Abstract
Type 2 diabetes (T2D) is a complicated systemic disease, and the exact pathogenetic molecular mechanism is unclear. Distinct histone modifications regulate gene expression in certain diseases, but little is known about histone epigenetics in diabetes. In the current study, C57BL/6 J mice were used to build T2D model, then treated with exendin-4 (10 μg/kg). Histone H3K9 and H3K23 acetylation, H3K4 monomethylation and H3K9 dimethylation were explored by Western blotting of liver histone extracts. Real-time polymerase chain reaction (PCR) was used to examine expression levels of diabetes-related genes, while chromatin immunoprecipitation (ChIP) was applied to analyze H3 and H3K9 acetylation, H3K4 monomethylation, and H3K9 dimethylation in the promoter of facilitated glucose transporter member 2 (Glut2) gene. The results showed that liver’s total H3K4 monomethylation and H3K9 dimethylation was increased in diabetic mice, which was abrogated with the treatment of exendin-4. In contrast, H3K9 and H3K23 acetylation were reduced in diabetic mice, while exendin-4 only alleviated the reduction of H3K9 acetylation. Our data indicated that the progression of type 2 diabetes mellitus (T2D) is associated with global liver histone H3K9 and H3K23 acetylation, H3K4 monomethylation, and H3K9 dimethylation. Exploiting exact histone modify enzyme inhibitors, which may represent a novel strategy to prevent T2D.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus (DM) is a chronic systemic metabolic disease, characterized by continuous high blood glucose, due to the deficiency of insulin or the low sensibility of target tissue to insulin [12]. A large portion of the patients is type 2 diabetes mellitus (T2D), and its pathogenic mechanisms still remain to be elucidated. For nearly a decade, scientists have studied the pathogenesis of T2D in-depth from different perspectives such as genetic predisposition [30], insulin resistance [22], signal transduction [1], the absorption and transportation of glucose [14, 29], the regeneration and differentiation of islet cells [9, 7, 5], etc. Recently, a large amount of evidence indicates that the interaction between genes and environmental factors plays an important role in diabetes and its complications, so change of epigenetic pattern may be an important influencing factor of diabetes. For instance, nutrition and functional foods have been demonstrated to affect epigenetic phenotypes by modulating DNA methylation enzymes or histone modifications [20, 26], indicating that epigenetic modification may be the bridge connecting genetic susceptibility with environmental exposures during the development of T2D.
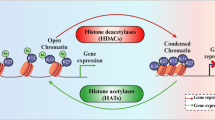
Histone modification is a key component of epigenetic modulation, which has a very important dynamic transcriptional regulatory function [28]. The accessibility of chromatin transcription factor binding sites is determined by the position and compaction of histones. Whether the nucleosomes are packed more or less tightly is determined by the activity of a number of enzymes, which regulate the covalent modification of distinct amino acids in histones such as acetylation, methylation, ubiquitination, and phosphorylation [3, 19]. Histone modifications affect the expression of target genes associated with disease progression. Histone H3 acetylation at lysines 9, 14, 18, and 23 activates chromatin for transcription factor binding, while histone H3 dimethylation or trimethylation at lysines 9 and 27 rather silences chromatin by inhibiting transcription factors accessibility [36].
Although an increasing number of reports describe the association of distinct histone modifications with autoimmunity [33] and carcinogenesis [34], little is known about its role in diabetes. Abnormal glucose metabolism is one of the main pathological factors for diabetes. The liver is the main organ that regulates blood sugar [13], where Glut2 is the carrier to assist the transmembrane transport of glucose [31]. Glucokinase (Gck) is a key enzyme that catalyzes the phosphorylation of glucose into a G-6-P, a prerequisite for glycogen synthesis. Pyruvate kinase (Pklr) is the last one of three key rate-limiting enzymes during glycolysis. Pklr messenger RNA (mRNA) level was decreased in the condition of diabetes, which was significantly increased after the intake of food in high sugar or insulin action [8]. Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha (Ppargc1a), plays an important regulatory role in gluconeogenesis, which affects the development of diabetes. It was reported that in Ppargc1a knockout mice, glycogen generation was lower significantly [6]. Insulin receptor (Insr) specifically allows the bounding with insulin or pro-insulin. It had been confirmed that maybe mutation, consequently, reduced the number of insulin receptors, thereby causing insulin resistance [18].
In this study, we hypothesized that T2D is associated with distinct histone H3 modifications, and preventing the progression of the disease could potentially revert abnormal histone modification patterns. To validate that, Western blotting of liver histone extracts obtained from mice was performed to evaluate histone H3K9 and H3K23 acetylation, H3K4 methylation, and H3K9 dimethylation, and the assumption was supported by our results.
Materials and methods
Animal studies
Thirty 3-week-old C57BL/6 J male mice (about 12 g body weight) were purchased from Laboratory Animal Center of the Academy of Military Medical Sciences of China (Animal license number: SCXK-2007-004). The experiments were approved and supervised by the local ethics committee. Mice were housed in 25 ± 3 °C, free to tap water and food, and 12 h light and dark cycle. After adaptive training for a week, 20 mice were fed high-fat and high-sucrose food, named HFS group, while the rest were fed normal food as the control group (n = 10). A month later, HFS mice were intraperitoneally injected with 100 mg/kg streptozotocin (STZ, Sigma, USA), together with a high-fat and high-sucrose diet to induce diabetes, while the control mice were given equal volume of citrate buffer. Fasting blood glucose (FBG) was measured after 1 month, 18 mice of FBG > 11.1 mmol/l were considered as type 2 diabetic mice, of which half the number were randomly picked to accept intraperitoneal injection of exendin-4 (10 μg/kg) once daily as Ex-4 group (n = 9). Exendin-4, as established diabetic inhibitor, reduces blood sugar by inhibition of apoptosis of islet β cell and promotion of β cell proliferation. The other diabetic mice were given equal volume of saline named T2D group (n = 9). After a month, the FBG level, fasting insulin (FINS) and triglycerides (TGs) levels, food and drink levels, and body weight were examined. HOMA-IR was calculated by formula: (FBG, mmol/l) × (FINS, mIU/l)/22.5.
Histone extraction and immunoblot
From each mouse, part of the liver was dissected, and histone extraction and immunoblot were performed as previously described [32]. Briefly, livers were homogenized and processed for nucleus isolation. Histone was extracted using epigenetic total histone extraction kit (Epigentek, USA). Protein was quantified using a BCA kit (Solarbio, China). Equal amounts of proteins (60 μg) from each sample were loaded and separated on a tricine-SDS-polyacrylamide gel and transferred to a 0.2 μm PVDF membrane (DingGuo, China). Antibodies against acetylated histone H3 at lysines 9 and 23 (rabbit, 1:1000 dilution), dimethylated histone H3 at lysine 9 (rabbit, 1:1000), and monomethylated histone H3 at lysine 4 (rabbit, 1:1000), and horseradish peroxidase-conjugated anti-rabbit secondary antibody were all purchased from Cell Signaling Technology (Danvers, MA, USA). Enhanced chemiluminescence (Tiangen, China) was employed for developing blots.
Real-time PCR
The RNA was extracted from the liver using Trizol (Invitrogen), and reverse-transcribed to complementary DNA (cDNA) using cn-transcriptor first-strand cDNA synthesis kit (Roche, Basel, Switzerland). Keeping house gene β-actin was used as internal reference. The primers for the target gene cross an intron so that contamination with genomic DNA can be detected. All primers were listed in Table 1. Each reaction mixture with a total volume of 25 μl contained 20 ng of cDNA, 12.5 μl of SYBR Premix Ex Taq (2×, Roche), 10 μM of each primer. PCR amplification conditions were as follows: 30 s at 95 °C followed by 40 cycles of 95 °C for 5 s, 60 °C for 20 s, and 72 °C for 20 s, then 72 °C for 2 min, melting 60–94 °C. The relative gene expression level was calculated by the 2−∆∆Ct method [21]. Gene expression was normalized to β-actin and presented as relative expression compared to a calibrator sample.
ChIP Assay and PCR
Chromatin immunoprecipitation (ChIP) was performed using the EpiQuik™ tissue chromatin immunoprecipitation kit (Epigentek) following the manufacturer’s instructions. Briefly, the liver tissue was mixed with formaldehyde at a final concentration of 1.0 % for 15 min at room temperature to cross-link protein to DNA. DNA cross-linked with protein was sonicated into fragments of 200–1,000 bp. One tenth of the sample was set aside as an input control, and the rest was added into the microwell immobilized with affinity antibodies equably. The antibodies used for immunoprecipitation were anti-acetyl-histone H3, histone H3-acetyl K9, histone H3-dimethy K9, and histone H3-methy K4 (Cell Signal Technology). All conventional PCRs were performed on a BioRad Gradient 96 (BioRad, USA) in a 20 μl reaction volume containing 1 μl template DNA, 2 μl 10 × Buffer, 0.3 μl dNTP (10 mM), 0.5 μl primers (10 μM), 0.2 μl Taq DNA polymerase (5 U/μl). The primers used were listed in Table 2. The PCR conditions were 10 min at 95 °C followed by 40 cycles of 1 min at 95 °C, 30 s at 50–55 °C, and 30 s at 72 °C. The PCR products obtained with the immunoprecipitated DNA were normalized to that of total input DNA. The bands were quantified using Quantity One imaging software (Bio-Rad) as a function of both band size and band intensity (intensity/mm2). The PCR products had the expected molecular size, and their identity was confirmed by sequencing.
Histopathological evaluation
From each mouse, parts of the liver were fixed in 10 % formalin in PBS for more than 24 h and dehydrated by gradient alcohol. The tissue was then embedded in paraffin and cut into slices with the section thickness of 5 μm. The sections were stained by HE staining with hematoxylin for 1 min, eosin for 7 min. E600 Nikon eclipse was applied to observe at high magnification microscopy and take photos.
Statistical analysis
Values are expressed as means ± SEM from at least three independent experiments. Student’s t test was used to determine differences between two groups. In all analyses, p values < 0.05 were considered as statistical significance.
Results
Therapeutic effects of exendin-4 on reducing plasma glucose in type 2 diabetic mice
After treatment with exendin-4 1 month later, the FBG was about 10.15 ± 0.18 mmol/l for Ex-4 group after intraperitoneal injection of exendin-4, lower than (p < 0.05) that of Ex-4 group before treatment which was 14.76 ± 1.74 mmol/l. On the other hand, FBG in T2D group treated with saline continued to rise to 18.00 ± 1.73 mmol/l, which was 21.8 % higher (p < 0.05) at the last day, and nearly three times that of normal group (4.56 ± 0.51 mmol/l, p < 0.01). As expected, the FBG of the control group exhibited no significant change over all the time. The FINS was 5.06 ± 0.63, 2.46 ± 0.38, and 3.78 ± 0.13 mIU/l in control, T2D, and Ex-4 groups. The FINS of diabetic mice was significantly lower than normal mice (p < 0.01), which was increased after exendin-4 treatment (p < 0.01). The body weight of T2D group and Ex-4 group showed a trend of increase to 24.03 ± 1.75 and 24.38 ± 0.56 g, respectively, higher (5 % and 3 %, respectively, p < 0.05) compared with that before treatment. The food intake of Ex-4 group was lower compared to that before treatment, by contrast, the T2D group had been increased to 3.85 ± 0.28 g, which was higher than that before treatment (13 %, p < 0.05). The drink level of T2D group continued to increase to 15.28 ± 3.86 ml (10 %, p < 0.01); however, the normal group and Ex-4 group presented no significantly change during the treatment. The TGs of control group, T2D group, and Ex-4 group mice were 1.70 ± 0.13, 3.16 ± 0.43, and 2.29 ± 0.46 mmol/l, respectively, which were increased in diabetic mice (p < 0.01) and decreased after treatment with exendin-4 (p < 0.05). After calculating, the HOMA-IR values in different groups were 1.03, 1.97 and 1.71, respectively. All data were shown in Table 3.
Increased global histone H3K9 dimethylation and H3K4 methylation and decreased histone H3K9 acetylation and H3K23 acetylation in liver of diabetic mice
The status of histone modifications was examined by Western-blotting approach using four histone antibodies: anti-acetyl histone H3 lysine 9 (H3K9ac), anti-acetyl histone H3 lysine 23 (H3K23ac), anti-dimethyl histone H3 lysine 9 (H3K9me2), and anti-methyl histone H3 lysine 4 (H3K4me). The results showed that in T2D group mice H3K9 dimethylation and H3K4 methylation levels were increased, but H3K9 acetylation and H3K23 acetylation were decreased as compared to age-matched control mice (Fig. 1a). The levels of H3K9 acetylation, H3K4 methylation, and H3K9 dimethylation were significantly reversed by exendin-4 administration (Fig. 1c, d, e), whereas exendin-4 had no effect on modifications at H3K23 acetylation which continued to decrease (Fig. 1b). Thus, advanced type 2 diabetic mouse is specifically associated with increased liver global H3K9 dimethylation and H3K4 methylation and decreased H3K23 acetylation and H3K9 acetylation.
Effects of diabetes and exendin-4 on histone H3 modifications in liver. H3 Acetylation (Lys 23 and 9), methylation (Lys 4), and dimethylation (Lys 9) were determined by Western blotting in total liver histone isolated from mice of control, T2D as well as Ex-4 group at the age of 4 months. a The blots shown are representative of three different blots from histone isolated from different mice of each group. For quantitative analysis, each band was normalized to the respective total histone H3 content (the lowest blot), and the result was shown in arbitrary units, where blots of control mice are set as 100 %. Quantitative analysis of b H3K23ac, c H3K9ac, d H3K4me, and e H3K9me2. The values in the histograms represent means ± SEM from three separate blots. * p < 0.05 and ** p < 0.01 vs. the control group; # p < 0.05 and ## p < 0.01 vs. the T2D group
Liver mRNA expression of several diabetes-related genes in mice
Type 2 diabetes may be associated with decreased expression of diabetes-related genes in the liver. The results showed that the mRNA levels of Glut 2, Gck, as well as Pklr were significantly reduced in liver of type 2 diabetic mice vs. control mice (Fig. 2a, b, c). Meanwhile, the expression levels of Ppargc1a and Insr were lower in T2D than control mice (Fig. 2d, e). The decreased gene expression in type 2 diabetic mice was restored by exendin-4 treatment. Except for Insr (Fig. 2e), the expression levels of all these genes were significantly increased as compared with age-matched T2D mice (Fig. 2a, b, c, d). These data would support that the decreased expression of diabetes-related genes in the liver may lead to type 2 diabetes.
The mRNA expression level of diabetes-related genes was quantified using RT-PCR as described in “Materials and methods.” mRNA expression of a Glut-2, b Gck, c Pklr, d Ppargc1a, and e Insr. All values represent means ± SEM. * p < 0.05 and ** p < 0.01, significantly different from control mice; # p < 0.05 and ## p < 0.01, significantly different from diabetic mice
The change of epigenetic modifications at the Glut2 gene promoter in mouse liver
To investigate if there were changes of epigenetic modifications at the Glut2 gene promoter in T2D diabetic mice, the ChIP assay was conducted using isolated mouse liver with anti-acetyl histone H3 or H3K9, anti-dimethyl histone H3K9, or anti-methyl histone H3K4 antibodies. The results showed that H3K9me2 and H3K4me in diabetic mice were increased significantly as compared to age-matched control mice (Fig. 3a, c). The level of acetylated histone H3 was decreased, while histone H3K9 acetylation increased in T2D mice compared to control mice (Fig. 3b, d). Increased H3K9me2 and H3K4me levels and decreased H3 acetylation basically were correlated with decreased Glut2 gene expression. It also can be seen that exendin-4 treatment reversed the H3 acetylation, H3K4 methylation, and H3K9 dimethylation and therefore prevented the progression of T2D (Fig. 3).
The level of acetyl histone H3 or H3K9 and dimethyl histone H3K9 or methyl histone H3K4 at the Glut2 gene promoter in isolated mouse liver. a, c The level of H3K9me2 and H3K4me epigenetic modifications associated with the mouse Glut2 gene promoter was analyzed in different groups using the ChIP assay with anti-dimethyl histone H3K9 or anti-methyl histone H3K4 antibodies. b, d H3ac and H3K9ac were analyzed with anti-acetyl histone H3 or H3K9. Rabbit IgG was used as negative control. The quantification of the data is shown as means ± SD, n = 3 (panel). * p < 0.05 and ** p < 0.01, significantly different from control mice; # p < 0.05 and ## p < 0.01, significantly different from diabetic mice
The change of histopathology in liver tissue
The pathologic morphology of liver tissue was different among the three groups (Fig. 4). In control group mice, the structure of liver tissue was clear and integrity. Liver cells were arranged regularly, with equal cytoplasm, clear nucleolus structure, and normal morphology, and there were no abnormal changes in the structure of hepatic sinusoid (Fig. 4a). However, in diabetic group mice, lobular boundary was ill-defined, with liver cell edema, disordered arrangement, and hepatic steatosis. Parts of the liver nuclei appeared as shrinkage in the surface, fragmentation, and dissolution (Fig. 4b). Compared with diabetic model group, the degree of disorder hepatic lobule was reduced, the liver cells were arranged regularly basically, and hepatic steatosis was alleviated significantly in exendin-4 treatment group (Fig. 4c).
Results of HE staining in liver tissue of different group mice (100×). a The HE-staining result of normal mouse liver: the structure of liver tissue was clear and normal morphology. b The HE-staining result of diabetic mouse: the liver cells presented edema, disordered arrangement, and hepatic steatosis. c The HE staining result of Ex-4 treatment group: liver cells were arranged regularly, and hepatic steatosis was alleviated significantly, which was nearly similar to control group
Discussion
Although scientists have been conducted a large number of researches on the pathogenesis of diabetes, so far the exact molecular mechanism was still not elucidated. We explored the pathogenesis of diabetes from a novel epigenetic visual angle. Our results showed that the pathogenesis of T2D is associated with distinct total histone H3 modifications, as well as its distinct modifications in Glut2 promoter in liver tissue, were all almost abrogated when the exendin-4 treatment prevented persistent hyperglycemia in the mice (Figs. 1 and 3); this indicated that histone H3K9 and H3K23 acetylation, H3K9 dimethylation, and H3K4 methylation were correlated with the pathogenesis of T2D in C57BL/6 J mice.
After Ex-4 treatment for a month, the physiological and biochemical indexes of mice were examined, as shown in Table 3. Previous studies have pointed to an effect of Ex-4 on decelerating food intake and weight gain in T2D mice models [35]. In our study, after treatment with Ex-4, the symptoms of type 2 diabetic mice were ameliorated significantly, especially FBG which decreased from 14.76 to 10.15 mmol/l. As FBG returned to normal level slowly, the symptoms of emaciation, polydipsia, and polyphagia in type 2 diabetic mice were alleviated. So Ex-4 was not able to decrease body weight; on the contrary, weight was increased a little.
The chromatin consists of histone proteins and DNA which control chromatin packaging and organization by histone modification and DNA methylation [17]. Covalent modifications of histone residues determine accessibility and availability of DNA for transcription via specific acetylation or methylation on histone and then regulate chromatin replication and gene transcription [4, 19]. Histone acetylation could make the chromatin process a state of relaxation, which was beneficial for combining with the transcription factor [11]. It has been reported that histone H3K4 dimethylation has a similar permissive effect and enhances NF-κB-dependent gene expression of inflammatory genes in macrophages of diabetic mice [23]. In contrast, H3K4 methylation and H3K9 dimethylation repress gene transcription [19]. Therefore, we analyzed the status of histone H3K9 (dimethylation and acetylation), H3K4 (methylation), and H3K23 (acetylation) levels in liver.
Although not much is known about histone epigenetics in the pathogenesis of T2D, a recent study demonstrated that high glucose level could increase H3 acetylation by activating the histone acetylases CBP and p/CAF which, in turn, led to an elevated transcription of inflammatory genes in cultured monocytes [24]. High glucose has been reported to alter gene expression by acetylation or methylation of lysine residues on histone H3 [10]. Reports recently showed an increased H3K9 dimethylation in lymphocytes from patients with type 1 diabetes and also considerable cell-type specificity in histone methylation [25, 27]. Our results showed that H3K9 dimethylation and H3K4 methylation levels were significantly higher, while H3K9 and H3K23 acetylation were significantly lower in liver of diabetic mice as compared to normal mice (Fig. 1), indicating that hyperglycemia may activate histone-modifying enzymes, thus change the expression levels of histone H3 modification. Our data generated first evidence that distinct H3 modifications pattern is consistently associated with the progression of diabetes in B6 mice.
Further analyses of diabetic-related genes showed that the gene expressions of Glut2, Gck, Pklr, Ppargc1a, and Insr were all decreased in the livers of diabetic mice. In order to verify the relationship between the gene expression and histone modification, Glut2 promoter was chosen as an example to perform the ChIP assay. The results indeed showed that H3K9 dimethylation and H3K4 methylation were increased, while H3 acetylation was decreased (Fig. 3), which was consistent with the outcome of decreased expression of Glut2 in diabetic mice. In contrast, Ex-4 treatment reversed the histone modifications except H3K9 acetylation. Actually, H3K9 acetylation in Glut2 promoter was different from total H3K9 acetylation in liver, and the anti-acetyl histone H3 antibody was first used to recognize acetylated H3 in Glut2 promoter. There are lots of acetylation loci of histone H3; for example, histone H3 could be acetylated at lysines 9, 14, 18, and 23 which activate chromatin for transcription factor binding [36]. Both histone H3K9 acetylation and H3K23 acetylation only represent a single site, but histone H3 acetylation could represent the total histone H3 acetylation level in the gene promoter. In cell, the level of H3 acetylation is more than H3K9 acetylation, and H3 acetylation may cover the role of H3K9 acetylation. In other words, H3 acetylation plays a more important role in regulating Glut2 expression than H3K9 acetylation. Therefore, in our ChIP assay, histone H3 acetylation, instead of histone H3K23 acetylation, was chosen for analysis. The acetylated H3K23 in the Glut2 promoter was detected afterwards, as shown in Fig. S1. Histone acetylation was often associated with active chromatin to promote gene expression [15]. The downregulation of Glut2 in diabetic mice was mainly the consequence of decreased H3 acetylation in Glut2 promoter, although H3K9 acetylation was increased. After exendin-4 treatment, the levels of H3 acetylation and H3K9 acetylation were both increased, so Glut2 mRNA expression level was upregulated (Fig. 2a). In general, Glut2 expression level was in accordance with histone H3 acetylation in the promoter. In addition, the increased H3K9 acetylation in Glut2 promoter, which was just opposite of total decreased liver histone H3K9 acetylation of T2D mice, indicated that the H3K9 acetylation may be decreased in the other genes’ promoters; this needed further experiments, because H3K9 acetylation of only one gene could not represent the whole H3K9 acetylation level of the liver.
Moreover, Lee et al. [16] had reported that Ex-4 treatment increased mRNA and protein expression of deacetylase SIRT1 in high-fat diet-induced obese mice. However, a contrary result showed that GLP-1 could suppress SIRT1 expression and activity to stimulate β-cell mass expansion [2]. The change of histone H3 epigenetic modification may be due to variation of histone-modifying enzymes levels, which needs further experiments. Additional experiments showed that the deacetylase SIRT1 mRNA and protein were decreased in T2D group mice and increased after Ex-4 treatment (Fig. S2), and the levels of other deacetylases were also needed to examine. In addition, the pathologic morphology of liver tissue was deteriorated in diabetic mice and was recovered after treatment with Ex-4 (Fig. 4).
Conclusions
Taken together, the increased global H3K9 dimethylation and H3K4 methylation, as well as the decreased H3K9 and H3K23 acetylation observed in livers of diabetic mice indicate that histone-modifying enzymes are possibly activated by hyperglycemia, which needs further experiments. The progression of T2D is associated with abnormal hepatic total histone H3K9 and H3K23 acetylation, H3K9 dimethylation, and H3K4 methylation. Yet it remains unclear whether these histone modifications only represent markers of T2D progression or play pathogenic roles. Future studies will have to define whether modifying histone epigenetics, for example, with specific enzyme inhibitors, may represent a novel strategy to prevent T2D.
References
Annicotte JS, Blanchet E, Chavey C, Iankova I, Costes S, Assou S, Teyssier J, Dalle S, Sardet C, Fajas L (2009) The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat Cell Biol 11:1017–1023
Bastien-Dionne PO, Valenti L, Kon N, Gu W, Buteau J (2011) Glucagon-like peptide 1 inhibits the sirtuin deacetylase SIRT1 to stimulate pancreatic β-cell mass expansion. Diabetes 60:3217–3222
Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447:407–412
Bhaumik SR, Smith E, Shilatifard A (2007) Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14:1008–1016
Brun T, Franklin I, St-Onge L, Biason-Louber A, Schoenle EJ, Wollheim CB, Gauthier BR (2004) The diabetes-linked transcription factor PAX4 promotes beta-cell proliferation and survival in rat and human islets. Cell Biol 167:1123–1135
Burgess SC, Leone TC, Wende AR, Croce MA, Chen ZJ, Sherry AD, Malloy CR, Finck BN (2006) Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1 alpha)-deficient mice. J Biol Chem 281:19000–19008
Cras-Meneur C, Elghazi L, Czernichow P, Scharfmann R (2001) Epidermal growth factor increases undifferentiated pancreatic embryonic cells in vitro — a balance between proliferation and differentiation. Diabetes 50:1571–1579
da Silva XG, Rutter GA, Diraison F, Andreolas C, Leclerc I (2006) ChREBP binding to fatty acid synthase and l-type pyruvate kinase genes is stimulated by glucose in pancreatic- beta cells. J Lipid Res 47:2482–2491
Dror V, Nguyen V, Walia P, Kalynyak TB, Hill JA, Johnson JD (2007) Notch signalling suppresses apoptosis in adult human and mouse pancreatic islet cells. Diabetologia 50:2504–2515
El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M (2008) Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205:2409–2417
Fukuda H, Sano N, Muto S, Horikoshi M (2006) Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic 5:190–208
Gavin JR, Alberti K, Davidson MB et al (1997) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 20:1183–1197
Gb Y (2004) Clinical hepatology. Shanghai Scientific and Technical Press, Shanghai, pp 28–30
Han S, Hagan DL, Taylor JR, Xin L, Meng W, Biller SA, Wetterau JR, Washburn WN, Whaley JM (2008) Dapagliflozin, a selective SGLT2 inhibitor, improves glucose homeostasis in normal and diabetic rats. Diabetes 57:1723–1729
Jaskelioff M, Peterson CL (2003) Chromatin and transcription: histones continue to make their marks. Nat Cell Biol 5:395–399
Lee J, Hong S-W, Chae SW, Kim DH, Choi JH, Bae JC, Park SE, Rhee EJ, Park CY, Oh KW, Park SW, Kim SW, Lee WY (2012) Exendin-4 improves steatohepatitis by increasing SIRT1 expression in high-fat diet-induced obese C57BL/6 J mice. PLoS One 7:e31394
Jones PL, Wolffe AP (1999) Relationships between chromatin organization and DNA methylation in determining gene expression. Semin Cancer Biol 9:339–347
Klinkhamer MP, Groen NA, van der Zon GC, Lindhout D, Sandkuyl LA, Krans HM, Moller W, Maassen JA (1989) A leucine-to-proline mutation in the insulin receptor in a family with insulin resistance. EMBO J 8:2503–2507
Kulkarni O, Pawar RD, Purschke W, Eulberg D, Selve N, Buchner K, Ninichuk V, Segerer S, Vielhauer V, Klussmann S, Anders HJ (2007) Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J Am Soc Nephrol 18:2350–2358
Li YP, Liu Y, Strickland FM, Richardson B (2010) Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp Gerontol 45:312–322
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Luan B, Zhao J, Wu H, Duan B, Shu G, Wang X, Li D, Jia W, Kang J, Pei G (2009) Deficiency of a beta-arrestin-2 signal complex contributes to insulin resistance. Nature 457:1146–1149
Miao F, Gonzalo IG, Lanting L, Natarajan R (2004) In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 279:18091–18097
Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R (2008) Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 57:3189–3198
Miao F, Wu XW, Zhang LX, Riggs AD, Natarajan R (2008) Histone methylation patterns are cell-type specific in human monocytes and lymphocytes and well maintained at core genes. J Immunol 180:2264–2269
Nian H, Delage B, Ho E, Dashwood RH (2009) Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen 50:213–221
Ninichuk V, Clauss S, Kulkarni O, Schmid H, Segerer S, Radomska E, Eulberg D, Buchner K, Selve N, Klussmann S, Anders HJ (2008) Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36-3ʹPEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am J Pathol 172:628–637
Sims RJ 3rd, Reinberg D (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 9:815–820
Tahrani AA, Barnett AH (2010) Dapagliflozin: a sodium glucose cotransporter 2 inhibitor in development for type 2 diabetes. Diabetes Ther 1:45–56
Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, Chang CC, Chen P, Chen CH, Fann CS, Chen YT, Wu JY (2010) A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 6:e1000847
Uldry M, Thorens B (2004) The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch 447:480–489
Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, Pinilla-Ibarz J, Seto E, Sotomayor EM (2009) The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol 10:92–100
Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R (2008) Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A 105:9047–9052
Wilson CB, Rowell E, Sekimata M (2009) Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 9:91–105
Young AA, Gedulin BR, Bhavsar S, Bodkin N, Jodka C, Hansen B, Denaro M (1999) Glucose-lowering and insulin-sensitizing actions of exendin-4:studies in obese diabetic(ob/ob, db/db)mice, diabetic fatty Zucker rats, and diabetic rhesus monkeys (Macaca mulatta). Diabetes 48:1026–1034
Zimmet P, Alberti KG, Shaw J (2001) Global and societal implications of the diabetes epidemic. Nature 414:782–787
Acknowledgments
This work was supported by the 863 Program of China (2008AA02Z205) and the Key Technologies R&D Program of Tianjin (14ZCZDSY00013).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Tu, P., Li, X., Ma, B. et al. Liver histone H3 methylation and acetylation may associate with type 2 diabetes development. J Physiol Biochem 71, 89–98 (2015). https://doi.org/10.1007/s13105-015-0385-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13105-015-0385-0