Abstract
In order to characterize the diversity of plant growth promoting rhizobia associated with legumes cultivated in Assam, 32 bacterial isolates were obtained from the root nodules of Cajanus cajan L and Lablab purpureus L. The isolates were investigated for their morphological, biochemical and plant growth promoting features. The isolates showed similar morphological features such as creamy, white colonies, gram negative staining, rod shape cells but showed variation in the results of biochemical tests. In addition, the isolates produced indole-3-acetic acid, ammonia, solubilized inorganic phosphate and showed varied level of tolerance to acidic pH and high salinity. Present study revealed the presence of nitrogen fixation (nifH) gene and nodulation (nodC) gene in the selected isolates. Restriction fragment length polymorphism (RFLP) analysis of 16S rDNA of the isolates and reference strains revealed a high genetic diversity among them. Basic local alignment search tool (BLAST) analysis of 16S rDNA sequences revealed that isolates from Cajanus cajan L were phylogenetically related to Burkholderia, Mesorhizobium, Rhizobium genera and isolates from Lablab purpureus L were phylogenetically related to Bradyrhizobium genus. The present study also revealed the nodulation and plant growth promoting ability of the isolates on their host plants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrogen is an important component of most biological compounds such as amino acids, nucleic acids, and proteins. Although 78% of earth’s atmosphere is made up of nitrogen (N2), lack of usable nitrogen in soil is considered as a major limiting factor in agricultural activities. Legume-rhizobium symbiosis is an important plant microbe interaction which plays an essential role in nitrogen cycle. Rhizobia are defined as nitrogen-fixing soil bacteria capable of forming root nodules on leguminous plants in which atmospheric nitrogen is reduced to ammonia for benefit of the plant [1]. In legume-rhizobium symbiosis, rhizobia supply ammonia to the plant and thus reduce the requirement for hazardous nitrogenous fertilizers during plant growth [2]. Biological nitrogen fixation by rhizobia provides an easy way to enhance soil fertility. A number of factors affect legume-rhizobium symbiosis which includes soil pH, salinity, moisture and extreme temperatures [3]. Thus, inoculation of legume plants with superior rhizobial strains which can tolerate the edaphic factors such as low pH, high salinity, is being widely practiced to increase the plant yield. Rhizobia are also included in a group of bacteria called plant growth promoting rhizobacteria or PGPR which increases the growth of plants due to different activities. Such beneficial activities include production of plant growth substances, solubilization of inorganic phosphates, production of siderophore, ammonia, antimicrobial compounds, etc. There have been several previous reports on plant growth promoting activities of rhizobia in leguminous and non-leguminous plants [4, 5]. In addition to plant growth promotion, rhizobia can produce enzymes, polysaccharides and antibiotics [6].
Previously rhizobial diversity was studied based on their morphological and biochemical characteristics, however in recent years the availability of advanced PCR-based genotyping methods has revealed the presence of many indigenous rhizobial strains in root nodules of different leguminous plants. 16S rDNA, which codes for 16S rRNA, is considered as the most authenticated marker for bacterial identification. RFLP analysis of PCR amplified 16S rDNA is a successful technique for studying the diversity of rhizobia [7]. Direct sequencing of 16S rDNA is also considered as an important technique for characterizing rhizobia [8]. Traditionally, the symbiotic root nodulating bacteria were believed to be composed of only alpha-proteobacteria viz. Rhizobium, Mesorhizobium, Sinorhizobium, Bradyrhizobium, and Allorhizobium. However, during last decade extensive rhizobial diversity study related to beta-proteobacteria viz. Burkhoderia, Cupriavidus and gama-proteobacteria from root nodules of different leguminous and non-leguminous plants using advanced PCR based methods have been reported [9].
Cajanus cajan L (pigeon pea) and Lablab purpureus L (hyacinthbean) belong to the plant family Fabaceae. Both the plants are important legumes cultivated for seed and green manure in different parts of the world. They are also used in co-cropping and intercropping systems where they enhance soil fertility through their symbiotic abilities [10]. Similarly to other legumes, both plant species develop nodules on their roots, which become populated with specific symbiotic strain(s). Biological nitrogen fixation plays an important role in the growth of pigeon pea plant and it was reported that about 88% of the nitrogen in pigeon pea was fixed from the atmosphere by biological nitrogen fixation [11]. Evaluation of diversity of indigenous rhizobial population from the cultivated legumes of Assam can play an important role in screening novel and effective rhizobial strains which can significantly contribute in agriculture.
The aim of the present study is isolation and characterization of indigenous rhizobia associated with root nodules of Cajanus cajan L and Lablab purpureus L plants collected from different locations of Assam and to assess their diversity based on different phenotypic and genotypic methods. The present study also aims to assess the plant growth promoting, stress tolerant and nodulation abilities of the isolates in order to select potential isolate which can be used as effective inoculants/biofertilizer for their respective host plants.
Material and Methods
Isolation of Bacteria
Fresh root nodules of C. cajan and L. purpureus plants were collected and sterilized with 75% ethanol for 3 min and 0.1% HgCl2 for 5 min. Nodules were then rinsed, crushed in sterile distilled water, streaked on yeast extract manitol agar (YEMA) plates and incubated at 28 °C for 3–5 days [12]. In order to compare and confirm the phenotypic and genotypic traits of the isolates, Rhizobium leguminosarum MTCC-99, Mesorhizobium thiogangeticum MTCC-7001 and Bradyrhizobium japonicum MTCC-120 were obtained as reference strains from the Institute of Microbial Technology (IMTECH), Chandigarh, India.
Phenotypic and Biochemical Characterization
All the isolates were investigated for their phenotypic and biochemical characteristics. For phenotypic study, the isolates were inoculated on YEMA plate and after three days of incubation, colony morphology of the isolates including size, color, shape were observed and gram staining was performed [1]. The isolates were also investigated for eight different biochemical tests, ketolactose, catalase, oxidase, nitrate reduction, starch hydrolysis, urease, citrate utilization and gelatin liquefaction as per Bergey’s manual of Determinative Bacteriology by following standard procedure [13].
Salt and pH Tolerance
The isolates were streaked and incubated on three different YEMA plates adjusted with three different concentration of NaCl (1.0, 2.0 and 3.0%) and three different pH (4.0, 5.0, 6.0) to determine their capability to grow in salt stress and acidic pH [14, 15]. Growth of isolates on standard YEMA media (pH-7.0; NaCl-0.1%) was used as control.
Plant Growth Promoting Activities
The ability of the isolates to solubilize inorganic phosphate was detected by spotting the isolates on Pikovskaya’s agar plates [16].The plates were observed for the formation of halo zone around the spot of inoculation. The diameter of solubilization zone is compared with colony diameter and the efficiency was expressed in terms of phosphate solubilization efficiency% or PSE% (solubilization diameter/colony diameter × 100). The isolates were also tested for their production of indole-3-acetic acid (IAA) on YEM broth medium [5]. To observe IAA production, isolates were incubated on YEM broth medium supplemented with l-tryptophan (0.1%) for 24 h. Supernatant of the isolates were collected and transferred separately to a fresh tube to which 4 ml of Salkowski(‘s) reagent (1 ml of 0.5 mM FeCl3 in 35% HClO4) was added. Mixtures were incubated at room temperature for 25 min and the production of IAA was observed spectrophotometrically by measuring the O.D. at 540 nm. The amount of IAA produced by the isolates was determined by comparing the data with a standard graph prepared by measuring the O.D. with different concentrations (10–100 µg/ml) of IAA [17]. For determining the ability of isolates to produce ammonia, the isolates were inoculated in peptone broth media and incubated for 48 h. Nessler(‘s) reagent was added to the grown bacterial culture and change in colour was observed [18].
Nitrogen Fixation (nifH) and Nodulation (nodC) Gene Amplification
Genomic DNA extraction of the isolates was performed using the standard Phenol–chloroform extraction procedure [19]. Primers used for nifH gene amplification reaction were zehrf- 5′TGCGACCCAAAAGCAGA3′ and zehr-5′AAAGCCATCATCTCACC3′ [20]. Amplification was performed with a total volume of 50 μl containing genomic DNA (30 ng), dNTP’s (2.5 mM), primers (10 pmol), PCR buffer (10×), Taq polymerase (1.5 U) and nuclease free water. The reaction conditions were, initial denaturation at 95 °C for 5 min, denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 45 s (30 cycles) and then final extension at 72 °C for 7 min. PCR amplification of nodC gene was carried out with primers nodC1- 5′GCCATAGTGGCAACCGTCGT3′ and nodC2- 5′TCACTCGCCGCTGCAAGTC3′ [5]. Composition of PCR reaction mixture and reaction condition for amplification of nodC gene is similar to nifH gene amplification except the annealing temperature (56 °C for 30 s). Amplified products of nifH and nodC genes were resolved on 1.5% agarose gel and documented in gel documentation system.
PCR–RFLP Analysis of 16S rDNA
Primers used for 16S rDNA amplification were 27f- 5′AGAGTTTGATCATGGCTCAG3′ and 1492r- 5′ACGGATACCTTGTTTACGACTT3′ [21]. Composition of PCR reaction mixture for 16S rDNA amplification is similar to nifH gene amplification. Amplifications were carried out with the following temperature profile, initial denaturation at 95 °C for 5 min, denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 2 min (30 cycles) and then final extension at 72 °C for 7 min. PCR amplified products were digested with restriction enzyme AluI [7] and the RFLP patterns obtained from the digestion were observed on 2% agarose gel. A simple matching coefficient was calculated to construct a similarity matrix and the UPGMA (Unweighted Pair Grouping with Mathematic Average) algorithm was used to perform hierarchical cluster analysis and to construct a dendrogram by using NTSYS-pc package [22].
16S rDNA Sequencing and Analysis
The PCR amplified 16S rDNA region of the isolates was sequenced with 3500 Genetic Analyzer at Genomics and DNA Bar-coding Lab, Department of Biotechnology, Assam University. The quality of the sequences was assessed and edited using BioEdit program [23]. NCBI-BLAST analysis was carried out with the 16S rDNA sequences of the isolates to determine their closest relatives and to predict their identity.
Assessment of Plant Growth
Seeds of C. cajan and L. purpureus were surface sterilized by gently shaking it in 70% ethanol for 2 min and HgCl2 for 3 min. The bacterial isolates were grown in YEM broth medium at 28 °C for 3–5 days (108 CFU/ml) and inoculated on the emerging seedlings of their original hosts. Seeds were sown in plastic pot containing vermiculite-quartz sand mixture and moistened with nitrogen free nutrient solution at regular intervals. The treatment was carried out in triplicate (three plants per pot) and nine plants were considered for each isolate. Uninoculated plants were kept as control. The plants were harvested after 45 days of planting and their vegetative parameters were measured. The experiment was performed in a completely randomized design with three replications. The parameters evaluated were presence and absence of nodules, shoot and root height, shoot and root dry weight. Statistical Analysis of Variance (ANOVA) was carried out to identify significant difference in each vegetative parameter between treated and control at 5% significance level by using SPSS 16.0 software.
Results and Discussion
Phenotypic and Biochemical Characterization
In the present study, a total of 32 bacterial isolates were obtained from the root nodules of the two plant species collected from 6 different sites of Assam. The isolates and reference strains showed white, creamy, raised colonies on YEMA media and colony size of the isolates were 1–3 mm in diameter after 4–5 days of incubation. Microscopic examination revealed that most of the isolates were Gram negative and rod shaped. Similarity phenotypic features of isolates and reference strains confirm the close relationship among them. However, the isolates showed variation in their biochemical characteristics. Most of the isolates showed positive result for catalase, oxidase; negative results for Ketolactose, starch, gelatin and variable results for nitrate, urease and citrate tests (Table 1). Based on the results of morphological, microscopic, biochemical features and their comparison with reference strains 20 isolates (10 isolates from each plant) were selected for further study. Out of 20 selected isolates, 9 isolates showed similarity with R. leguminosarum MTCC-99 and M. thiogangeticum MTCC-7001, 7 isolates showed similarity with B. japonicum MTCC-120 and 4 isolates showed no similarity with the reference strains in their biochemical features.
Salt and pH Tolerance
Soil pH and salinity play an important role in limiting the growth of microorganisms. Isolation and characterization of indigenous rhizobial strains which can tolerate acidic pH and high salinity can play an important role in selection of superior rhizobial strain. Isolates from both plant species grew well at pH 6, showed variable growth at pH 5 but were unable to grow at pH 4 (Table 2). Similar to the present study, Choudhury et al. [24] reported the presence of acidic pH tolerant Rhizobium strains from different legumes cultivated in lower Brahmaputra valley of Assam. In salinity tolerance test, most of the isolates grew well at 1 and 2% NaCl but were unable to grow at 3% NaCl (Table 2).
Plant Growth Promoting Activities
Plant growth promoting microorganisms play a key role in the growth and nutrition of plants by performing different direct and indirect activities [25]. In the present study, the isolates were investigated for their ability to produce IAA, ammonia and solubilization of inorganic phosphate. IAA is an important plant hormone and plays a significant role in plant growth. Production of IAA by Rhizobia sp isolated from C. cajan and L. purpureus has been previously reported by many researchers [5]. The present study revealed that all the isolates except (KCC3 and HCC2) were capable of producing IAA, however the amount of IAA produced varied greatly among the isolates (Table 2). Among the 20 isolates investigated, nine isolates produced higher amount of IAA than the reference strain R. leguminosarum MTCC-99 and the isolate SDL1 produced maximum amount of IAA (77 µg/ml).
All the isolates and reference strains were capable of phosphate solubilization which is confirmed by the formation of solubilization zone around the spot of inoculation of isolates on Pikovskaya’s agar plates. PSE of the isolates was determined by comparing the colony diameter of the isolates with the diameter of the solubilization zone. Seven isolates showed higher PSE than the reference strain B. japonicum MTCC-120 (Table 2). The maximum PSE was shown by IDL1 (181%) isolated from L. purpureus. The present study also revealed that 15 isolates were capable of producing ammonia. Development of faint yellow to dark brown color on bacterial inoculated peptone broth indicated the production of ammonia.
Nitrogen Fixation (nifH) and Nodulation (nodC) Gene Amplification
PCR amplification of nifH and nodC genes were carried out for determining nitrogen fixation and nodulation potential of the isolates. nifH Gene encodes Fe-protein subunit of nitrogenase enzyme, which is an important enzyme of biological nitrogen fixation pathway. nodC Gene encodes N-acetylglucosaminyltransferase enzyme that synthesizes the N-acetyl glucosamine backbone of the nod factor. The presence of nifH and nodC genes in rhizobial isolates of C. cajan and L. purpureus was previously reported by Dubey et al. [5] and Chang et al. [26], respectively. Results of the current study revealed the presence of nifH and nodC genes in 6 isolates of C. cajan and 7 isolates of L. purpureus (Table 2). PCR amplification reaction using nifH and nodC primers generated specific bands at 360 and 500 bp, confirming the presence of nifH and nodC gene in the isolates, respectively. In both the amplification reactions, reference strain R. leguminosarum MTCC-99 was used as a positive control.
PCR–RFLP Analysis of 16S rDNA
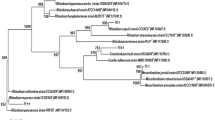
PCR–RFLP analysis of 16S rDNA is a useful technique for studying genetic diversity of rhizobia. Characterization of rhizobial strains based on 16S rDNA PCR–RFLP analysis was previously reported by many researchers. Costa et al. [27] characterized rhizobia from C. cajan based on PCR–RFLP analysis by employing three restriction enzymes DdeI, MspI, and HinfI. Likewise Chang et al. [26] employed PCR–RFLP technique for characterizing rhizobial isolates of L. purpureus. Previously, Dubey et al. [5] reported that AluI was the most efficient restriction enzyme for discriminating rhizobial isolates, so instead of choosing multiple restriction enzymes only AluI enzyme was used for restriction analysis. On the basis of restriction pattern of 16S rDNA gene an UPGMA dendrogram is generated for isolates of both the plant species. Dendrogram derived from the PCR–RFLP analysis grouped the isolates of C. cajan into 3 different clusters (Fig. 1). Cluster I was comprised of 4 isolates with reference strain B. japonicum MTCC-120. Cluster II was comprised of 6 isolates with reference strain R. leguminosarum MTCC-99. Reference strain M. thiogangeticum MTCC-7001 makes a separate cluster III. Similarly dendrogram derived from the PCR–RFLP analysis grouped the isolates of L. purpureus into 2 different clusters (Fig. 2). Cluster I was comprised of 7 isolates with reference strains R. leguminosarum MTCC-99 and B. japonicum MTCC-120. Cluster II was comprised of 3 isolates with reference strain M. thiogangeticum MTCC-7001.
Sequence Analysis of 16S rDNA
Many previous reports suggested the presence of fast and slow growing rhizobia in the root nodules of C. cajan. Ramsubhag et al. [28] reported the isolation of slow growing B. elkanii from C. cajan root nodules. Likewise, Dubey et al. [5] reported the presence of plant growth promoting strains of Sinorhizobium freedi from root nodules of C. cajan. In the present study, four isolates of C. cajan (ICC1, ICC3, DCC2 and SICC1) representing the three clusters of PCR–RFLP analysis were selected for 16S rDNA gene sequencing. In BLAST analysis, the isolate DCC2 showed closest similarity with Rhizobium mayense strain CCGE526. Previously Rhizobium mayense was reported from Calliandra grandiflora root nodules growing in Mexico [29]. Likewise, BLAST result of ICC1 showed its closest similarity with Mesorhizobium loti strain MAFF 303099 which is a common legume symbiont (Table 3).
The present study also revealed that 2 isolates of C. cajan belong to Burkholderia genus. The genus Burkholderia is one of the largest groups of species within the class Betaproteobacteria and distributed in diverse habitats such as plant rhizosphere, in plant root as endophytes, in root nodules as symbiont or as an opportunistic pathogen on plants and humans. In the present study, isolate ICC3 from Cachar and SICC1 from Hailakandi showed close similarity with Burkholderia cenocepacia strain AU 1054 and Burkholderia anthina strain W92B, respectively, both of which belongs to a group called Burkholderia cepacia complex. Although Burkholderia cepacia complex was considered as human pathogen but it also includes free-living soil bacteria and nitrogen fixing symbiotic bacteria [30]. Recently, Lu et al. [31] isolated strains of Burkholderia stabilis and Burkholderia cepacia both of which are included in Burkholderia cepacia complex from root nodules of Dalbergia odorifera growing in Hainan Island of southern China. ICC3 and SICC1 showed no amplification of nifH and nodC genes, so the presence of Burkholderia cenocepacia and Burkholderia anthina in the nodules of C. cajan may likely to be considered as an opportunistic interaction by the microorganisms. The acquired 16S rDNA sequences of the four C. cajan isolates were deposited in GenBank database and the strain were named as Mesorhizobium loti ICC1, Burkholderia cenocepacia ICC3, Rhizobium mayense DCC2 and Burkholderia anthina SICC1.
Similarly, 16S rDNA gene sequencing of 4 L. purpureus isolates (HIDL1, SDL1, SIDL1, DDL2) was carried out which represents the two PCR–RFLP clusters. In contrast to different PCR–RFLP patterns, all the L. purpureus isolates belong to Bradyrhizobium genus. In BLAST analysis, the isolates HIDL1, SDL1, SIDL1 and DDL2 showed closest similarity with Bradyrhizobium elkanii, Bradyrhizobium lablabi, Bradyrhizobium pachyrhizi and Bradyrhizobium elkanii respectively. Similar to the present study, symbiotic bacteria belonging to the genera Bradyrhizobium was reported as the most common symbiont of L. purpureus in many previous studies [32]. The result of the current study is in agreement with the findings of Chang et al. [26] who reported the presence of Bradyrhizobium elkanii, Bradyrhizobium lablabi, Bradyrhizobium pachyrhizi, Bradyrhizobium yuanmingense, Bradyrhizobium jicamae as a symbiont of L. purpureus. However, the presence of bacterial isolates from Rhizobium and Mesorhizobium genera in the root nodules of L. purpureus has also been reported in previous studies [27]. The acquired 16S rDNA sequences of the four L. purpureus isolates were also deposited to GenBank database and the strain were named as Bradyrhizobium elkanii HIDL1, Bradyrhizobium lablabi SDL1, Bradyrhizobium pachyrhizi SIDL1 and Bradyrhizobium elkanii DDL2.
Assessment of Plant Growth
Many previous reports suggested that inoculation of leguminous and non-leguminous plants with rhizobia can significantly increase the growth and activity of plants [33]. Recently, Silva et al. [34, 35] reported that inoculation of Capsicum annuum (L) with Rhizobium strains have significant effect on the metabolic activities of the plant. In the present study, a pot experiment was carried out to evaluate the effect of isolates on C. cajan and L. purpureus plants. Two isolates of C. cajan, ICC3 and SICC1 were excluded from the study as they were known as human pathogens. The results showed that the isolates significantly increased the vegetative parameters of C. cajan and L. purpureus indicating that the indigenous inoculants improve the growth of their host plants. The isolates ICC1, DCC2 and reference strain R. leguminosarum MTCC-99 successfully induced root nodules on their host plant C. cajan (Table 4). Inoculation of DCC2 to the host plant significantly increased shoot length and shoot dry weight as compared to the un-inoculated control. However, there is no significant change in root height and root dry weight after treating with DCC2. No significant change was observed in vegetative parameters of C. cajan when treated with ICC1. Reference strain R. leguminosarum MTCC-99 significantly increased shoot length, root length and shoot dry weight as compared to the un-inoculated control.
The four isolates of L. purpureus also induced root nodules on their host plant. Inoculation of HIDL1 significantly increases the shoot dry weight and inoculation of DDL2 significantly increased root length of L. purpureus when compared with un-inoculated control (Table 5). However, the other two isolates SDL1 and SIDL1 have no significant effect on the vegetative parameters of the host plant when compared with un-inoculated controls. There is no clear correlation between the PGPR abilities and the effect on vegetative parameter which is evident by the fact that the highest producer of IAA showed no significant effect on the vegetative parameter of its host plant. Reference strain R. leguminosarum MTCC-99 significantly increased root length and root dry weight of L. purpureus plant as compared to the un-inoculated control.
Conclusion
Both C. cajan and L. purpureus are important vegetative legume plants widely cultivated and consumed in different parts of Assam. They are also used in co-cropping and intercropping systems for enhancing soil fertility through their symbiotic association with rhizobia. The present study revealed the presence of plant growth promoting and stress tolerant rhizobial strains in the root nodules of C. cajan and L. purpureus plants growing in Assam. Phenotypic and genotypic characterization of isolated strains revealed a considerable diversity among them. 16S rDNA sequence analysis of isolates revealed that rhizobial strains isolated from C. cajan belong to three different genera: Rhizobium (R. mayense), Mesorhizobium (M. loti) and Burkholderia (B. cenocepacia, B. anthina). Interestingly, the two Burkholderia strains isolated from C. cajan were pathogenic in nature and rarely identified from legume plants. Rhizobial strains isolated from L. purpureus plant belong to single genus Bradyrhizobium (B. elkanii, B. lablabi, B. pachyrhizi and B. elkanii). The isolates have the ability to produce IAA, solubilzation of inorganic phosphate and production of ammonia. Present study also revealed that few isolates can tolerate low pH (pH-5) and high salinity (2% NaCl). Successful amplification of nifH and nodC genes confirms that the native isolates can synthesize nitrogenase enzyme for fixing atmospheric nitrogen and nod factors for forming an effective symbiosis. Nodulation study revealed that the selected isolates were capable of forming effective nodules on their host plants C. cajan and L. purpureus. In addition, the isolates significantly increased the vegetative parameters viz. shoot dry weight, and root dry weight of their respective host plants as compared to un-inoculated controls which indicated that the native isolates could be used as inoculants to improve the yield of legume plants. In future, the isolates could be subjected to further greenhouse and field trials to ascertain their stability under different environmental conditions.
References
Somasegaran P, Hoben HJ (1994) Handbook for rhizobia. Springer, New York
Lodwig EM, Hosie AHF, Bourdes A, Findlay K, Allaway D (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726
Brockwell J, Bottomly PJ, Thies JE (1995) Manipulation of rhizobia microflora for improving legume productivity and soil fertility: a critical assessment. Plant Soil 174:143–150
Deshwal VK, Dubey RC, Maheshwari DK (2003) Isolation of plant growth promoting strains of Bradyrhizobium (Arachis sp.) with biocontrol potential against Macrophomina causing charcoal rot of peanut. Curr Sci 84:443–448
Dubey RC, Maheshwari DK, Kumar H, Choure K (2010) Assessment of diversity and plant growth promoting attributes of rhizobia isolated from Cajanus cajan L. Afr J Biotechnol 9:8619–8629
Deshwal VK, Pandey P, Kang SC, Maheshwari DK (2003) Rhizobia as a biological control agent against soil borne plant pathogenic fungi. Indian J Exp Biol 41:1160–1164
Laguerre G, Allard MR, Revoy F, Amarger N (1994) Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol 60:56–63
Young JP, Downer HL, Eardly BD (1991) Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol 173:2271–2277
Chen WM, deFaria SM, Straliotto Pitard RM, Simoes JL, Chou YJ (2005) Proof that Burkholderia forms effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl Environ Microbiol 71:7461–7471
Peoples MB, Herridge DF, Ladha JK (1995) Biological nitrogen fixation: an efficient source of nitrogen for sustainable agricultural production. Plant Soil 174:3–28
Rao JVDKK, Thompson JA, Sastry PVSS, Giller KE, Day JM (1987) Measurement of N2-fixation in field-grown pigeonpea [Cajanus cajan (L.) Mill sp.] using 15N-labelled fertilizer. Plant Soil 101:107–113
Vincent JM (1970) A manual for the practical study of the root nodule bacteria. Blackwell Scientific, Oxford
Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) In: Bergey’s manual of determinative bacteriology, 9th edn. Williams and Wilkins Press, Baltimore, pp 71–101
Romdhane S, Nasr H, Samba-Mbaye R, Neyra M, Ghorbel MH (2006) Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J Appl Microbiol 100:436–445
Kucuk C, Kivanc M, Kinaci E (2006) Characterization of Rhizobium sp. isolated from bean. Turk J Biol 30:127–132
Pikovskaya RI (1948) Mobilization of phosphorous in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:362–370
Gordon SA, Weber RP (1951) Colorimeteric estimation of indole acetic acid. Plant Physiol 26:192–195
Kumar A, Devi S, Patil S, Payal C, Negi S (2012) Isolation, screening and characterization of bacteria from rhizospheric soils for different plant growth promotion (PGP) activities: an in vitro study. Recent Res Sci Techol 4:01–05
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York
Zehr JP, McReynolds LA (1989) Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55:2522–2526
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Rohlf FJ (1998) NTSYS-PC: numerical taxonomy and multivariate analysis system version 2.02f. Exeter Software. New York, USA
Hall TA (1999) BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Choudhury B, Azad P, Kalita MC (2010) Variability in symbiotic effectiveness of native rhizobia in acid stress. Curr Microbiol 61:85–91
Flores-Felix JD, Marcos-Garcia M, Silva LR, Menendez E, Martinez-Molina E, Mateos PF, Velazquez E, Garcia-Fraile P, Andrade P, Rivas R (2015) Rhizobium as plant probiotic for strawberry production under microcosm conditions. Symbiosis 67:25–32
Chang YL, Wang ET, Sui XH, Zhang XX, Chen WX (2011) Molecular diversity and phylogeny of rhizobia associated with Lablab purpureus (Linn.) grown in Southern China. Syst Appl Microbiol 34:276–284
Costa FM, Schiavo JA, Brasil MS, Leite J, Xavier GR, Fernandes PI (2014) Phenotypic and molecular fingerprinting of fast growing rhizobia of field-grown pigeonpea from the eastern edge of the Brazilian Pantanal. Genet Mol Res 13:469–482
Ramsubhag A, Umaharan P, Donawa A (2002) Partial 16S rRNA gene sequence diversity and numerical taxonomy of slow growing pigeonpea (Cajanus cajan L. Mill sp.) nodulating rhizobia. FEMS Microbiol Lett 216:139–144
Rincon-Rosales R, Villalobos-Escobedo JM, Rogel MA, Martinez J, Ormeno-Orrillo E, Martinez-Romero E (2013) Rhizobium calliandrae sp. nov., Rhizobium mayense sp. nov. and Rhizobium jaguaris sp. nov., rhizobial species nodulating the medicinal legume Calliandra grandiflora. Int J Syst Evol Microbiol 63:3423–3429
Miller SCM, LiPuma JJ, Parke JL (2002) Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl Environ Microbiol 68:3750–3758
Lu JK, He XH, Huang LB, Kang LH, Xu DP (2012) Two Burkholderia strains from nodules of Dalbergia odorifera T. Chen in Hainan Island, southern China. New For 43:397–409
Trinick MJ (1980) Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J Appl Microbiol 49:39–53
Couto C, Silva LR, Valentao P, Velázquez E, Peix A, Andrade PB (2011) Effects induced by the nodulation with Bradyrhizobium japonicum on Glycine max (soybean) metabolism and antioxidant potential. Food Chem 127:1487–1495
Silva LR, Azevedo J, Pereira MJ, Carro L, Velazquez E, Peix A, Valentao P, Andrade PB (2014) Inoculation of the nonlegume Capsicum annuum (L.) with Rhizobium Strains. 1. Effect on bioactive compounds, antioxidant activity, and fruit ripeness. J Agric Food Chem 62:557–564
Silva LR, Azevedo J, Pereira MJ, Carro L, Velazquez E, Peix A, Valentao P, Andrade PB (2014) Inoculation of the nonlegume Capsicum annuum (L.) with Rhizobium Strains. 2. Changes in sterols, triterpenes, fatty acids, and volatile compounds. J Agric Food Chem 62:565–573
Acknowledgements
The authors are thankful to Department of Biotechnology (DBT), Government of India for financial support under Major Research Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singha, B., Mazumder, P.B. & Pandey, P. Characterization of Plant Growth Promoting Rhizobia from Root Nodule of Two Legume Species Cultivated in Assam, India. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 88, 1007–1016 (2018). https://doi.org/10.1007/s40011-016-0836-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-016-0836-6