Abstract
Forty rhizobial strains were isolated from root nodules of Medicago littoralis Rhode and Melilotus indicus (L.) harvested from the sandy soils of Touggourt’s oases in the Oued Righ Valley, Algerian Sahara. The isolates were studied for their cultural, biochemical and symbiotic effectiveness. All of them were fast-growing bacteria; utilized a wide range of carbon sources, produced abundant extracellular polysaccharides, tolerated high concentrations of NaCl (up to 2.5 %), grew at temperatures between 28 and 45 °C and at pH values between 4.5 and 9. The isolates were sensitive to the antibiotics kanamycin, tetracycline and rifampicin but showed resistance to neomycin and erythromycin. All the isolates induced the formation of effective nodules on their host plants. On the basis of the physiological, biochemical and symbiotic effectiveness, we selected six strains MD05, MD09, MD12, ML08, ML17 and ML22 for genotypic characterization. Phylogenetic analysis of the selected strains based on 16S ribosomal RNA gene showed that these strains of bacteria were affiliated to the Ensifer meliloti group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In Algeria, arid lands with a Saharan climate represent about 90 % of the total area of the country (Le Houérou 1975). The Algerian Sahara is characterized by the presence of particular regions called “oases” that represent biological diversity hotspots (Riou 1990; Chouaki et al. 2006). The most famous oases are those of Oued Righ Valley (Fig. 1) where more than 47 oases are located between 32°54′ of latitude and 34°09′ longitude. This valley has a hyper arid climate, with gritty soil characterized by a high salinity due to irrigation with artesian salty water (3 g/l). The chemical composition of these soils is variable but is generally very poor in nitrogen, phosphorous and potassium (Toutain 1974; Le Houérou 1975; Koull et al. 2013). Agriculture at these oases is characterized by small production units (Ferry and Toutain 1990) and the integration of fruit trees and tall crops to protect herbal plants against the strong sunlight. The oases of Oued Righ Valley have a beneficial microclimate due to the shade and the constant moisture provided by irrigation. These decrease the effect of the very hot Saharan climate (Riou 1990; Messar 1996).
The Leguminosae are one of three important families which are predominant in these agriculture units; the others being the Poaceae and the Asteraceae. Medicago sativa is the main annual fodder crop of the oases and occupies about 80 % of the land surface devoted to such crips and a third of the irrigated area (Janati 1990; Abdelguerfi et al. 1996; Chaabena and Abdelguerfi 2007; Chouaki et al. 2006; Chafi and Bensoltane 2009; Chaabena et al. 2011).
Plant productivity in these arid and semi-arid regions is low because of poor soil fertility (Toutain 1974; Le Houérou 1975; Djennane 1990; Zahran 1999; Priefer et al. 2001). Nitrogen is one of the main factors limiting plant growth, and soils are generally very poor on organic matter (Fitouri Dhane et al. 2012; Palma et al. 2013). Biological Nitrogen Fixation (BNF) is the sustainable source of nitrogen in the cropping systems (Serraj et al. 2004) and Rhizobium-legume symbioses provide soil enrichment (Zahran 1999, 2001). In Algeria, studies on rhizobia-legume symbiosis have only been completed in northern regions of the country while others have been carried out in the arid regions of Tunisia (Zakhia et al. 2004; Mnasri et al. 2007a, b; Djedidi et al. 2011; Rejii et al. 2014), Morocco (Maâtallah et al. 2002; Thami-Alami et al. 2010) and Egypt (Zahran 1997; Abdel-Salam et al. 2010).
Rhizobia associated with wild legumes in the oases of Touggourt have not previously been investigated and the present work was carried out to study rhizobial strains associated with two indigenous legumes; Medicago littoralis and Melilotus indicus growing in salty soils of Touggourt’s oases. The phenotypic characterization of these strains was conducted to evaluate their capacity to grow under environmental stresses which include high temperature, salinity, drought and pH. The phylogenetic positions of our strains were determined by an analysis of the 16S rRNA gene.
2 Materials and methods
2.1 Plant material
The wild legumes Medicago littoralis and Melilotus indicus were collected from the oases of Touggourt in Oued Righ Valley.
2.2 Isolation of rhizobia
The nodules were surface-sterilized with sodium hypochlorite (2 %) for 4 min and rinsed five times with sterile water; then the nodules were crushed with glass rod in a sterile test tube. One loopful of the nodule content suspension was streaked on yeast extract mannitol agar medium (YEMA) containing Congo red dye and incubated at 28 °C until colonies achieved a maximum size (Somasegaran and Hoben 1985).
After incubation for 3 to 5 days at 28 °C; single colonies were selected and transferred several times on YEMA medium. Isolates were purified and maintained on YEMA medium and conserved at −20 °C in YEMB-glycerol (yeast extract mannitol broth supplemented with 20 % glycerol) until use (Somasegaran and Hoben 1985; Shetta et al. 2011).
2.3 Rhizobia presumptive tests
The absorption or not of the Congo Red dye (CR) and the production of acid or alkali by the isolates was estimated after 3 days of growth respectively on YEMA supplemented with CR (0.0025 %) and on YEMA supplemented with Bromothymol blue (BTB 0.0025 %) (Somasegaran and Hoben 1985; Shetta et al. 2011). Ensifer meliloti strain 1021, Mesorhizobium ciceri (M4) and Agrobacterium tumefaciens C58 were utilized as reference strains for the phenotypic tests. All the tests were done with suspension of isolates at the exponential growth phase.
2.4 Temperature and salt tolerance assays
The effect of the temperature and the tolerance to different concentrations of NaCl were determined by inoculating the isolates on YEMA medium supplemented with NaCl at different concentrations (w/v), 2.5; 3.5; 4.5 and 6.0 %. The plates were incubated at four different temperatures (28; 37; 40 and 45 °C) for 7 days (Maâtallah et al. 2002; Djedidi et al. 2011).
2.5 pH tolerance
The tolerance of isolates to large range of pHs (4.5; 5.5; 6.8; 8 and 9) was determined on YEMA medium. The plates were incubated for 3 days at 28 °C (Thami-Alami et al. 2010).
2.6 Tolerance to drought stress
The isolates were suspended in 1 ml of PEG 4000 solutions at final concentrations: 0; 10; 15; 20 and 25 % (w/v) and then 10 μl of each suspension were used for inoculating YEMA plates. The plates were incubated at 28 °C for 3 days; the tolerance was estimated by measuring the growth of isolates on plates at different concentrations of PEG 4000.
2.7 Intrinsic antibiotic resistance
In a first protocol, the test was performed by using antibiotics discs. A volume of 0.5 ml of an exponential culture of the isolates was spread on the surface of the YEMA plates; the plates were dried for 30 min and we put the antibiotics discs on the surface of the plates : Streptomycin (30 μg); ampicillin (30 μg); chloramphenicol (30 μg); tetracycline (30 μg); nalidixic acid (30 μg); kanamycin (30 μg). After 3 to 5 days of incubation at 28 °C, the inhibition zones were measured (Rome et al. 1996; Elbanna et al. 2009).
In a second protocol, filter-sterilized antibiotics were added aseptically to sterile YEM medium to give a final concentrations of rifampicin (30 μg/ml); erythromycin (30 μg/ml) and neomycin (30 μg/ml) (Somasegaran and Hoben 1985). Plates were incubated at 28 °C, after 3 days, results were recorded by the absence or the presence of bacterial growth (Shetta et al. 2011; Thami-Alami et al. 2010; Zhang et al. 1991).
2.8 Plant nodulation test
The ability of isolates to nodulate their host plants was tested. Seeds were hand-sorted for size uniformity and for absence of damages, surface-disinfected by soaking in sodium hypochlorite (2.5 %) for 10 min and rinsed several times with sterile distilled water, then left to germinate in the dark (Somasegaran and Hoben 1985; Amrani et al. 2010).
The nodulation tests were done using pots pre-filled with sterile sand. At least three seeds were used per pot for each isolate tested (Abdel-Salam et al. 2010). The suspensions were prepared using isolates at the exponential growth phase. One ml of each suspension was used to inoculate young plants of Medicago and Melilotus. The plants were kept under natural conditions and watered with the solution of Rigaud and Puppo (Priefer et al. 2001; Djedidi et al. 2011). After 6 to 8 weeks of plant growth, the presence or absence of nodules on the roots was recorded. The efficiency of the nodules was estimated by the presence of red pigment (leghemoglobin) inside the nodules (Djedidi et al. 2011). The plants were dried at 50 °C after separating roots from aerial parts.
2.9 Test of 3-ketolactose
The determination of 3-ketolactase enzyme activity allows distinguishing between Agrobacterium and other rhizobial strains (Murugesan et al. 2010). Isolates were cultivated on nutrient glucose agar medium (NGA) for 48 to 72 h (Bouzar et al. 1995) and then suspensions were prepared with colonies grown on NGA. Drops of 10 μl were putted on lactose agar medium (LAM) with lactose (1 %), yeast extract (0.1 %), and agar (2 %). Three isolates per plate were tested. After incubation at 28 °C for 48 to 72 h, the plates were flooded with a thin layer of Benedict’s reagent (Bouzar et al. 1995). The presence of 3-ketolactose in the medium was assessed by the formation of a yellow ring of cuprous oxide around the rhizobial colony after 15 to 20 min.
2.10 Biochemical and auxanographic tests
API 20NE and API 50CH are standardized systems, API 20 NE system combining 20 tests for identification of gram-negative rod and API 50CH gallery consists of 50 tests used for the study of carbohydrate metabolism. API 20 NE tests, the inoculation was done as recommended by the manufacturers (Shetta et al. 2011). The ability of isolates to metabolize carbohydrates was tested by using API 50 CH gallery (BioMérieux, France) as described by Kersters et al. (1984). After inoculation, galleries were incubated at 28 °C; the results were scored every day for 7 days.
2.11 Statistical analysis
The Principal Component Analysis (PCA) was performed to examine the relationships between the tolerance of isolates in vitro to both high temperature and salinity. Computations and graphical display were made using the XLSTATTM software package (version 2004. 7. 5. 2, Addinsoft, Paris, France, http://www.xlstat.com). A computer cluster analysis of 206 phenotypic variables was carried out using STATISTICA 7.0 software package and a phenogram was constructed by the Unweighted Pair Group Method with Average (UPGMA) clustering method.
2.12 Genomic DNA extraction and phylogenetic analysis of rRNA genes
Genomic DNA was extracted using QIAamp DNA Mini Kit (QIAGEN, Düsseldorf, Germany) according to the manufacturer’s instructions. The bacterial 16S rRNA gene fragment was amplified using the bacterial universal primers fDl (5′ AGAGTTTGATCCTGGCTCAG-3′) and rDl (5′-AAGGAGGTGATCCAGCC-3′) (Weisburg et al. 1991). The amplification reaction mixture consisted of 1x PCR reaction buffer, 100 nM of each primer, 200 mM of each deoxyribonucleotide triphosphate, 0.25 Units Hot Star Taq DNA polymerase (Qiagen, Hilden, Germany) and 20 ng of bacterial DNA.
The PCR program consisted of an initial step activation of the enzyme at 95 °C for 15 min, followed by a step of denaturation at 95 °C for 15 min; 30 cycles of denaturation for 1 min at 95 °C, annealing for 1 min at 55 °C and elongation for 1.5 min at 72 °C, followed by a final elongation step at 72 °C for 7 min. The amplicons were gel-purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and bidirectionally sequenced using primers rD1 and fD1. A single strand sequence was obtained for the strain ML08.
The 16S rRNA gene sequences were subjected to a NCBI nucleotide and 16S rRNA gene BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify sequences of the highest similarity. Phylogenetic analysis was inferred using the Maximum-likelihood method and bootstrap support for each node was evaluated with 1000 replicates. The multiple sequence alignment of nucleotide sequences for creating Maximum-likelihood was inferred using free software MEGA 5.2.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene are from KR476464 to KR476469.
3 Results
3.1 Rhizobial isolation
In this study, 40 strains were isolated from nodules of the wild legumes Medicago littoralis Rhode and Melilotus indicus (L.) All. plants harvested from the field of Touggourt’s Oases. Respectively, 15 isolates were from nodules of M. littoralis Rhode (MD1 to MD15) and 25 were from nodules of M. indicus (L.) All. (ML1 to ML25). All these isolates were fast growing bacteria (72 h), with smooth, semi-translucent circular colonies and produced abundant quantity of polysaccharide. The diameter of the colonies varied between 3 and 4 mm after 3 days of incubation. The colonies of two isolates from M. indicus were yellow, smooth with 1 to 2 mm of diameter after 3 days of incubation and did not produce polysaccharide.
3.2 Phenotypic characterization
3.2.1 Rhizobia presumptive tests
None of the isolates absorbed Congo red dye and nearly all (97 % ) caused a reduction in the pH of the medium during growth. The growth of isolates ML11, ML12, ML15 and MD07 slightly increased the pH of the medium.
3.2.2 Salt and high temperature tolerance
All of the isolates grew at temperatures between 28 and 40 °C and 98 % of them tolerated temperatures up to 45 °C. The reference strain isolates Ensifer meliloti strain 1021; Mesorhizobium ciceri and Agrobacterium tumefaciens C58 were also able to grow at these temperatures.
The ability of isolates to tolerate NaCl was examined and the results show that 100 % of the isolates grow at low concentrations of NaCl (0.01 % w/v). Ninety eight percent of the isolates tolerated 1 % NaCl, 87 % 2.5 %, 75 % at 3.5 % and 52.5 % at 4.5 %. Only 45 % of the isolates tolerated high salinity (6 %). In comparison, A. tumefaciens C58, E. meliloti 1021 and M. ciceri did not tolerate more than 1 % of NaCl. The Principal Component Analysis graphs (PCA), showed that the direction of axis variables reflected tolerance to different concentrations of NaCl (Figs. 2, 3, 4 and 5).
Analysis of the combined effects of temperature and NaCl showed that temperatures between 30 and 37 °C improve the salt tolerance of isolates. Figures 2 and 3 show that the number of tolerant isolates varied, from 13 at 28 °C to 19 at 37 °C. Indeed, the optimal temperature of growth for our isolates was about 30 °C. It was interesting that incubation of isolates at high temperatures (40 and 45 °C) decreased the salt tolerance of isolates. At 40 °C, only 8 isolates showed a weak growth at 4 % of NaCl (Figs. 4 and 5), while at 45 °C isolates were only able to grow at low concentrations of salt. Only the strain ML12 had a remarkable tolerance to high salinity and high temperature.
3.2.3 pH and drought stress tests
Thirty-nine isolates and the references strains were able to grow on medium with pH values from 4.5 to 9.5. Only isolate ML15 did not grow at pH < 5.5. All the isolates tolerate 10 to 25 % of PEG4000 in the culture medium.
3.2.4 Antibiotics tests
Antibiotic resistance or sensitivity was performed using nine different antibiotics. All the isolates as well as E. meliloti 1021 and A. tumefaciens C58 showed a strong sensitivity to kanamycin, tetracycline and rifampicin. e M. ciceri was only resistant to kanamycin and tetracycline. Ninety eight percent of the isolates were resistant to neomycin, 87 % to ampicillin, 85 % to the nalidixic acid, 80 % to erythromycin and 68 % to chloramphenicol but resistance to other antibiotics varied from isolate to isolate (Fig. 6).
3.2.5 Nodulation test
After 8 weeks of plant growth, root nodules were collected and recorded and we observe that 87 % of MD isolates and 100 % of ML isolates induced nodulation of the hosts’ plants. All formed pinky indeterminate nodules. The number of nodules obtained per host on M. littoralis plants inoculated with MD isolates varied from 0 to 21. In comparison, an average number of 26 nodules per plant were recorded for E. meliloti 1021 (no nodules were detected on negative controls of Medicago or Melilotus plants). In the case of M. indicus plants inoculated by ML isolates, the number of nodules per plant varied from 3 to 26. An average of 17 nodules were recorded on inoculated M. cicero plants. Figure 7 shows the relationship between dry matter (dry weight of the aerial parts of the nodulated plants) and the number of nodules. We observed that the dry matter increased proportionally with the number of nodules and that dry matter and nodule numbers were higher for Melilotus plants than for Medicago plants.
3.2.6 Test of 3-ketolactose
The 3-ketolactose enzyme activity distinguishes between Agrobacterium species and other rhizobial strains. We did not observe any 3-ketolactose enzyme activity in any of the isolates tested and concluded that the MD and ML isolates do not belong to the Agrobacterium group.
3.2.7 Carbohydrates assimilation
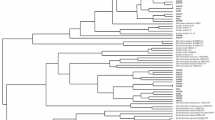
Fifteen isolates, nine ML and six MD, and E. meliloti 1021, M. ciceri and A. tumefaciens C58 were selected for phenotypic characterization by API20NE and API50CH tests. A total of 49 substrates were tested for the study of the carbohydrate metabolism. The analysis of phenotypic characters is presented as a dendrogram of dissimilarity in Fig. 8 which shows that the isolates comprise four groups. Group 1 includes ML1, MD3 and MD4; group 2 includes ML3, ML5, ML8, MD9 and MD12; group 3 includes ML17 and ML22 and the group 4 includes ML4, ML7 and MD13. The reference strains E. meliloti 1021, M. ciceri, A. tumefaciens C58 as well as the isolate ML12 constitute a single group and were used to confirm the phenotypic characterization.
UPGMA dendrogram of phenotypic characters of MD and ML root-nodulating isolates and their resistance/sensitivity profile to antibiotics: K Kanamycin, TE tetracycline, RIF rifampicin, AMP ampicillin, NA nalidixic acid, C chloramphenicol, S streptomycin, N neomycin, E erythromycin, S sensitive, R resistant, V variable
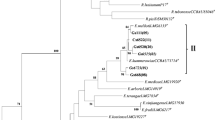
3.3 Phylogenetic analysis of partial 16S rRNA genes
Based on the physiological characters and results of symbiotic effectiveness tests, six isolates (MD05, MD09 and MD12 from Medicago littoralis, ML08, ML17 and ML22 from Melilotus indicus) were selected for further genotypic characterization (Table 1). Genomic DNA extract were amplified by PCR using universal primers fD1 and rD1 and the amplicons obtained were sequenced. The sequences were BLAST analyzed and the results showed clearly that our isolates belong to the Ensifer group with 99 % identity (Table 2). The phylogenetic tree built from 16S rRNA genes sequences (Fig. 9) showed that all the six isolates were clustered with the Ensifer meliloti type strain and neighbor species.
4 Discussion
Legume species, within the genera Medicago and Melilotus, are important as foliage and as a source of green manure, they are naturally distributed or cultivated in many regions of the world within arid regions (Yan et al. 2000; Chouaki et al. 2006; Bruning and Rozema 2012; Palma et al. 2013). Wild legumes often have a higher nitrogen fixing activity and are more tolerant to stress conditions than crop legumes. They are also better able to establish symbiotic relationships (Zahran 2001; Serraj et al. 2004; Bruning and Rozema 2012; Palma et al. 2013).
Our objective was to characterize the nitrogen fixing bacteria associated with two wild legumes Medicago littoralis Rhode and Melilotus indicus (L.) All., collected in the arid regions in Algeria, for which there are few data in the literature. We studied 40 isolates from saline soils of Touggourt oases in the Algerian Sahara. All our rhizobia strains isolated from Medicago nodules, and most from Melilotus nodules had typical colony and growth characteristics, when grown on standard YEMA medium (Somasegaran and Hoben 1985). Rhizobia generally do not absorb Congo red dye when plates are incubated in the dark. The fast-growing rhizobia that nodulated Medicago and Melilotus also showed the usual acidification of the culture medium. (Somasegaran and Hoben 1985; Shetta et al. 2011).
In arid regions, high soil temperature affects both the free-living and symbiotic rhizobia (Zahran 1999). For most of rhizobia, the optimum temperature range for growth is 28 to 31 °C. Many are unable to grow at 38 °C (Zahran 2001; Priefer et al. 2001; Cacciari et al. 2003; Thami-Alami et al. 2010; Shetta et al. 2011). In the present study 98 % of our isolates were able to grow at 45 °C. These strains were isolated from a hot arid region which suggests that temperature tolerance is correlated with the isolation site. Indeed, Rejii et al. (2014) identified Ensifer species from wild lotus nodules in arid Tunisian soil that were able to grow between 35° and 42 °C.
Salinity is a serious threat to agriculture in arid and semi-arid regions (Le Houérou 1975; Djennane 1990; Zahran 2001; Palma et al. 2013). Nearly 40 % of the world’s land surface can be categorized as having potential salinity problems, most of these areas are confined to the tropics and Mediterranean region (Toutain 1974; Le Houérou 1975; Djennane 1990; Zahran 1997; Koull et al. 2013). Successful rhizobia-legumes symbiosis under salt stress requires the selection of salt tolerant rhizobia. It has been found that fast-growing strains are more tolerant than slow-growing rhizobia (Zahran 1999; Cacciari et al. 2003; Thami-Alami et al. 2010). Our results showed a strong relationship between the tolerance (in vitro) of isolates to salt and high temperature conditions which was clearly confirmed by the PCA study. Most of our strains appear to be tolerant to more than 2 % NaCl, the highest level of tolerance reported for most strains by Priefer et al. (2001); Maâtallah et al. (2002); Cacciari et al. (2003); Mnasri et al. (2007a) and Rejii et al. (2014). This may be different from the level of tolerance exhibited in nature when plants are irrigated by water with increased salt levels, (Djennane 1990; Tabouche and Achour 2004; Koull et al. 2013). Rhizobia associated with Medicago and Melilotus are usually able to grow in presence of more than 2 % NaCl (Rome et al. 1996; Yan et al. 2000; Thami-Alami et al. 2010; Djedidi et al. 2011). However, the in vitro tolerance of rhizobial strains to NaCl in pure culture was not correlated with the salinity of the soils from which they were isolated (Boukhatem et al. 2012). There was also no correlation between laboratory assays and isolation conditions. This could be due to the fact that rhizobia are located in microniches at the rhizospheric level and are inside nodules, and thus are protected.
Analysis of the combination between temperature and salt stress results showed that only one isolate, ML12, survived at both of 45 °C and more than 3 % of NaCl. This may reflect the fact that oases have a distinctly different temperature and humidity from the arid hot surrounding areas. Djedidi et al. (2011) found that the frequency of resistance to both stresses is low among isolates. With respect to water stress, we found that all of the ML and MD isolates were drought-tolerant. Amrani et al. (2010) reported the good correlation between salt and drought stress, and that halotolerant strains were also tolerant to PEG, suggesting common osmo-adaptation mechanisms.
In the present study, ML and MD isolates were tolerant of pH and grew on media ranging from 4.5to 9. This agrees with Thami-Alami et al. (2010) who reported that some fast-growing E. meliloti strains, isolated from semi-arid soils of Morocco were able to grow normally at pH 3.5. Low pH appears to be a major factor limiting survival and activity of microorganisms in soils (Maâtallah et al. 2002; Shetta et al. 2011). Fast-growing rhizobia strains are generally considered to be less tolerant to acidic pH than the slow growing strains (Yan et al. 2000; Priefer et al. 2001) although there are some exceptions (Zahran 1999). In addition, Somasegaran and Hoben (1985) reported that fast-growing rhizobia could metabolize a wider range of carbon substrates than slow-growing strains. Carbohydrate utilization assays indicated that our isolates are able to use 44 of 49 carbohydrate sources tested, and may be able to produce important enzymes. The utilization of L-glutamine was a characteristic that can distinguish between strains of Rhizobium and Ensifer (Hartmann et al. 2006; Shetta et al. 2011).
Fast-growing rhizobia are more sensitive to antibiotics (Jordan 1984). In the present study, ML and MD isolates which are fast-growers and showed sensitivity tetracycline and rifampicin. However, they showed a high resistance to chloramphenicol, nalidixic acid, ampicillin, neomycin and erythromycin. Bromfield et al. (2010) report that E. meliloti strains are susceptible to about 5 μg of kanamycin and tetracycline, but could resist more than 50 μg of erythromycin and 20 μg of nalidixic acid. Elboutahiri et al. (2010) noted that E. meliloti and E. medicae from Morocco soils were resistance to chloramphenicol (up than 25 μg/l).
ML and MD isolates were able to infect their host plant and to fix atmospheric nitrogen, but we observed that the number of nodules formed by our strains are relatively low than Ensifer meliloti 1021, Mesorhizobium ciceri. Zahran (2001) suggested that rhizobia strains surviving under heat stress may lose their infectivity, due to plasmid curing or to alterations in cellular polysaccharides necessary for infection. High soil temperature (35–40 °C) usually results in the formation of ineffective nodules. Jebara et al. (2001) reported that plant growth and symbiotic efficiency in sand culture, were 5 to 8 time lower than that in hydro-aeroponic culture. The establishment and activity of legumes-rhizobia symbiosis in natura is particularly sensitive to environmental constraints (Zahran 1999, 2001; Drevon et al. 2004; Serraj et al. 2004; Bruning and Rozema 2012; Palma et al. 2013).
Remarkably, E. meliloti isolates showed a broader host-range among wild legumes species and we found that Melilotus indicus represent one of the best host traps to isolate rhizobial strains which display high abiotic stress resistance in oases soils. The osmotolerance, fast growth and nitrogen fixation efficiency suggests that ML and MD isolates may be good candidates as biofertilizers and provide promising strains for developing inoculants.
Analysis of 16S rDNA sequences is widely used to define the taxonomic position and trace the evolutionary history of nodule bacteria (Shamseldin et al. 2013; Gnat et al. 2014). In order to clarify the taxonomic status and the phylogenetic relationship of M. littoralis and M. indicus nodules isolates and other rhizobia, PCR amplification and sequencing of nearly full-length 16S rRNA gene (about 1400 bp) of six symbionts were performed. The comparative 16S rRNA gene sequence analysis of M. littoralis and M. indicus nodule isolates indicate that they are grouped on the phylogram into a separate cluster with known E. meliloti strains with more than 95 % of similarity The type strains E. meliloti USDA 1002 and E. kummerowiae were localized on the phylogram between our strains suggesting that they belong to one of this species. The preliminary results the blast shows clearly that our isolates belong to the Ensifer meliloti but they could also be a new species. More research is needed to confirm this result. Our strains clustered near to the Ensifer strains that have been isolated from neighboring regions with similar environments, e.g. E. numidicus ORS 1407 and E. garamanticus ORS 1400, which were isolated from Tunisian soil (Merabet et al. 2010). Rome et al. (1996); Yan et al. (2000); Zahran (2001); Zribi et al. (2004); Badri et al. (2007). It appears that the majority of wild legumes are nodulated by strains belonging to Ensifer and Rhizobium genus and few by Mesorhizobium and Bradyrhizobium (Thami-Alami et al. 2010; Rejii et al. 2014). The wild legumes particularly Medicago and Melilotus, show a great variation in nodulation and nitrogen fixation ability in relation to location (Zahran 2001).
In conclusion, we showed that Melilotus indicus and Medicago littoralis nodule bacteria, from the oases of Touggourt, were grouped within the Ensifer meliloti cluster. Our study provides the basis for further research on the phylogeny of rhizobial strains nodulating wild legumes in Sahara oases of Algeria and we suggest that these strains could be used as inoculants to improve plant growth and nitrogen fixation in arid lands. These strains may represent new genospecies that need further characterization to assess their taxonomical status.
References
Abdelguerfi A, Ait Ouada M, Kies N, Si Ziani Y (1996) From autoecology to variability of medics in Algeria: synthesis trial of works realized at the National Agronomic Institute - El Harrach. Options Méditerr Sér A Mediterr Semin 18:39–52
Abdel-Salam MS, Ibrahim SA, Abd-El-Halim MM, Badawy FM, Abo-Aba SEM (2010) phenotypic characterization of indigenous Egyptian Rhizobial strains for abiotic stresses performance. J Am Sci 6(9):498–501
Amrani S, Nazhat-Ezzaman N, Tej Bhatnagar T, Argandonña M, Nieto JJ, Vargas C (2010) Phenotypic and genotypic characterization of rhizobia associated with Acacia saligna (Labill.) Wendl. In nurseries from Algeria. Syst Appl Microbiol 33:44–51
Badri Y, Zribi K, Badri M, Huguet T, van Berkum P, Aouani ME (2007) Comparison of rhizobia that nodulate Medicago laciniata and Medicago truncatula present in a single Tunisian arid soil. Can J Microbiol 53:277–283
Boukhatem ZF, Domergue O, Bekki A, Chahinez Merabet C, Sekkour S, Bouazza F, Duponnois R, de Lajudie P, Galiana A (2012) Symbiotic characterization and diversity of rhizobia associated with native and introduced acacias in arid and semi-arid regions in Algeria. FEMS Microbiol Ecol:1–14
Bouzar H, Jones JB, Bishop AL (1995) Simple Cultural Tests for Identification of Agrobacterium Biovars: Methods in Molecular Biology. In: Gartland KMA, Davey Humana Totowa MR (eds) Agrobacterium Protocols, vol 44. Inc Press, NJ, pp 9–13
Bromfield ESP, Tambong JT, Cloutier S, Prévost D, Laguerre G, van Berkum P, Tran-Thi TV, Assabgui R, Barran LR (2010) Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156:505–520
Bruning B, Rozema J (2012) Symbiotic nitrogen fixation in legumes: perspectives for saline agriculture. Environ Exp Bot 2587:1–10
Cacciari I, Di Mattia E, Quatrini P, Moscatelli MC, Grego S, Lippi D, De Paolis MR (2003) Réponses adaptatives des isolats de Rhizobium aux stress. In: Grouzis M, Le Floc’h E (eds) Un arbre au désert, Acacia raddiana. IRD, Paris, pp 183–200
Chaabena A, Abdelguerfi A (2007) Aperçu sur les cultures fourragères au sahara septentrional est. Annu Fac Sci Sci Ingé 1(2):13–20
Chaabena A, Laouar M, Guediri O, Benmoussa A, Abdelguerfi A (2011) Quelques populations sahariennes de luzerne pérenne (Medicago sativa L.) face à un stress hydrique. Rev Bio Res 1(2):36–48
Chafi MH, Bensoltane A (2009) Vicia faba (L), a source of organic and biological manure for the Algerian arid regions. World J Agric Sci 5(6):698–706
Chouaki S, Bessedik F, Chebouti A, Maamri F, Oumata S, Khaldoun S, Hamana M-F, Douzene M, Bellah F, Kheldoun A (2006) Deuxième rapport national sur l’état des ressources phytogénétiques. Institut national de la recherche agronomique d’Algérie (INRA) et l’Organisation des nations unies pour l’alimentation et l’agriculture (FAO), pp 9–10
Djedidi S, Yokoyama T, Ohkama-Ohtsu N, Chandra-Prasad-Risal C, Abdelly C, Hitoshi-Sekimoto H (2011) Stress tolerance and symbiotic and phylogenic features of root nodule bacteria associated with Medicago species in different bioclimatic regions of Tunisia. Microbes Environ 26(1):36–45. doi:10.1264/jsme2.ME10138
Djennane A (1990) Les systèmes agricoles oasiens: constat de situation des zones sud des oasis algériennes. Options Méditerr Sér A Mediterr Semin 11:29–32
Drevon JJ, Abdelly C, Amarger N, Aouani EA, Aurag J, Jebara M, Gherbi H, Liuch C, Payre H, Schump O, Sifi B, Trabelsi M (2004) FABAMED interdisciplinary strategy to improve symbiotic nitrogen fixation of legumes in the Mediterranean basin. In: Serraj R (ed) Symbiotic nitrogen fixation prospects for enhanced application in tropical agriculture. Oxford and IBH Publishing, New Delhi, pp 223–232
Elbanna K, Elbadry M, Gamal-Eldin H (2009) Genotypic and phenotypic characterization of rhizobia that nodulate snap bean (Phaseolus vulgaris L.) in Egyptian soils. Syst Appl Microbiol 32(7):522–530
Elboutahiri N, Thami-Alami I, Udupa SM (2010) Phenotypic and genetic diversity in Sinorhizobium meliloti and S. medicae from drought and salt affected regions of Morocco. BMC Microbiol 10(15):1–13
Ferry M, Toutain M (1990) Concurrence et complémentarité des espèces végétales dans les oasis. Options Méditerr Sér A Mediterr Semin 11:261–270
Fitouri Dhane S, Ben Jeddi F, Zribi K, Rezgui S, Mhamdi R (2012) Effet de l’inoculation par une souche osmotolerante de Rhizobium sullae sur la croissance et la production en proteine du sulla (Sulla coronarium L.) sous déficit hydrique. J Appl Biosci 51:3642–3651
Gnat S, Wójcik M, Wdowiak-Wróbel S, Kalita M, Ptaszyńska A, Małek W (2014) Phenotypic characterization of Astragalus glycyphyllos symbionts and their phylogeny based on the 16S rDNA sequences and RFLP of 16S rRNA gene. Antonie Van Leeuwenhoek 105:1033–1048. doi:10.1007/s10482-014-0163-y
Hartmann AM, Rodiguez-Navarro DN, Temprano Vera F, Cleyet-Marel J-C, Priny G, Fernández-López M, Toro N, Moënne-Loccoz Y (2006) Nodulating Symbiotic Bacteria and Soil Quality. In: Bloem J, Hopkins D W, Benedetti A (eds) Microbiological methods for assessing soil quality. pp 231–234
Janati A (1990) Les cultures fourragères dans les oasis. Options Méditérr A1:165–167
Jebara M, Drevon JJ, Aouani ME (2001) Effects of hydroponic culture system and NaCl on interactions between common bean lines and native rhizobia from Tunisian soils. Agronomy 21:601–605
Jordan DC (1984) Rhizobiaceae. In: Krieg N R, Holt J G (Eds) Bergey’s Manual of systematic bacteriology. The Williams and Wilkins Baltimore, pp 234–242
Kersters K, Hinz KH, Hertle A, Segers P, Lievens A, Siegmann O, De Ley J (1984) Bordetella avium sp. Nov., Isolated from the respiratory tracts of Turkeys and other birds. Int J Syst Bacteriol 34(1):56–70
Koull K, Kherraze MH, Lakhdari K, Benzaoui T, Helimi S, Laouissat MS, Kherfi Y, Bougafla A, Mimouni F, Lakhdari K, Mezrag M, Benazzouz MT (2013) Eaux d’irrigation et salinisation des sols des perimetres irrigues dans la vallee de l’oued righ. J Alg Rég Arid 12:97–102
Le Houérou HN (1975) Problèmes et potentialités des terres arides de l’Afrique du Nord. Options Méditerr Sér A Mediterr Semin 26:17–32
Maâtallah J, Berraho EB, Muñoz S, Sanjuan J, Lluch C (2002) Phenotypic and molecular characterization of chickpea rhizobia isolated from different areas of morocco. J Appl Microbiol 93:531–540
Merabet C, Martens M, Mahdhi M, Zakhia F, Sy A, Le Roux C, Domergue O, Coopman R, Bekki A, Mars M, Willems A, de Lajudie P (2010) Multilocus sequence analysis of root nodule isolates from Lotus arabicus (Senegal), Lotus creticus, Argyrolobium uniflorum and Medicago sativa (Tunisia) and description of Ensifer numidicus sp. nov. and Ensifer garamanticus sp. nov. Int J Syst Evol Microbiol 60:664–674
Messar EM (1996) Le secteur phœnicicole algérien. Options méditerranéennes CIHEM and Estación Phoenix, 24
Mnasri B, Aouani ME, Mhamdi R (2007a) Nodulation and growth of common bean (Phaseolus vulgaris) under water deficiency. Soil Biol Biochem 39:1744–1750
Mnasri B, Mrabet M, Laguerre G, Aouani ME, Mhamdi R (2007b) Salt-tolerant rhizobia isolated from a Tunisian oasis that are highly effective for symbiotic N2-fixation with Phaseolus vulgaris constitute a novel biovar (bv. mediterranense) of Sinorhizobium meliloti. Arch Microbiol 187(1):79–85. doi:10.1007/s00203-006-0173-x
Murugesan S, Manoharan C, Vijayakumar R, Panneerselvam A (2010) Isolation and characterization of Agrobacterium rhizogenes from the root nodules of some leguminous plants. Int J Microbiol Res 1(3):92–96
Palma F, Tejera NA, Lluch C (2013) Nodule carbohydrate metabolism and polyols involvement in the response of Medicago Sativa to salt stress. Environ Exp Bot 85:43–49
Priefer UB, Aurag J, Boesten B, Bouhmouch I, Defezy R, Filali-Mltouf A, Miklis M, Moawad H, Mouhsin B, Prell J, Schlüter A, Senatore B (2001) Characterization of Phaseolus symbionts isolated from Mediterranean soils and analysis of genetic factors related to pH tolerance. J Biotechnol 91:223–236
Rejii M, Mahdhi M, Domínguez-Núñez JA, Mars M (2014) the phenotypic, phylogenetic and symbiotic characterization of rhizobia nodulating Lotus sp. in Tunisian arid soils. Ann Microbiol 64:355–362. doi:10.1007/s13213-013-0670-5
Riou C (1990) Bioclimatologie des oasis. Options Méditerr Sér A Mediterr Semin 11:207–208
Rome S, Fernandez MP, Brunel B, Normand P, Cleyet-Marel J-C (1996) Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol 46(4):972–980
Serraj R, Adu-Gyamfi J, Rupela O P, Drevon J J (2004) Symbiotic Nitrogen Fixation: Prospects for Enhanced Application in Tropical Agriculture. In: Serraj R (ed) International Crops Research Institue for the Semi-Arid Tropics. Oxford and IBH Publishing, pp 67–97
Shamseldin A, Moawad H, Abd El-Rahim WM, Sadowsky MJ (2013) Near-full length sequencing of 16S rDNA and RFLP indicates that Rhizobium etli is the dominant species nodulating Egyptian winter Berseem clover (Trifolium alexandrinum L.). Syst Appl Microbiol. doi:10.1016/j.syapm.2013.08.002
Shetta ND, Al-Shaharani TS, Abdel-Aal M (2011) Identification and characterization of Rhizobium associated with woody legume trees grown under Saudi Arabia condition. Am J Agric Environ Sci 10(3):410–418
Somasegaran P, Hoben HG (1985) Methods in Legume Rhizobium Technology. United States Agency for International Development (USAID), pp 69–70
Tabouche N, Achour S (2004) Etude de la qualité des eaux souterraines de la région orientale du sahara septentrional algérien. Larhyss J 03:99–113
Thami-Alami I, Elboutahiri N, Udupa SM (2010) Variability in natural population of Sinorhizobium meliloti in Morocco. Options Méditerr Sér A Mediterr Semin 92:265–269
Toutain G (1974) Conservation des sols en palmeraies sahariennes et bordières au Sahara. Options Méditerr Sér A Mediterr Semin 25:65–69
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Yan AM, Wang ET, Kan FL, Tan ZY, Sui XH, Reinhold-Hurek B, Chen WX (2000) Sinorhizobium meliloti associated with Medicago sativa and Melilotus spp. In arid saline soils in Xinjiang, China. Int J Syst Evol Microbiol 50:1887–1891
Zahran HH (1997) Diversity, adaptation and activity of the bacterial flora in saline environments. Biol Fertil Soils 25:211–223
Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63(4):968–989
Zahran HH (2001) Rhizobia from wild legumes: diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J Biotechnol 91:143–153
Zakhia F, Jeder H, Domergue O, Willems A, Cleyet-Marel J-C, Gillis M, Dreyfus B, de Lajudie P (2004) Characterisation of wild legume nodulating bacteria (LNB) in the infra-arid zone of Tunisia. Syst Appl Microbiol 27:380–395
Zhang X, Harper R, Karsist M, Lindstrom K (1991) Diversity of rhizobium bacteria isolated from the root nodules of leguminous trees. Int J Syst Bacteriol 41(1):104–113
Zribi K, Jeidi N, Mhamdi R, Aouani ME, Huguet T (2004) Diversité génétique et polymorphisme symbiotique de Sinorhizobium meliloti nodulant Medicago truncatula en sols tunisiens des régions arides. Options Méditerr Sér A Mediterr Semin 62:149–152
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baba Arbi, S., Chekireb, D., Quatrini, P. et al. Phenotypic and genotypic characterization of root nodules rhizobia of Medicago littoralis Rhode and Melilotus indicus (L.) All. growing in the Oasis of Touggourt, Oued Righ Valley, in the Algerian Sahara. Symbiosis 66, 75–87 (2015). https://doi.org/10.1007/s13199-015-0336-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-015-0336-0