNA restriction analysis (ARDRA). The 16S rRNA was amplified with the universal primers 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (TACGGYTACCTTGTTACGG) (Weisburg et al. 1991) and digested separately with the endonucleases AluI and MspI (Thermo Scientific, USA) overnight at 37 °C according to the manufacturer’s instructions. The digested PCR product was subjected to horizontal electrophoresis (TBE 0.5X buffer) in agarose 1.5% (w v−1) gel for 3 h at 90 V. The primer sequences and PCR conditions are detailed in Table S1. The gel pictures were taken in an UVDoc-400i UV chamber (Delpho, São Carlos, Brazil) and analyzed with BioNumerics 7.6 software (Applied Maths, Belgium) for the normalization of gel images and the construction of dendrograms applying the UPGMA clustering method and the Dice coefficient of similarity.
2.3 Influence of bacteria on the germination and initial development of maize seeds and 16S gene sequencing
All bacterial strains obtained were assessed according to their influence on the germination of maize seeds and young seedlings. The maize seeds were surface disinfected, as described above. The bacteria were grown in Bushnell and Hass mineral medium (BHMM) (Bushnell and Haas 1941) supplied with glucose (5 g), mannitol (5 g), and malic acid (5 g) with the pH adjusted to 6.8. The optical density of the bacterial broth was verified spectrophotometrically at 540 nm and adjusted to 0.3 (OD540 = 0.3). Seeds were microbiolized by imbibition in the bacterial broth for 15 min. The inoculated seeds were carefully laid in agar-water 1% (w v-1) dishes (10 seeds per dish) and incubated in a growth chamber in the dark for seven days at 26 °C. On the 7th day of incubation, the experiment was harvested, and the following variables were assessed: germination (radicle emergence), length of the primary root, and overall development of the roots [by attributing the scores from "1" (to the worst treatments) to “10” (to the best treatments)] . In addition to the treatments inoculated with the 51 SEB, a reference strain control was added using Azospirillum brasilense Ab-V5. An uninoculated control was also evaluated.
The selected bacterial strains were used in the next steps and deposited in the Culture Collection of Microorganisms with Agricultural Interests of Embrapa Semiárido (CMISA). This experiment was conducted in a completely randomized design with three replications and conducted twice, to confirm the results.
The sequencing of the 16S rRNA of 29 bacterial strains with plant-growth-promotion ability was also conducted. The PCR amplification was conducted as described above, and the PCR products were purified with the commercial kit Wizard® SV Gel and PCR Clean-up System (Promega, USA). The fragments were sequenced at Macrogen Inc. (Seoul, South Korea) in an ABI 3037 xl platform (Applied Biosystems, USA). The quality of sequence was verified by Sequence Scanner Software v. 2.0 (Applied Biosystems, USA), and the almost complete 16S rRNA gene sequence was assembled. The fragments were compared to those of type strains available in the GenBank database using the BLASTn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were deposited in the GenBank database of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MT482555 to MT482583.
2.4 In vitro plant growth-promoting traits: auxin, siderophore, and biofilm biosynthesis, calcium phosphate solubilization and nifH gene amplification
For the evaluation of auxin production, the colorimetric procedure described by Sarwar and Kremer (1995) was adapted. The bacteria grew in liquid BHMM medium supplied with 100 mg L−1 of L-tryptophan (L-Trp) for seven days under constant stirring of 120 rotations per minute at room temperature. The optical density of each culture was adjusted to OD540 = 0.3, to standardize the cell concentration.
Aliquots of 1.0 mL of the adjusted cultures were centrifuged for 3 min at 6000 g, after which 150 μL of supernatant was added to 96-well ELISA microplates with 100 μL of Salkowski solution (1.0 mL of 0.5 M FeCl3.6H2O and 50 mL of 35% (v v−1) HClO4). The microplates were stored in the dark at room temperature for 30 min. Thereafter, the intensity of the reddish coloration was determined spectrophotometrically at 530 nm. The auxin concentration was estimated using a standard curve with some known concentration of indole acetic acid (Sigma Aldrich, USA).
For siderophore production, we adopted the quantitative approach described by Ribeiro and Cardoso (2012) with adaptations. The bacteria grew in the BHMM medium for four days, as described above. Afterward, 1 mL of the culture was centrifuged, and 150 μL of the supernatant was added to 96-well ELISA microplates with 150 μL of CAS reagent [6.0 mL of hexadecyltrimethylammonium bromide (HDTMA); 1.5 mL of a FeCl3.6H2O solution; 4.307 g of piperazine; and 6.25 mL of 33% (v v−1) HCl] (Schwyn and Neilands 1987). The plates were incubated in the dark at room temperature for 30 min. The yellowish coloration of the wells was evaluated at spectrophotometrically at 420 nm. To quantification, a standard curve was made with known concentrations of ethylenediaminetetraacetic acid (EDTA).
The bacterial strains were evaluated for tricalcium phosphate solubilization in solid medium (Sylvester-Bradley et al. 1982). The bacteria were cultured in liquid BHMM medium for four days, centrifuged and OD540 adjusted to 0.3 as described above. Aliquots of 10 μL of the cultures were inoculated into plates containing GL (glucose-yeast extract) medium [10 g L−1 glucose, 2 g L−1 yeast extract with 50 mL of K2HPO4 and 100 mL f CaCl2 (both 10% w v−1)], and incubated at room temperature for six days. After the incubation period, the presence of a translucent zone surrounding the colonies was observed.
Biofilm production was evaluated following the method developed by Nostro et al. (2007), with the adaptation. In 96-well ELISA microplate, 150 μL of liquid BHMM medium was added into each well. An aliquot of 5 μL of the OD540 = 0.3 adjusted bacterial broth was added to the medium. The microplates were incubated in the dark at room temperature for 8 days, and afterwards, the culture broths were discarded, and each microplate well was washed three times, with 200 μL of DAW. Therefore, the plates were dried at room temperature, and 100 μL of 0.25% (w v−1) gentian violet [Tris(4-(dimethylamino)phenyl) methylium chloride] was added and incubated for five minutes. The plates were rewashed with DAW, an ethanol/acetone solution (80:20 v v−1) was added. The biofilm formation was quantified by measuring the intensity of the purple-blue color spectrophotometrically at 620 nm. A standard curve was constructed using different dilutions of 0.25% (w v−1) gentian violet and an ethanol/acetone (80:20) solution, with the following concentrations of gentian violet: 0.0 (blank); 13.7; 24.6; 41.4; 55.1; 68.9 and 82.7 μ mol L−1. The experiment was implemented with four replications in a completely randomized design.
A fragment of nifH gene was amplified using the primers pair NifHfor (ACCCGCCTGATCCTGGACGC) and NifHrev (ACGATGTAGATTTCCTGGGC) (Soares et al. 2006). The PCR product was submitted to horizontal electrophoresis (TBE 0.5X buffer) in agarose 1.0% (w v−1) gel for 1 h at 120 V. The bacteria were considered positive for nifH amplification when a clear amplicon of 300–350 bp was observed. The negative bacteria were repeated twice to assure the absence of the target gene. In all PCR we used the known diazotrophic strain A. brasilense Ab-V5 as the positive control.
2.5 Maize growth promotion experiment
Twenty-nine bacterial strains were assayed in pot experiments to evaluate their plant-growth promotion abilities. In addition to the treatments inoculated with 29 SEB, a reference strain control was added using Azospirillum brasilense Ab-V5. The uninoculated control without bacterial inoculation was also evaluated.
The experiment was performed in 5 L pots filled with the surface layer of a red-yellow Ultisol. A soil sample was subjected to soil chemical analysis, according to Teixeira et al. (2017). The results showed the following chemical characteristics: pH (water) 5.5, electrical conductivity 0.92 mS cm−1, C (total) 9.3 g kg−1, P 20.44 cmolc dm−3, K+ 0.32 cmolc dm−3, Na+ 0.12 cmolc dm−3, Ca2+ 2.2 cmolc dm−3, Mg2+ 0.9 cmolc dm−3, Al3+ 0.05 cmolc dm−3, H++Al3+ 0.7 cmolc dm−3, sum of bases 3.5 cmolc dm−3, cation exchange capacity 4.3 cmolc dm−3 and base saturation 83.1%. The maize genotype BRS Gorutuba was used. A completely randomized block design was used with four replications per treatment.
The disinfection of seed surface and inoculation were conducted as described above. The experimental treatments included single inoculations of 29 new bacterial strains and the reference strain Azospirillum brasilense Ab-V5. Also, an uninoculated treatment was tested (uninoculated control). The soil fertility was not corrected, and all plants received a small nitrogen supply of 300 mg N plant−1 (NH4NO3) five days after sowing. The N fertilization rate is equivalent to 20 kg N ha−1.
At seven days after emergence (DAE), the plants were thinned, and a single plant was left in each pot. The plants received 500 mL of tap water daily and were harvested at 48 DAE. The roots were separated from the shoots and carefully washed with running tap water, after which they were separately dried at 65 °C in an oven and weighed.
For mineral composition, the shoots were milled, and separate aliquots (100 mg) of the tissues were submitted to sulfuric and nitroperchloric digestion. The total N concentration in the sulfuric digested product was evaluated by the semi-micro Kjeldahl method (Liao 1981). The P concentration in the nitroperchloric digested product was assessed spectrophotometrically through the colorimetric ammonium vanadate-molybdate method at 420 nm (Teixeira et al. 2017). The Ca, Mg, K, Fe, Mn, Cu, and Zn contents of the nitroperchloric digested product were assessed by atomic absorption in a 900H device (PerkinElmer, USA).
2.6 Statistical analysis
All quantitative data were analyzed thought variance analysis using appropriated data transformation to meet the ANOVA requirements. The data of in vitro plant growth promotion traits, early root growth, and germination compared by the Scott-Knott’s mean range test (p > 0.05) using the software Sisvar 5.0 (Ferreira 2011). The data of maize growth promotion were submitted to the Dunnett’s mean comparison test (p > 0.05) against the uninoculated control, using the software Statistica 7.0 (TIBCO Software Inc., USA).
3 Results
3.1 Bacterial isolation and genetic variability within the collection
A total of 51 bacterial strians were obtained in all plant compartments and culture media (Table 1). Twenty-six, 10, 9, and 6 bacterial strains were obtained from the seeds, seedling roots, seedling shoots and “rhizosphere”, respectively. NA was the best culture medium since 25 SEB bacterial strains were retrieved from it (roots-7, shoots-3, “rhizosphere”-2 and seeds-13). Dyg’s and YMA were the sources of 17 (roots-2, shoots-4, “rhizosphere”-2 and seeds-9) and 9 (roots-1, shoots-2, “rhizosphere”-2 and seeds-4) bacteria, respectively.
The combined analysis of the polymorphism generated by the reactions with the BOX-PCR, (GAC)5 and (GTG)5 molecular markers showed that there were no clones within the culture collection since none of the strains clustered with 100% similarity to each other (Fig. 1), indicating the high diversity of maize seed-borne bacterial strains.
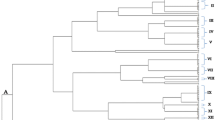
The analysis of ARDRA profiles grouped all strains with 35% similarity. Six clusters were observed, primarily based on the origin of the SEB (Fig. 2). In cluster I, 2 bacteria isolated from seeds and 1 from seedling roots were observed. In the cluster II, 18 out of 21 strains were isolated from maize seeds, and there were 2 and 1 bacteria from the “rhizosphere” and seedling roots, respectively. Cluster III showed 3 bacteria from seeds, 3 from seedling roots, and 1 from seedling shoots. In cluster IV, there were no bacteria isolated from seeds, in this cluster the seedling roots, seedling shoots, and the “rhizosphere” had 5, 5, and 2 representatives, respectively. Cluster V had 3 bacteria isolated from maize seeds, and cluster VI had 3 bacteria from seedling shoots and 2 from the “rhizosphere”.
3.2 Seed germination and early seedling development
Bacterial inoculation influenced the germination of the maize seeds and the seedlings development . Seeds inoculated with 29 SEB derived seedlings that were classified in the two highest statistical clusters according to the Scott-Knott mean range test, considering the length of the primary root. Thirty-six bacteria improved seed germination (Fig. 3). The qualitative evaluation of the development of the roots showed that the 16 inoculation treatments (ESA 640, ESA 641, ESA 647, ESA 649, ESA 650, ESA 651, ESA 652, ESA 653, ESA 654, ESA 655, ESA 658, ESA 659, ESA 660, ESA 661, ESA 675, and ESA 677) had the higher score (10) in all replications.
Overall, 32 inoculation treatments (including A. brasilense Ab-V5) increased root development, and 20 had the same as or worse results than the uninoculated control. The lowest score (1) was attributed to 11 bacteria that also inhibited maize germination and primary root development. In Fig. 4, the influence of inoculation can be observed on the bacteria ESA 641, ESA 677, ESA 652, ESA 663 (scores 9–10), Ab-V5 (score 7) and M1 (score 1).
Early root development of 7-day-old germinated maize seeds inoculated with seed-borne bacteria and Azospirillum brasilense Ab-V5. 4 = ESA 641 (score 10), 36 = ESA 677 (score 10); 32 = ESA 652 (score 9); 38 = ESA 655 (score 10); Test. = uninoculated control (score 5); ABV5 = Azospirillum brasilense Ab-V5 (score 7); 42 = M1 (score 1). Ruler scale = 1 cm
3.3 Identification of bacteria by 16S rRNA sequencing
The 29 SEBs were classified within three genera: Bacillus (22 bacteria), Paenibacillus (2 bacteria), and Acinetobacter (5 bacteria) (Fig. 5). Among the Bacillus spp., 12, 5, 3 and 2 strains were obtained from seeds, seedling roots, seedling shoots, and the “rhizosphere”, respectively. The Acinetobacter strains were obtained from the seedling shoots (3 strains) and “rhizosphere” (2 strains). One Paenibacillus strain was isolated from the seedling shoots, and the other was obtained from the seeds.
Neighbor-joining phylogenetic tree (Jukes-Cantor model) based on partial 16S rRNA gene sequences (1211 nt) of 29 maize seed-borne bacterial strains (“ESA” strains) and related type strains. Numbers in the nodes of branches are the bootstrap values (%) with 1000 pseudoreplications (values <60% are not shown). Symbols represent the origins of the isolates - circles: seeds; squares: seedling shoots; triangles: seedling roots and diamonds: “rhizosphere”
3.4 In vitro plant growth promotion traits
Considering all bacteria assessed, 21 out of 29 strains evaluated produced detectable amounts of auxin in the culture medium supplied with L-Trp (Table 2). Bacillus sp. ESA 676 and Acinetobacter sp. ESA 660, along with strain Azospirillum brasilense Ab-V5, stood out when compared to the other strains and were grouped in a higher cluster by the Scott-Knott test (p < 0.05). All bacteria produced detectable amounts of siderophores and biofilm in these assays.
Bacillus spp. ESA 645, ESA 650, and ESA 651 stood out as producing more siderophores than the other strains assayed. Acinetobacter sp. ESA 662 and Bacillus sp. ESA 656 produced the highest amounts of biofilm compared to the other strains. Bacillus spp. ESA 649, ESA 651, and ESA 653 as well as Acinetobacter sp. ESA 655 produced less biofilm than ESA 656 and ESA 662 but still produced more biofilm than the other bacteria evaluated in this assay. Calcium phosphate solubilization was observed in Bacillus spp. ESA 643, ESA 644, ESA 676, ESA 649, and ESA 650 but not in the other strains. The PCR for the nifH gene was positive for Bacillus spp. ESA 642, ESA 643, ESA 647, and ESA 657, Acinetobacter sp. ESA 658 and Paenibacillus ESA 664, along with the known diazotrophic bacteria Azospirillum brasilense Ab-V5.
3.5 Maize growth promotion experiment
Considering the dry mass of maize plants, eight strains (Bacillus spp. ESA 640, ESA 641, ESA 643, ESA 647, ESA 652, ESA 659, and ESA 674, Acinetobacter sp. ESA 660) induced shoot growth compared to that in the uninoculated control (Table 3). The inoculation of six SEB strains induced root growth (Bacillus spp. ESA 642, ESA 648, ESA 657 and ESA 658, Acinetobacter spp. ESA 661 and ESA 662) in addition to the reference strain Ab-V5. None of the strains induced both root and shoot growth.
Regarding nutritional aspects, N accumulation in the shoots was increased in 27 out of 29 inoculation treatments with SEB, compared that to the uninoculated control. Ca accumulation was promoted by inoculation with Bacillus sp. ESA 652. Bacillus sp. ESA 674, ESA 644, ESA 652, ESA 662, and Acinetobacter spp. ESA 654 and ESA 661 promoted Mg uptake in maize plants. The P levels of the plants inoculated with Bacillus spp. ESA 641 and ESA 674 were higher than those in uninoculated control.
For the micronutrients, the inoculation of Bacillus spp. ESA 641, ESA 645, ESA 647, and ESA 674 along with Acinetobacter sp. ESA 655 induced Cu accumulation in maize shoots. The contents of Mn were improved by inoculation with Bacillus sp. ESA 674 and Acinetobacter spp. ESA 654 and ESA 662. Zn accumulation was improved by Bacillus spp. ESA 641, ESA 652, ESA 653, and ESA 674, and Acinetobacter spp. ESA 660, ESA 662, and Azospirillum brasilense Ab-V5. For plant height and Fe accumulation, none of the inoculated treatments differed from the uninoculated control.
4 Discussion
Maize growth-promoting bacteria are usually isolated from tissues of adult plant, not from seeds. The isolation of bacteria from roots and stems of field-grown maize has been efficient in obtaining maize growth promoters and developing valuable biotechnological products (Hungria et al. 2010). Studies assessing the diversity of endophytic bacteria in maize seeds have revealed a plethora of microbial taxa (Mitter et al. 2017), but their potential to promote plant growth remains unknown.
In this study, maize seeds were colonized by a primordial bacterial inoculum that spread through the whole plant, under axenic conditions, since we isolated SEB from different plant compartments. The spread of seed-borne bacteria to whole plants, in non-sterilized substrate, was already shown by Hardoim et al. (2012), who obtained many bacteria from rice in two consecutive generations. The authors showed that in the second generation, 45% of the bacterial community (at the genus or species level) was derived from the first generation, indicating that seed endophytes can be transfered vertically and they play an essential role in whole-plant colonization.
Most of the bacteria isolated were obtained from the maize seeds. Diverse SEB has been obtained from seeds of Poaceae crops such as rice (Mano et al. 2006; Kaga et al. 2009; Hardoim et al. 2012), wheat (Ringelberg et al. 2012) and maize (Rijavec et al. 2007; Liu et al. 2012, 2013; Johnston-Monje et al. 2016; Bodhankar et al. 2017). The acquisition of bacterial endophytes from seeds is dependent on the composition of the community inside the mother maize seeds but also on-field management, mainly in the flowering stage; hence, maize seed endophytes can be acquired horizontally or vertically (Truyens et al. 2015; Mitter et al. 2017; Chowdhury et al. 2019). Due to axenic growth conditions in this study, the seed bacterial community spread to the whole plant, diluting the composition in the other compartments.
The isolation of bacteria from the “rhizosphere”, an external region surrounding the roots, indicated that the maize seeds could be a source of bacteria for soil colonization. Several studies have indicated that root exudates change the chemical composition of the rhizosphere and influence the rhizospheric microbial community (Haichar et al. 2008; Dennis et al. 2010; Rolfe et al. 2019; Pathan et al. 2020). However, recent results have proven that the bacterial community of wheat seeds change the rhizospheric bacterial community in non-sterile arable soils (Kavamura et al. 2019). Our findings are convergent to those observed by Kavamura et al. (2019), considering the different experimental conditions of both studies. In addition to shaping the rhizosphere bacterial community through root exudates, the rhizospheric bacterial community can also be derived from the primordial inoculum confined in the endophytic tissues of the seeds, influencing the soil bacterial community actively.
The dendrogram generated through the ARDRA assay (Fig. 2) shows the presence of some groups where strains from seeds were prevalent; for example, cluster II, the largest cluster. However, in clusters IV and VI, the bacteria isolated from the seeds were absent. These results indicate that although all strains originated from the seeds, the bacteria spread differentially through the whole plant, probably due to a preference for different environments (found in the plant compartments), occupying other ecological niches. The ability to colonize the whole plant can partially explain why 45% of the rice seed endophytes were different from those present in the mother seeds, according to the findings observed by Hardoim et al. (2012).
Twenty-nine out of 51 bacteria (57%) were considered potential plant-growth promoters. The analysis of their molecular fingerprints showed that all SEB from this study were different from each other, highlighting the high diversity of cultivated bacteria within the maize seeds, most of which were potential PGPB. At the species level, a high diversity of cultivable SEB has already been shown in Poaceae species such as maize (Liu et al. 2012; Johnston-Monje et al. 2016) and rice (Kaga et al. 2009; Hardoim et al. 2012). However, data on the diversity of cultivable SEB at the intraspecific level have not been available until the current study. The data in the present study amplify the results available in the literature, showing a higher diversity of SEB at the intraspecific level.
Nevertheless, the diversity at genus level in this study was very low, with 23 out of 29 identified strains classified as Firmicutes and 21 as Bacillus genus. Bacillus spp. has already been identified as prevalent cultivated bacteria within maize seeds (Bodhankar et al. 2017). However, the diversity of total bacterial species within maize seeds is dependent on the plant genotype (Johnston-Monje et al. 2016). The predominance of Bacillus spp. within the culture collections are probably related to the plant material used as a source of bacteria (i.e., the harsh seed environment).
In addition to Bacillus, Paenibacillus and Acinetobacter were isolated in the present study. All genera were already reported as PGPB (Molina-Romero et al. 2017; Antunes et al. 2019; Cavalcanti et al. 2020). Paenibacillus has already been isolated as endophyte from seeds of rice (Mano et al. 2006), but Acinetobacter is a new occurrence for studies on SEB. All five Acinetobacter spp. were not isolated from seeds but from seedling shoots and “rhizosphere”. These bacteria are likely present at low densities in the seeds but can spread to the whole plant during its development.
Early root development is crucial for good plant development and nutrient acquisition (Bewley et al. 2012), and microbial inoculants can help plants develop their roots and improve nutrient absorption (Calvo et al. 2017). Therefore, all 29 SEB selected as potential plant growth promoters are candidates to be effective PGPB.
A total of 20 SEB produced detectable amounts of auxin. This hormone plays a pivotal role in root growth during germination and early seedling development (Miransari and Smith 2014). Some of the bacteria that increased initial root development were not the best auxin producers but produced detectable amounts of this hormone and also showed other growth promotion mechanisms, such as biofilm biosynthesis. For example, Bacillus sp. ESA 652, isolated from seedling shoots, produced auxin and siderophores and was ranked in the highest cluster for biofilm biosynthesis. Inoculation with this bacterium induced good early root development and promoted six out of the 12 parameters assessed in the plant-growth promotion pot experiment. Bacteria showing multiple plant-growth-promoting traits are desirable in inoculant selection (de Souza et al. 2015; Santos et al. 2019) and can promote plant growth in pot experiments (da Silva et al. 2018; Antunes et al. 2019; Santana et al. 2020) and under field conditions (Alves et al. 2014; dos Santos et al. 2017; Cavalcanti et al. 2020).
The ability to promote the growth and increase the uptake of different nutrients, indicates that several maize bacteria display different plant-growth-promoting mechanisms in addition to those assessed in the present study. The increase of the contents of zinc, manganese, and copper in the maize shoots provided by the inoculation of different Bacillus, and Acinetobacter bacterial strains points to the ability of some of them to solubilize these insoluble elements in the rhizosphere or increase the plant uptake. This feature is commonly described for Bacillus spp. (Luo et al. 2012; Khande et al. 2017; Bhatt and Maheshwari 2020), but it is a novelty for Acinetobacter. The same pattern could be observed to potassium, a macronutrient with low mobility in soils, since a Bacillus sp. (ESA 652), and an Acinetobacter sp. (ESA 661) increased the maize K content. As highlighted to the micronutrients, potassium solubilization is a feature observed in Bacillus (Pramanik et al. 2019), and other bacteria, but is a new feature to Acinetobacter.
Inoculation with PGPB can result in more efficient N absorption in maize (Araújo et al. 2015). In our pot experiment, 27 out of 29 inoculated treatments efficiently assimilated the low amount of N applied as fertilizer, since all treatments showed higher nitrogen in the shoots than the uninoculated control treatment. In the present study, this feature was not extended to other nutrients, since few bacteria stood out as accumulating more than two other macro and/or micronutrients. The inoculation of a given bacterial isolate can have an impact on the interactions of the host plant with other members of the soil microbiota, such as mycorrhizae (Adesemoye et al. 2008). The inoculation of some bacteria, such as Bacillus ap. ESA 674 and Acinetobacter sp. ESA 662, in addition to improving plant establishment, may influence other plant-microbial interactions, resulting in better nutrient accumulation in the shoots.
In the plant-growth-promotion experiment, the strains used for inoculation overgrew the bacteria present within the seeds and those present in the soil to promote plant growth. The performance of some SEB inoculated on maize seeds increased plant growth, and the nutritional parameters showed the great potential of the bacteria isolated in the present study to promote maize growth and be used as inoculants for maize in Brazilian drylands.
Some authors have argued that seeds are small “Noah’s arks” for plant colonization (Hardoim 2019; Verma and White Jr 2019). Our data are convergent to this statement and showed, for the first time, the ability of SEB from Brazilian drylands to occupy different niches in maize seedlings under axenic conditions. This study also showed that some of these bacteria, mainly Bacillus spp. ESA 674, ESA 652, and Acinetobacter sp. ESA 662, are potential biotechnological tools to alter the diversity of the maize SEB community composition and produce maize seeds enriched with PGPB. Further colonization and field trials are needed to ensure the potential of these bacteria to be used as inoculants for maize.
References
Adesemoye AO, Torbert HA, Kloepper JW (2008) Enhanced plant nutrient use efficiency with PGPR and AMF in an integrated nutrient management system. Can J Microbiol 54:876–886. https://doi.org/10.1139/W08-081
Aeron A, Khare E, Kumar C et al (2020) Revisiting the plant growth - promoting rhizobacteria : lessons from the past and objectives for the future. Arch Microbiol 202:665–676. https://doi.org/10.1007/s00203-019-01779-w
Alves GC, Videira SS, Urquiaga S, Reis VM (2014) Differential plant growth promotion and nitrogen fixation in two genotypes of maize by several Herbaspirillum inoculants. Plant Soil 387:307–321. https://doi.org/10.1007/s11104-014-2295-2
Ambrosini A, Stefanski T, Lisboa BB, Beneduzi A, Vargas LK, Passaglia LMP (2016) Diazotrophic bacilli isolated from the sunflower rhizosphere and the potential of Bacillus mycoides B38V as biofertiliser. Ann Appl Biol 168:93–110. https://doi.org/10.1111/aab.12245
Antunes G dos R, Santana SRA, Escobar IEC, et al (2019) Associative diazotrophic bacteria from forage grasses in the Brazilian semiarid region are effective plant growth promoters. Crop Pasture Sci 70:899–907. doi: https://doi.org/10.1071/CP19076
Araújo ÉDO, Martins MR, Vitorino ACT et al (2015) Effect of nitrogen fertilization associated with diazotrophic bacteria inoculation on nitrogen use efficiency and its biological fixation by corn determined using 15 N. African J Microbiol Res 9:643–650. https://doi.org/10.5897/AJMR2014.7072
Bacilio-Jiménez M, Aguilar-Flores S, Del Valle MV et al (2001) Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol Biochem 33:167–172. https://doi.org/10.1016/S0038-0717(00)00126-7
Bewley JD, Bradford K, Hilhorst H (2012) Seeds: physiology of development, germination and dormancy. Springer Science & Business Media
Bhatt K, Maheshwari DK (2020) Zinc solubilizing bacteria (Bacillus megaterium) with multifarious plant growth promoting activities alleviates growth in Capsicum annuum L. 3 Biotech 10: https://doi.org/10.1007/s13205-019-2033-9
Bodhankar S, Grover M, Hemanth S et al (2017) Maize seed endophytic bacteria: dominance of antagonistic, lytic enzyme-producing Bacillus spp. 3 Biotech 7:232. https://doi.org/10.1007/s13205-017-0860-0
Bodhankar S, Grover M, Reddy G (2019) In Planta screening of maize seed endophytic bacteria for potential applications under dryland conditions. Indian J Dryl Agric Res Dev 34:53. https://doi.org/10.5958/2231-6701.2019.00009.5
Bushnell LD, Haas HF (1941) The utilization of certain hydrocarbons by microorganisms. J Bacteriol 41:653–673
Calvo P, Watts DB, Kloepper JW, Torbert HA (2017) Effect of microbial-based inoculants on nutrient concentrations and early root morphology of corn (Zea mays). Z Pflanzenernähr Bodenkd 180:56–70. https://doi.org/10.1002/jpln.201500616
Cavalcanti MIP, Nascimento R de C, Rodrigues DR, et al (2020) Maize growth and yield promoting endophytes isolated into a legume root nodule by a cross-over approach. Rhizosphere 15:100211. https://doi: 10.1016/j.rhisph.2020.100211
Chowdhury S, Lata R, Kharwar RN, Gond SK (2019) Microbial Endophytes of Maize Seeds and Their Application in Crop Improvements BT - Seed Endophytes: Biology and Biotechnology. In: Verma SK, White Jr JF (eds). Springer International Publishing, Cham, pp 449–463. https://doi.org/10.1007/978-3-030-10504-4_21
CONAB (2019) Séries históricas. https://www.conab.gov.br/conteudos.php?a=1252&t=&Pagina_objcmsconteudos=3#A_objcmsconteudos. Accessed 10 Feb 2020
da Silva JF, da Silva TR, Escobar IEC, Fraiz ACR, dos Santos JWM, do Nascimento TR, dos Santos JMR, Peters SJW, de Melo RF, Signor D, Fernandes-Júnior PI (2018) Screening of plant growth promotion ability among bacteria isolated from field-grown sorghum under different managements in Brazilian drylands. World J Microbiol Biotechnol 34:186. https://doi.org/10.1007/s11274-018-2568-7
de Souza R, Ambrosini A, Passaglia LMP (2015) Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol Biol 38:401–419. https://doi.org/10.1590/S1415-475738420150053
Ferreira DF (2011) Sisvar: a computer statistical analysis system. Ciência e Agrotecnologia 35 (6):1039-1042
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol 72:313–327
dos Santos CLR, Alves GC, de Matos Macedo AV, Giori FG, Pereira W, Urquiaga S, Reis VM (2017) Contribution of a mixed inoculant containing strains of Burkholderia spp. and Herbaspirillum ssp. to the growth of three sorghum genotypes under increased nitrogen fertilization levels. Appl Soil Ecol 113:96–106. https://doi.org/10.1016/J.APSOIL.2017.02.008
Gadanho M, Sampaio JP (2002) Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. dairenensis comb. nov. FEMS Yeast Res 2:47–58. https://doi.org/10.1016/S1567-1356(01)00062-9
García-Fraile P, Menéndez E, Rivas R (2015) Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng 2:183–205. https://doi.org/10.3934/bioeng.2015.3.183
Haichar FEZ, Marol C, Berge O et al (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. https://doi.org/10.1038/ismej.2008.80
Hardoim P. (2019) The Ecology of Seed Microbiota. In: Verma S., White Jr JF. (eds) Seed Endophytes: Biology and Biotechnology. Springer International Publishing, Cham, pp 103–125. https://doi.org/10.1007/978-3-030-10504-4_6
Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD (2012) Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7:e30438. https://doi.org/10.1371/journal.pone.0030438
Hungria M, Campo RJ, Souza EM, Pedrosa FO (2010) Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331:413–425. https://doi.org/10.1007/s11104-009-0262-0
Hungria M, Nogueira MA, Araujo RS (2016) Inoculation of Brachiaria spp. with the plant growth-promoting bacterium Azospirillum brasilense: an environment-friendly component in the reclamation of degraded pastures in the tropics. Agric Ecosyst Environ 221:125–131. https://doi.org/10.1016/j.agee.2016.01.024
Ikeda AC, Savi DC, Hungria M, Kava V (2020) Bioprospecting of elite plant growth-promoting bacteria for the maize crop. Acta Sci - Agron 42:e44364. https://doi.org/10.4025/actasciagron.v42i1.44364
Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN (2016) Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 405:337–355. https://doi.org/10.1007/s11104-016-2826-0
Kaga H, Mano H, Tanaka F, Watanabe A, Kaneko S, Morisaki H (2009) Rice seeds as sources of endophytic bacteria. Microbes Environ 24:154–162. https://doi.org/10.1264/jsme2.ME09113
Kavamura VN, Robinson RJ, Hayat R, Clark IM, Hughes D, Rossmann M, Hirsch PR, Mendes R, Mauchline TH (2019) Land management and microbial seed load effect on rhizosphere and endosphere bacterial community assembly in wheat. Front Microbiol 10:1–11. https://doi.org/10.3389/fmicb.2019.02625
Khande R, Sharma SK, Ramesh A, Sharma MP (2017) Zinc solubilizing Bacillus strains that modulate growth, yield and zinc biofortification of soybean and wheat. Rhizosphere 4:126–138. https://doi.org/10.1016/j.rhisph.2017.09.002
Liao CFH (1981) Devarda’s alloy method for total nitrogen determination. Soil Sci Soc Am J 45:852–855. https://doi.org/10.2136/sssaj1981.03615995004500050005x
Liu Y, Zuo S, Xu L, Zou Y, Song W (2012) Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch Microbiol 194:1001–1012. https://doi.org/10.1007/s00203-012-0836-8
Liu Y, Zuo S, Zou Y, Wang J, Song W (2013) Investigation on diversity and population succession dynamics of endophytic bacteria from seeds of maize (Zea mays L., Nongda108) at different growth stages. Ann Microbiol 63:71–79. https://doi.org/10.1007/s13213-012-0446-3
Luo S, Xu T, Chen L, Chen J, Rao C, Xiao X, Wan Y, Zeng G, Long F, Liu C, Liu Y (2012) Endophyte-assisted promotion of biomass production and metal-uptake of energy crop sweet sorghum by plant-growth-promoting endophyte Bacillus sp. SLS18. Appl Microbiol Biotechnol 93:1745–1753. https://doi.org/10.1007/s00253-011-3483-0
Mano H, Tanaka F, Watanabe A, Kaga H, Okunishi S, Morisaki H (2006) Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ 21:86–100. https://doi.org/10.1264/jsme2.21.86
Miransari M, Smith DL (2014) Plant hormones and seed germination. Environ Exp Bot 99:110–121. http://dx.doi.org/10.1016/j.envexpbot.2013.11.005
Mitter B, Pfaffenbichler N, Flavell R, Compant S, Antonielli L, Petric A, Berninger T, Naveed M, Sheibani-Tezerji R, von Maltzahn G, Sessitsch A (2017) A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol 8. https://doi.org/10.3389/fmicb.2017.00011
Molina-Romero D, Baez A, Quintero-Hernández V, Castañeda-Lucio M, Fuentes-Ramírez LE, Bustillos-Cristales MR, Rodríguez-Andrade O, Morales-García YE, Munive A, Muñoz-Rojas J (2017) Compatible bacterial mixture, tolerant to desiccation, improves maize plant growth. PLoS One 12:1–21. https://doi.org/10.1371/journal.pone.0187913
Nostro A, Procopio F, Pizzimenti FC et al (2007) Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Med Microbiol 56:519–523. https://doi.org/10.1099/jmm.0.46804-0
Pathan SI, Ceccherini MT, Sunseri F, Lupini A (2020) Rhizosphere as hotspot for plant-soil-microbe interaction BT - carbon and nitrogen cycling in soil. In: Meena RS, Pathan SI, Ceccherini MT (eds) Datta R. Springer Singapore, Singapore, pp 17–43
Pramanik P, Goswami AJ, Ghosh S, Kalita C (2019) An indigenous strain of potassium-solubilizing bacteria Bacillus pseudomycoides enhanced potassium uptake in tea plants by increasing potassium availability in the mica waste-treated soil of North-East India. J Appl Microbiol 126:215–222. https://doi.org/10.1111/jam.14130
Puente ME, Li CY, Bashan Y (2009) Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ Exp Bot 66:402–408. https://doi.org/10.1016/j.envexpbot.2009.04.007
Ribeiro CM, Cardoso EJBN (2012) Isolation, selection and characterization of root-associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia). Microbiol Res 167:69–78. https://doi.org/10.1016/j.micres.2011.03.003
Rijavec T, Lapanje A, Dermastia M, Rupnik M (2007) Isolation of bacterial endophytes from germinated maize kernels. Can J Microbiol 53:802–808. https://doi.org/10.1139/W07-048
Ringelberg D, Foley K, Reynolds CM (2012) Bacterial endophyte communities of two wheatgrass varieties following propagation in different growing media. Can J Microbiol 58:67–80. https://doi.org/10.1139/W11-122
Rodrigues Neto J, Malavolta VA Jr, Victor O (1986) Meio simples para o isolamento e cultivo de Xanthomonas campestris pv. citri tipo B. Summa Phytopathol 12:32
Rolfe SA, Griffiths J, Ton J (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr Opin Microbiol 49:73–82
Santana SRA, Voltolini TV, Antunes G dos R, et al. (2020) Inoculation of plant growth-promoting bacteria attenuates the negative effects of drought on sorghum. Arch Microbiol 202:1015–1024. https://doi.org/10.1007/s00203-020-01810-5
Santos MS, Nogueira MA, Hungria M (2019) Microbial inoculants : reviewing the past , discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 9:205. https://doi.org/10.1186/s13568-019-0932-0
Sarwar M, Kremer RJ (1995) Determination of bacterially derived auxins using a microplate method. Lett Appl Microbiol 20:282–285. https://doi.org/10.1111/j.1472-765X.1995.tb00446.x
Schlemper TR, Dimitrov MR, Silva Gutierrez FAO, van Veen JA, Silveira APD, Kuramae EE (2018) Effect of Burkholderia tropica and Herbaspirillum frisingense strains on sorghum growth is plant genotype dependent. PeerJ 6:e5346. https://doi.org/10.7717/peerj.5346
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Seido SL, Sousa LP, Silva MJ, Donzeli VP, Queiroz SOP (2019) Melon growth-promoting rhizobacteria under saline stress. Rev Bras Ciências Agrárias 14:1–9. https://doi.org/10.5039/agraria.v14i1a5623
Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF, van Elsas JD, Faure D, Reiter B, Glick BR, Wang-Pruski G, Nowak J (2005) Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol 55:1187–1192. https://doi.org/10.1099/ijs.0.63149-0
Soares RA, Roesch LFW, Zanatta G, de Oliveira Camargo FA, Passaglia LMP (2006) Occurrence and distribution of nitrogen fixing bacterial community associated with oat (Avena sativa) assessed by molecular and microbiological techniques. Appl Soil Ecol 33:221–234. https://doi.org/10.1016/j.apsoil.2006.01.001
Švec P, Vancanneyt M, Seman M et al (2005) Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol Lett 247:59–63. https://doi.org/10.1016/j.femsle.2005.04.030
Sylvester-Bradley R, Asakawa N, La Torraca S et al (1982) Levantamento quantitativo de microrganismos solubilizadores de fosfatos na rizosfera de gramíneas e leguminosas forrageiras na Amazônia. Acta Amaz 12:15–22
Teixeira PC, Donagemma GK, Fontana A, Teixeira WG (eds) (2017) Manual de métodos de análise de solo, 3rd edn. Brasilia, Empresa Brasileira de Pesquisa Agropecuária
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7:40–50. https://doi.org/10.1111/1758-2229.12181
Verma SK, White Jr JF (2019) Seed Endophytes. Springer International Publishing
Versalovic J, Schneider M, De Bruijn FJ, Lupski JR (1994) Genomic fingerpriting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. [published for the] international biological Programme [by] Blackwell scientific
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Acknowledgments
Our acknowledgments are given to Brazilian Agricultural Research Corporation (Embrapa 23.13.08.003.00.00) and Brazilian Council for Scientific and Technological Development (CNPq 485168/2013–8), INCT–Plant Growth-Promoting Microorganisms for Agricultural Sustainability and Environmental Responsibility (CNPq/Fundação Araucária STI/CAPES INCT-MPCPAgro 465133/2014-4) for financial support. We also thank the Science foundation of the Pernambuco State (FACEPE) and CNPq for awarding scholarships to the first and sixth authors, respectively. The last two authors are research fellows of CNPq (306812/2018-5 and 311218/2017-2, respectively).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Ethical approval
This paper does not contain any studies with human participants or animals performed by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Bomfim, C.S.G., da Silva, V.B., Cursino, L.H.S. et al. Endophytic bacteria naturally inhabiting commercial maize seeds occupy different niches and are efficient plant growth-promoting agents. Symbiosis 81, 255–269 (2020). https://doi.org/10.1007/s13199-020-00701-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-020-00701-z