Abstract
Post-translational protein modifications present an elegant and energy efficient way to dynamically reprogram cellular protein properties and functions in response to homeostatic imbalance. One such protein modification is the tagging of proteins with the small modifier ubiquitin that can have an impact on protein stability, localization, interaction dynamics, and function. Ubiquitination is vital to any eukaryotic cell under physiological conditions, but even more important under stress including oxidative, genotoxic, and heat stress, where ubiquitination levels are drastically increased. Elevated levels of ubiquitin-protein conjugates are also observed in the brain after focal and global cerebral ischemia. Post-ischemic ubiquitination is immediately induced with reperfusion and transiently detected in neurons with survival potential located in the peri-infarct area. This review aims to critically discuss current knowledge and controversies on protein ubiquitination after cerebral ischemia, with special emphasis on potential mechanisms leading to elevated ubiquitination and on target identification. Further, possible functional implications of post-ischemic ubiquitination, including a relationship to SUMOylation, a neuroprotective modification, will be highlighted. The elevation in ubiquitinated proteins following cerebral ischemia is a greatly under-explored research area, the better understanding of which may contribute to the development of novel stroke therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction to Ubiquitin
Ubiquitination, a Multifaceted and Complex Post-translational Modification
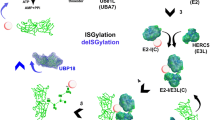
Ubiquitination is a highly versatile post-translational protein modification in all eukaryotic cells that occurs when the small 76 amino acid polypeptide ubiquitin is covalently attached to a target protein. It is estimated that there are more than 5000 proteins ubiquitinated in a cell at any given time, which makes up about one quarter of the encoded human proteome [1]. Therefore, the implications of ubiquitination on cell homeostasis are predicted to be enormous. Upon its discovery in the early 1980s, it was believed that ubiquitination solely serves as protein degradation signal [2, 3]. However, over the years, it became clear that it is essential for a wide variety of cellular processes, including cell cycle control, enzyme activation, signal transduction, transcription and DNA repair, receptor trafficking, immune response, inflammation, and apoptosis (reviewed in [4,5,6,7,8]). The remarkable ability of ubiquitin to influence protein function in seemingly endless ways stems on the one hand from the complexity of the conjugation system, and on the other hand from ubiquitin’s ability to form polymers with different topologies (Fig. 1).
The ubiquitin conjugation system is complex, influencing proteins in many different ways. a Mammalian cells, including neurons, contain four ubiquitin genes (Uba52, Uba80, Ubb, and Ubc) that contribute to the cellular ubiquitin protein pool. Ubiquitin is synthesized either fused to large (LRS) and small (SRS) ribosomal subunits, or as chains. Single ubiquitin molecules are obtained from these precursors by cleavage with ubiquitin-specific proteases (USPs). In order to be attached to a substrate, ubiquitin needs to be first activated in an ATP-dependent manner by an E1 activating enzyme, before it is transferred to an E2 conjugating and E3 ligating enzyme, with all of which it forms thiolester bondages (S). Finally, ubiquitin is covalently linked to a substrate by an isopeptide bond involving a lysine residue (K) in a substrate and a C-terminal glycine on ubiquitin. Whereas there are only two E1 enzymes and a restricted number of E2 enzymes (about 40), ubiquitin ligases are abundant (more than 600). An intimate interplay between E2 and E3 enzymes guarantees specific substrate recognition. b Ubiquitin can be attached to a target either as monomer, multiple monomers, or as chain that are formed through internal lysine (K) or the N-terminal methionine (M) residues in the ubiquitin molecule itself. Depending on the type of ubiquitin attachment, the functional outcome involving the substrate is entirely different (see list on the left). AMP adenosine monophosphate, ATP adenosine triphosphate, CC cell cycle, PPi pyrophosphate, Ub ubiquitin
For the conjugation of ubiquitin to substrates with few exceptions, the consecutive action of three enzymes is required. In the first step, an E1-activating enzyme carries out the ATP-dependent activation of ubiquitin by forming a thiolester bond with its C-terminal glycine. Ubiquitin is then sequentially transferred to E2-conjugating and E3-ligating enzymes, which catalyze the attachment of ubiquitin via an isopeptide bond to one or more lysine (K) residues in a protein substrate (reviewed in [9]) (Fig. 1a). While there are only two E1 enzymes encoded in mammalian cells, there are about 40 E2s and more than 600 E3s that have the potential to work together in a “mix and match” fashion to create about 25,000 different conjugation pairs that drive substrate specificity. In comparison, phosphorylation is directed by only about 520 kinases [10, 11]. Further, more than 100 deubiquitinating enzymes (DUBs) that can remove ubiquitin from a substrate have also been identified [12] (Fig. 2).
The 26S proteasome, ubiquitin de-conjugation, and ubiquitin recycling. After serving their purpose, ubiquitinated proteins can be de-ubiquitinated by one of the approximately 100 known deubiquitinases (DUBs). On the other hand, if the attached ubiquitin chain signals for degradation, substrates are recognized and degraded by the 26S proteasome. The 26S proteasome consists of a 20S barrel-shaped core (α and β subunits) that has chymotryptic, tryptic, and caspase-like protease activities. The core is flanked on one or both sides with 19S lids that a recognize ubiquitinated proteins through ubiquitin-binding domains (UBDs), b remove ubiquitin from substrates via DUB activity, and c unfold substrates with AAA ATPase activity. De-ubiquitination by proteasome-dependent and -independent DUBs feeds ubiquitin molecules back into the free ubiquitin pool, awaiting a new conjugation round. AMP adenosine monophosphate, ATP adenosine triphosphate, K lysine residue, PPi pyrophosphate, Ub ubiquitin
Apart from the increased number of conjugation options, ubiquitination has another additional layer of complexity compared with phosphorylation that greatly increases its versatility: it can form chains that are connected by each of its seven internal lysine residues (K6-, 11-, 27-, 29-, 33-, 48-, and 63-chains) and its N-terminal α-amino group (linear chains), resulting in at least eight different linkages [13, 14] (Fig. 1b). Although less commonly, mixed linkages as well as branched mixed chains, where different lysines in a single ubiquitin molecule are used for chain extension, are also observed [15,16,17,18]. Finally, ubiquitin can also be attached as a monomer on one or multiple target sites within a substrate (reviewed in [19]) (Fig. 1b). The fate of an ubiquitinated protein is critically dependent on the configuration of the associated ubiquitin (Fig. 1b). Well-characterized modifications in this regard are monomers as well as linear and K11, 48, and 63-linked chains. K48 chains with a few exceptions always serve as degradation signal, and there is some evidence that K11 and 29-linked chains serve as proteasomal recognition signals as well [20, 21]. On the contrary, monomers, K63-linked, and linear chains are completely non-degradative (reviewed in [22]). This is made possible by the entirely different three-dimensional shape of K11, 29, and 48 polymers compared to linear and 63-type chains [23,24,25,26], and naturally monomers. With this in mind, monomeric ubiquitination as well as ubiquitination via different chains may be seen as entirely distinct modifications that dictate a proteinsʼ fate in very different ways (Fig. 1b). This diversity of effects on proteins should also be considered when assessing ubiquitin’s impact on the post-ischemic brain.
Ubiquitin-Mediated Degradation and the 26S Proteasome
Quantitative analysis revealed that K11, 29, and 48 ubiquitin chains represent about 60% of all ubiquitin linkages in vivo [21]. Although this analysis was carried out in yeast and the percentage may slightly vary in other eukaryotic cells, it can be predicted that about 50% or more of ubiquitinated proteins are tagged for degradation. The main protease responsible for intracellular protein degradation is a large 2.5 MD complex termed the 26S proteasome (Fig. 2). It consists of a 20S proteolytic core that is capped on one or both ends by a 19S/PA700 regulatory activator complex. The 20S core is built as a barrel-shaped ring structure containing two different types of subunits that have distinct structural and functional roles [27, 28]. The outer two rings are composed of α subunits that ensure structural integrity of the core, while the β subunits are forming the inner rings that confer proteolytic activity (Fig. 2). The 20S core by itself is essentially inactive as its structure sequesters the catalytic sites in an internal chamber that can only be reached through a narrow opening in the α subunits [27]. Although a role for the 20S core in the degradation of damaged or unfolded proteins has often been suggested especially in the context of oxidative stress [29, 30], it remains unclear as to how polypeptides manage to enter the uncapped 20S particles in vivo. Access to the 20S core is gated by the 19S/PA700 regulatory activator that confers specificity of the protease for ubiquitinated proteins by binding to ubiquitin chains and tethering the ubiquitinated substrate to the proteasome [31, 32]. The 19S particle contains a lid composed of subunits with ubiquitin-binding domain (UBD) and deubiquitinase (DUB) activities, and a base that contacts with the core particle and is composed of six ATPases, which belong to the AAA ATPase family (Fig. 2). Before delivering proteins for degradation to the 20S core, 19S/PA700 subunits deubiquitinate [33, 34] and then unfold substrates in a highly ATP-dependent manner [35]. It should be mentioned that the 20S core can also associate with two alternative activators, the 11S/PA28 and PA200 complexes; however, these assemblies are unlikely to play a role in ubiquitin-mediated degradation as neither PA28 nor PA200 recognize ubiquitinated proteins or use ATP [36, 37].
Rather than being an unregulated destructive machine that uniformly degrades ubiquitinated substrate proteins, the activity of the 26S proteasome can be modulated by metabolic and inflammatory factors. Exposure to immune mediators or oxidative stress can initiate the exchange of classical proteolytic subunits of the 20S core with unconventional subunits that alter proteolytic capacity [38, 39]. This gives rise to the so-called immunoproteasome. This specialized form of the proteasome is mainly present in immune cells; however, it is also detected in non-immune cells, including the brain. However, while its role in immune cells with the generation of antigens for MHC class I presentation is well described (reviewed in [40]), it is not clear how it affects for example neurons or glia. Notably, there is indication that induction of the immunoproteasome in brain cells is associated with neurodegeneration [41,42,43]; however, it is not known whether this is a cause or effect of degeneration. Another means of altering 26S proteasomal activity is the post-translational modification of both 20S and 19S/PA700 subunits. In the brain, phosphorylation of Rpt6, an AAA ATPase member of the 19S complex, by CaMKII boosts proteasome activity to establish synaptic plasticity and memory [44,45,46]. Phosphorylation of the same subunit by PKA increases brain proteasome activity, and its associated down-regulation was proposed to be critical for Huntington’s disease pathogenesis [47, 48]. An increase of O-GlcNAcylation and reactive oxygen species (ROS) in the brain is connected with reduction in proteasomal activity through modification of Rpt2 and Rpt3 proteasomal subunits [49,50,51]. Further, proteasomal activity is activated by poly-ADP ribosylation and acetylation of unidentified core subunits [52, 53], while ubiquitination of Rpn10 and Rpn13 19S subunits reduces proteolysis [54, 55]. Whether these modifications play a role in brain cells under stress, particularly after ischemia is yet to be determined.
Non-degradative Functions of Ubiquitin
Approximately 50% of ubiquitinated proteins are estimated to be associated with non-degradative ubiquitin chains or ubiquitin monomers [1] that control a variety of cellular functions primarily by modifying protein interaction profiles and protein activity. First, attached ubiquitin can promote the recruitment of new binding partners that are not at all or only weakly bound by the non-modified protein. An example for this in neurons is the ubiquitination of PSD95 by Mdm2, which does not affect PSD95 protein levels, but rather increases its interaction with β-adaptin to regulate AMPA receptor endocytosis [56]. Conversely, ubiquitin can also impede interactions. Monoubiquitination of the transcription factor Smad4, whose activity is important for neuronal development, blocks its association with the transcriptional co-activator Smad2 leading to blunted transcription [57]. Ubiquitination of receptors guides their endocytosis, sorting, recycling, and degradation by serving as docking signal for adaptor proteins. Prominent examples for this regulation in neurons are the AMPA and GABA receptors that require ubiquitination to initiate internalization and lysosomal degradation [58, 59]. Ubiquitination has also gained special attention as platform for the formation of signaling complexes and regulation of kinase function, which has been extensively shown in pro-inflammatory NF-κB signal activation [60,61,62,63]. In summary, in addition to playing a major role in protein degradation, ubiquitin serves also many important non-degradative functions that need to be kept in mind when considering the role of ubiquitination in brain cells under stress, e.g., after ischemia.
Ubiquitin Is Essential for Neuronal Development and Function
Ubiquitination is crucial for health and function of neurons. Synaptic plasticity and transmission, maintenance of synaptic strength, as well as memory formation are only a few examples that rely on a functional ubiquitination system [64,65,66]. It has been shown that increases and decreases in neuronal activity correlate with the amount of ubiquitinated proteins in dendritic spines [64]. Many key molecules involved in synaptic function are regulated by ubiquitination. These include structural components of the postsynaptic density, for example PSD95 [56, 67], GKAP [68], and Shank [64], or receptors like the AMPAR [58, 69], NMDAR [70], and GABAR [59, 71]. The effect of ubiquitination on the synapse is multifaceted. On the one hand, ubiquitination of scaffolding proteins strengthens the synapse, maintains synaptic morphology, and leads to reorganization of signaling complexes [67, 68]. On the other hand, ubiquitination of receptors leads to changes in receptor surface abundance, ensuring proper propagation of stimulatory and inhibitory signals [58, 70, 71]. In addition to its role in mature neurons, the ubiquitin system is also important during neurodevelopmental processes, such as dendrite pruning and axon guidance [72, 73].
Ubiquitin and the Stress Response
Apart from controlling important physiological functions, the ubiquitin system takes on essential roles in response to cellular stress. Indeed, an increase in the level of ubiquitin conjugates occurs with virtually all types of stressors, e.g., heat shock, oxidants, heavy metals, DNA damaging agents, etc. This is in part made possible by an increased expression of ubiquitin itself. Mammalian cells, including neurons, contain four genes that feed into the cellular ubiquitin protein pool. The transcription of two of them, Ubb and Ubc, is heavily induced by stresses [74, 75], which provides an increased amount of ubiquitin necessary for survival under these conditions. Also, the activity and expression of E1 and some E2 and E3 enzymes are elevated as stress response [76,77,78,79]. The protection of cells via ubiquitin is mediated by degradative and non-degradative mechanisms. A stressed cell contains an increased amount of unfolded proteins due to changed energy demands that, if chaperones are overwhelmed with refolding, are tagged with predominantly K48-linked ubiquitin and delivered for degradation by the 26S proteasome. The role of K48-linked ubiquitination and proteasomal degradation in protein quality control is the topic of many expert reviews and will therefore here not be further discussed (reviewed in [80,81,82,83]). On the other hand, a vastly under-studied area is the role of non-degradative ubiquitin chains in stress response. Although not fully appreciated, it was already shown in 1994 that over-expression of ubiquitin unable to form K63 chains as opposed to all other chains sensitizes yeast cells for heat, indicating a specific role of non-degradative K63-linked chains in cells under stress [84]. Recently, K63-ubiquitination was rediscovered in this regard, when it was shown that lack of K63 ubiquitin diminishes the cellular resistance to oxidative stress [85]. The authors identified several protein groups as K63-ubiqutination targets, the largest group associated with translation. The effect of ubiquitination on these proteins, however, remains entirely unknown and will be of interest to investigate in the future. Considering that oxidative stress is an integral part of the ischemic cascade, this regulation could undoubtedly be also important in neurons exposed to ischemia.
Ubiquitin and Cerebral Ischemia-Reperfusion Injury
Ubiquitination Is Induced in the Brain by Ischemia-Reperfusion
Cerebral ischemia puts an immense stress on cells in the affected brain area, and it seems reasonable that brain cells react to this stress with an increase in ubiquitination. Indeed, it was first shown by Hayashi et al. in 1991 that sublethal forebrain ischemia leads to elevated ubiquitin immunoreactivity as well as appearance of high molecular weight ubiquitin conjugates in the rat hippocampus [86]. In a follow-up study, the same authors reported that ubiquitination was increased in the early reperfusion period after global ischemia in the gerbil cortex and hippocampus with a peak at 30 min to 1 h and tended to be resolved by 12–24 h [87]. A similar pattern of ubiquitination was also observed after focal cerebral ischemia in rat and mouse cortices [88, 89]. In both ischemia models, reperfusion was absolutely required for ubiquitination [87, 89], presumably because ATP, which is essential for ubiquitin conjugation, is limiting in ischemic tissue prior to reperfusion. This has not been experimentally tested yet and needs further assessment. Another possibility would be that reperfusion provides the inducing signal for ubiquitination, e.g., by production of free radicals or the activation of signaling cascades associated with excitotoxicity and beyond. Interestingly, pure hypoxic insults in rats did not increase ubiquitination in the post-hypoxia recovery period [90, 91], possibly indicating that lack and re-establishment of blood flow (and the associated availability of glucose) may be essential for activation of ubiquitination. However, additional research will be required to confirm this, since one study used mild hypoxia (15% oxygen) that may not induce sufficient stress [90], while the other study was performed in immature brains [91].
Prevalence of Post-ischemic Ubiquitination in Neuronal Detergent-Insoluble Fractions
Since neurons appear to be most vulnerable to ischemic stress, they are assumed to be the primary source of increased ubiquitination post-ischemia. Indeed, immunocytochemistry of brain sections after ischemia shows a drastic change in ubiquitin immunoreactivity preferentially in cells with neuronal morphology at different times of reperfusion [88, 92,93,94]. However, it has not been systematically assessed whether glial cells are also affected, thus warranting further investigation.
Where does elevated ubiquitination occur in neurons? As determined by immunocytochemistry of brain sections, most studies found that both global and focal ischemia leads to a redistribution of ubiquitin from an even staining covering the entire cell to a selected perinuclear and dendritic localization [88, 91, 92, 94, 95] (Fig. 3). One study found ubiquitin in the nucleus of hippocampal pyramidal neurons after ischemia/hypoxia [90]. This discrepancy may be the result of the different ischemia model used, or, more likely, due to staining with different ubiquitin antibodies. In fact, it is well established that dependent on their preferential recognition of diverse chains, free or conjugated ubiquitin, or epitopes in the ubiquitin protein, ubiquitin antibodies show great variability when used in immunohistochemistry (examples in the brain after ischemia include [94, 96]). To avoid this drawback, the localization of ubiquitin after ischemia can also be assessed biochemically using subcellular fractionation. Hayashi et al. performed in 1992 the first fractionation experiment where they showed that ubiquitin elevation is prominent in mitochondrial, synaptic membrane, and microsome fractions, but not in nuclei or cytosol [97] (Fig. 3). They also discovered that ubiquitin conjugates are almost exclusively formed in the Triton X100 (Tx100) insoluble fraction, which was confirmed by multiple studies in different ischemia models thereafter [88, 89, 98, 99]. It remains unclear to date what this insolubility refers to. On the one hand, the resistance of ubiquitin to high concentrations (up to 2%) of Tx100 could indicate an accumulation of ubiquitin-modified proteins in insoluble aggregates analogous to neurodegenerative diseases. Although this is not entirely impossible, it seems unlikely that such an aggregation would occur on the large scale seen within the short timeframe (minutes) observed after ischemia [89], especially as there is virtually no time when ubiquitin is found in a soluble state.
Cerebral ischemia-reperfusion leads to a redistribution of ubiquitin in neurons. Schematic representation of ubiquitin distribution in pre- and post-ischemic neurons. Under physiological conditions, ubiquitin is evenly distributed throughout the nucleus and cytoplasm showing little to no association with any organelles or microsomal structures (upper panel). After ischemia, ubiquitin is found strongly associated with the postsynape, mitochondria, and microsomes. In addition, redistribution to perinuclear regions is apparent (lower panel). Main accumulation areas of ubiquitin after ischemia-reperfusion are marked with boxes and arrows. Regions with darker blue shades indicate areas containing more ubiquitin. Ub ubiquitin
Alternatively, some intracellular components in neurons are by default largely insoluble and could preferentially be targeted by ubiquitination. One such compartment is the postsynaptic density (PSD), which is only minimally soluble in detergents like Tx100 and only efficiently solubilized with high concentrations of sodium dodecyl sulfate (SDS) or urea. Indeed, it was shown in one report that ubiquitinated proteins are highly enriched in the PSD after ischemia [93]. Supportive of this idea, global post-ischemic ubiquitinated proteins found in the Tx100-insoluble brain fraction by mass spectrometry include many proteins that show a tight connection to the PSD. Among the highly ubiquitinated proteins were CaMKIIα, β, γ and δ, the vesicle-fusing ATPase NSF, PKCβ and γ, NMDA receptor subunits epsilon-1 and 2 (NR2A and NR2B), disk large-associated proteins 1 and 2 (GKAP and DLG2), the calcium-binding protein hippocalcin, Ras GTPase-activating protein (SynGAP), and leucine-rich repeat-containing protein 7 (LRRC7/densin-180) [98]. Although PSD proteins seem to make up much of the ubiquitinated proteome after ischemia, it is entirely unclear what functional consequence this ubiquitination has on synaptic activity. This important question needs to be addressed in future studies.
Ubiquitination Types Induced by Cerebral Ischemia Are Diverse
In order to predict the impact of ubiquitination on target proteins, it is useful to assess the associated ubiquitin type. As determined by mass spectrometry, prevalent ubiquitin chains found after global forebrain ischemia include K6, 11, 48, and 63 linkages [98] (Fig. 4). Likewise, after focal ischemia, we detected with lysine specific antibodies both K48 and K63 chains [100]. The presence of degradative and non-degradative ubiquitin chains suggests that opposed to popular assumption, proteasomal blockade is likely not the only reason for post-ischemic poly-ubiquitin enrichment. Neither the conditional neuronal knockout of proteasomes in mice [101] nor proteasomal inhibition in yeast [85] is capable of inducing K63 poly-ubiquitination, pointing to alternative mechanisms that contribute to increased ubiquitination levels after ischemia.
Post-ischemic changes in the ubiquitin-proteasome system in early reperfusion. A 6-, 14-, 40-, and 11-fold induction in K6-, K11-, K48-, and K63-linked ubiquitinated substrates that is accompanied by decreased free ubiquitin has been found 30–180 min after ischemia. While ubiquitin gene synthesis remains unaffected early after ischemia, there are some reported changes in the ubiquitin conjugation, de-conjugation, and degradation systems that have not been well defined yet and are summarized in this figure. The increased ubiquitination after ischemia is likely a consequence of these converging mechanisms. Known up- or down-regulation of ubiquitin-proteasome components is marked with upward- and downward-facing arrows. AMP adenosine monophosphate, ATP adenosine triphosphate, DUB deubiquitinase, E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, E3 ubiquitin-ligating enzyme, K lysine residue, LRS large ribosomal subunit, PPi pyrophosphate, S thiolester, SRS small ribosomal subunit, Ub ubiquitin, UBD ubiquitin-binding domain, USP ubiquitin-specific protease
Factors Contributing to Elevated Post-ischemic Ubiquitination
Ischemia-Related Changes in Ubiquitin Conjugation
Elevated ubiquitination after ischemia could be caused by an up-regulation of ubiquitin gene expression leading to greater availability of ubiquitin for conjugation. However, as the increase in conjugates is accompanied by the depletion of free ubiquitin [88, 89, 96], the elevated ubiquitination is likely not a result of increased ubiquitin synthesis. This hypothesis is confirmed by the differential expression analysis of ubiquitin genes before and after transient forebrain ischemia in rats, which shows that Uba levels remain entirely unchanged and production of Ubb and Ubc is not activated earlier than 4–6 h into reperfusion [102]. Such increased gene expression occurs too late to account for the increased post-ischemic ubiquitination (Fig. 4).
It is reasonable to hypothesize that augmented activity of certain ubiquitination enzymes is responsible for the vast increase in ubiquitin conjugation after stroke. However, owing to the complexity of the ubiquitination system, this hypothesis has not been tested experimentally. As mentioned before, up to 25,000 different enzyme combinations can mediate target ubiquitination, all or only one of which could potentially be involved in ubiquitination regulation after ischemia. Proteome-wide screens with an easy readout will have to be created to address this issue in the future. In the meantime with the identification of specific ubiquitination targets, a more biased approach can be undertaken to study potentially involved ligases. For example, ubiquitin ligases for some NMDAR subunits (Nedd4–1 and Fbx2) and GKAP (Trim3) under physiological conditions have been identified and could be examined for their ubiquitination capacity after ischemia [103,104,105]. Another option is to consider ubiquitin ligases that are classically involved in stress regulation and neuronal degeneration. In this regard, the increased activity and level of a few specific ubiquitin ligases have been associated with neuroprotection after cerebral ischemia (Fig. 4). CHIP, an ubiquitin ligase that controls chaperone activity and levels during stress, prevents oxygen/glucose deprivation (OGD)-induced neuronal death when it is overexpressed in vitro in hippocampal slices and in vivo in rats after global ischemia [106]. CHIP overexpression also leads to a reduction in ubiquitinated proteins after OGD; however, this could be the result of decreased neuronal injury. The ubiquitin ligases Nedd4–2 and Itch are induced by focal ischemia in rats in surviving neurons, suggesting that they may be associated with neuroprotection [107]. Also, the Nedd4 adaptor protein Ndfip1 is protective in stroke, since knockout mice exhibit larger infarcts [108]. Since up-regulation of these proteins was measured beginning at 12 h of reperfusion, and maximal regulation was detected at 24 to 48 h post-ischemia, the relevance for early ubiquitination after ischemia is elusive. Another ubiquitin ligase with altered protein level after ischemia is Parkin. Both OGD in neuroblastoma cells as well as focal ischemia in mice was reported to essentially reduce Parkin levels, which leads to mitochondrial fragmentation and potentially ER stress [109, 110]. Like the other ubiquitin ligases, also Parkin protein is neuroprotective, but again, how Parkin levels relate to early ubiquitination after ischemia is not known. In summary, the elevated ubiquitin levels after ischemia cannot be attributed to an increase in ubiquitin protein levels, but are likely caused by an up-regulation of components of the ubiquitin conjugation machinery, the identity of which is entirely unknown to date (Fig. 4).
Ischemia-Related Changes in Ubiquitin De-conjugation
No deubiquitinase (DUB) has been studied for a specific involvement in elevated ubiquitination after stroke, but the activity of two DUBs has been linked to the ischemic outcome (Fig. 4). UCH-L1 is a DUB that is highly expressed in neurons [111] and has gained attention for its protective role in Parkinson’s and Alzheimer’s disease [112, 113]. Analogous to neurodegenerative diseases, overexpression of UCH-L1 protects neurons from hypoxic injury [114] and down-regulation of UCH-L1 activity in neuroblastoma cells increases OGD-induced cell death [115]. In neurodegeneration, the protective effect of UCH-L1 has been mostly attributed to its potential to prevent Aβ and α-synuclein aggregation by promoting APP and α-synuclein degradation [112, 113]. UCH-L1 potently recycles and stabilizes free ubiquitin derived from proteasomal and non-proteasomal sources and contains a DUB-independent ubiquitin ligase activity, both of which have been shown to contribute to the efficient degradation of disease proteins [113, 116, 117]. In ischemia, the mechanism of protection has not been elucidated. However, it was shown that after hypoxia pharmacological inhibition of UCH-L1 for 24 h increases ubiquitinated proteins in primary neurons [114], which could contribute to hypoxic injury, although direct evidence for this is lacking. In any case, earlier time points need to be studied to clarify the impact of UCH-L1 on the immediate increase in ubiquitination after ischemia. Another DUB with a probable role in stroke and neurodegeneration is USP14, which negatively regulates the proteasomal degradation of ubiquitinated proteins by trimming K48 ubiquitin chains [118]. Inhibition of USP14 was shown to be protective in Alzheimer’s disease cell models by facilitating Tau degradation through enhancing proteasomal activity [119]. Injection of a specific inhibitor of USP14 (IU1) [118] attenuated ischemic neuronal injury in a focal ischemia mouse model [120]. The protection correlated with an enhancement of proteasomal activity and reduction of ubiquitinated proteins measured 24 h after ischemia [120]. It is not clear if the decrease in ubiquitinated proteins is neuroprotective per se, or only a measure of less injury at this time-point. As with UCH-L1, more studies looking at earlier times of reperfusion will be necessary to causally connect USP14 with ubiquitination after ischemia.
Ischemia-Related Changes in Ubiquitin Conjugate Clearance
The Proteasome
Augmented presence of ubiquitination could be caused by a blockade in degradation of modified proteins, as one would observe with the use of proteasome inhibitors. The proteasome has been extensively studied after cerebral ischemia, and the consensus is that classical 26S proteasomal activity is indeed reduced during ischemia and also in the reperfusion period, where it recovers gradually (Fig. 4) [89, 121,122,123,124,125,126,127]. The mechanism of down-regulation of proteasomal activity after ischemia is not clear, especially whether it is actively repressed, or the repression is a consequence of ischemic damage. It should be noted here that proteasomal activity was mainly measured by its ability to cleave fluorogenic tri- or tetrapeptides containing consensus cleavage sites in vitro. Since these peptides are small enough to enter the proteasomal 20S core regardless of 19S presence, they are only suitable for measuring catalytic core activity, and limited conclusion can be drawn regarding ATP requirements and necessity for a complete 26S assembly [128]. For example, it was shown that 19S and 20S particles partly separate after ischemia [123]; however, the mode of measurement does not capture this change. In fact, oxidative stress was repeatedly reported to result in the dissociation of 20S and 19S subunits in other models, which was protective and necessary for the 20S proteasomes to clear oxidized proteins [30, 129]. In addition, calpain cleavage of the Rpn10 19S subunit was observed in cultured neurons with mitochondrial stress, leading to proteasome inactivation [130], which could also be relevant after ischemia.
There is some evidence that proteasome subunits are targeted by ROS after ischemia that may inhibit its activity. Proteasomal subunits persistently stain positive for HNE and co-immunoprecipitate with DNP from 1 to 24 h after focal ischemia [125, 126]. The same was observed after myocardical infarction in the heart [131]. Whether the modification with ROS is causative for proteasomal inhibition is not clarified. First, proteasomal inhibition seems to precede ROS production, and ROS modification of the proteasome persists even when activity is recovering [89, 121, 125, 126]. Second, at least in the heart, proteasomal activity is extremely resistant to ROS levels, and mapping of oxidized subunits detected a predominant modification of α, not β subunits that actually carry the catalytic activities [132]. Modified proteasomal subunits are still to be identified in the ischemic brain.
Regardless of how proteasomal core activity is reduced after ischemia, does this cause the accumulation of ubiquitinated proteins? On the one hand, inhibition of the proteasome is detected at early reperfusion times before ubiquitin accumulation starts after ischemia; therefore, it is possible. On the other hand, detected K63 ubiquitin chains after ischemia [98, 100] imply an alternative mechanism for accumulation, at least in part. Preservation of proteasomal activity after stroke is often linked with a reduction in ubiquitinated proteins. For instance, protein ubiquitination after transient global ischemia was reduced by pretreatment of rats with trehalose that maintained post-ischemic proteasomal activity [133]. Likewise, USP14 inhibition enhanced proteasomal activity after stroke and led to less ubiquitination [120]. Mice with transgenic over-expression of Ubqln-1, a proteasomal adaptor protein that enhances proteasomal degradation, showed less accumulation of ubiquitin [134]. However, a causal relationship between proteasomal inhibition and ubiquitination has not been determined and lower levels of ubiquitination might just reflect less insult, which was observed in all studies. More research needs to be carried out to properly address whether enrichment of ubiquitinated proteins after cerebral ischemia is a direct result of proteasomal impairment.
Another interesting question is what the reduced proteasome activity means for stroke outcome. Experimental evidence points to a down-regulation of the UPS in brain ischemia that correlates with neuronal death and incubation of neurons with proteasome inhibitors was shown to induce cell death. However, inhibition of the proteasome after ischemia is only between 20 and 50%, and it has been shown that a reduction of 80% is necessary to have an effect on cellular homeostasis [135]. Yet, a lower degree of inhibition may impair cell viability during stress conditions, and this needs to be still assessed.
Seemingly opposing a detrimental effect of proteasomal inhibition, proteasome inhibitors were shown to provide neuroprotection in various models of stroke. Intravenous, intraperitoneal, and intrastriatal injections of proteasome inhibitors MLN519, Bortezomib/Velcade, CVT-634, and BSc2118 after ischemia lead to neuroprotection [136,137,138,139,140,141]. Except for Doeppner et al. in 2012, inhibition of brain proteasomal activity was not verified in these studies and it is unclear whether the inhibitors can cross the blood-brain barrier. It has been suggested that inhibitors likely act via a brain-independent mechanism by attenuating neuroinflammation and blood-brain barrier breakdown after ischemia. In fact, proteasome inhibitors when administered at high concentrations and long durations induce neuronal cell death and do not protect isolated neurons from in vitro ischemia [142, 143].
Autophagy
Alternatively to the proteasome, clearance of ubiquitinated proteins can be executed by autophagy, which is mainly responsible for recycling organelles and large cytoplasmic structures, but may also digest larger ubiquitinated aggregates. Post-ischemic autophagy regulation is poorly studied, and there are conflicting reports on whether autophagy is impaired or activated after cerebral ischemia. One study showed that autophagy is persistently up-regulated in reperfusion after in vivo and in vitro ischemia, and that blockage of autophagy with 3-MA in reperfusion increases infarct volume and cell death [144]. In contrast, in another study, the autophagic flux was decreased after ischemia specifically in neurons destined to die [145]. Whether this leads to ubiquitinated protein accumulation was not specifically assessed. The measurements were carried out from 4-h reperfusion; therefore, it is hard to say whether this could be relevant for the early increase in ubiquitinated proteins.
Ubiquitination After Ischemia—Good or Bad?
Post-ischemic ubiquitination is specifically elevated in brain regions that are susceptible to delayed cell death, as opposed to areas where cells die immediately. Focal ischemia induces ubiquitination particularly in cortical regions that are not directly part of the infarct center, while there is only little ubiquitination found within the infarct core [88, 89]. In global ischemia, ubiquitination is highly increased in the hippocampal CA1 layer and cortex [92] that are selectively vulnerable to ischemic stress. Of note, the elevated ubiquitination is detected transiently in neurons that return to a normal morphology after the insult, whereas the ubiquitin stain remains high in neurons that appear to die [88, 92]. Several treatments were shown to alleviate ubiquitination levels after ischemia. Hypothermia preserved ubiquitin immune reactivity in the CA1 region after global ischemia [146] and diminished initiation of ubiquitination after focal ischemia [94]. Ischemic preconditioning led to a decrease in ubiquitination in the otherwise heavily ubiquitinated CA1 area in early and delayed tolerance in global ischemia models [95, 147]. Ischemic post-conditioning reduced ubiquitination as measured 12–72 h after ischemia [148], earlier time-points were not studied. Administration of geldanamycin, an Hsp70 inducer, prior to OGD blocks ubiquitin redistribution from nucleus to cytoplasm in hippocampal slices after insult [149]. Collectively, these observations suggest that ubiquitination contributes to the delayed neuronal cell death after stroke, which has been commonly proposed. However, the presence of ubiquitination in dying cells may not be causative and only be secondary to the cell death trigger, or may even be part of a compensatory protective mechanism that prevents immediate death of these neurons. The reduced ubiquitination in these models is likely due to milder ischemia that causes less stress in neurons. Ubiquitination is elevated immediately and transiently after onset of reperfusion, before any cell death occurs and is in our hands also apparent after insults that do not cause any damage [89]. Alternatively, ubiquitination may indeed increase with ischemic length as shown in a global ischemia model [150], but that may simply reflect higher stress levels. Interestingly, ubiquitination levels were also increased in the liver of squirrels during hibernation [151]. There it was suggested that during suppression of protein synthesis, which is also observed after ischemia, a shut down of proteolysis is required to maintain pools of key regulatory enzymes. Protein ubiquitination may still go on under these conditions.
Ultimately, the important question of whether post-ischemic ubiquitination is neurotoxic or neuroprotective can only be answered by actively manipulating the ubiquitin system after ischemia. But this is technically hard to achieve due to the essential role of ubiquitination in cell homeostasis and the time-window after ischemia in which that would have to be done. A global reduction in ubiquitination by targeting E1 enzymes seems a possibility, but currently, available inhibitors are suboptimal due to irreversibility. Transient knock down of ubiquitin genes with CRISPR or siRNA techniques may be challenging due to the existence of four ubiquitin genes, and efficacy is not guaranteed since mRNA expression of ubiquitin genes does not change with ischemia [102]. There are too many E2/E3 combinations involved to make any approach of targeting them feasible without knowing more about substrates and activation of specific enzymes after stroke.
The Relationship Between Ubiquitin and Sumo After Ischemia
SUMO, Ubiquitin’s Cousin
The small ubiquitin-related modifier (SUMO) has in contrast to ubiquitin no direct role in protein degradation, but regulates various other cellular processes, most notably nuclear transport and organization, ribosomal biogenesis and transcription (reviewed in [152,153,154]). SUMO, like ubiquitin is attached to lysines in substrate proteins through the consecutive action of E1, E2, and E3 enzymes; however, there are some important differences to note. First, the conjugation system is far less complex with one E1 (AOS1/Uba2), one E2 (Ubc9), and about 15 known E3s that are sometimes even dispensable [155]. This is a major advantage over the ubiquitin system when it comes to manipulation of SUMO conjugation. Second, the substrate recognition is carried out independently of the E3 by the E2 enzyme Ubc9 that recognizes a well-defined consensus motif in about 75% of targets [156, 157]. This simplifies the identification of potential SUMOylation substrates compared to ubiquitin. Third, in contrast to a single ubiquitin protein, there are at least three SUMO proteins in mammalian cells [158, 159]. SUMO2 and 3 are identical except for 3 amino acids at the N-terminus, cannot be functionally distinguished, and are consequently referred to as SUMO2/3. SUMO1 shares about 50% identity with SUMO2/3. Both use the same E1, E2, and certain E3 enzymes and have overlapping and individual substrates [160]. SUMO conjugation, like ubiquitin, is essential to cell survival, as deletion of Ubc9 that is used by both SUMO types is lethal in mice [161]. Notably, SUMO2/3 conjugation is heavily induced by oxidative, hypoxic, osmotic, and heat stress [162, 163], whereas SUMO1 modification is generally not affected, with the exception of hypoxia [164, 165], a regulation that is also relevant to the brain [166].
Although ubiquitin and SUMO use different enzymes for conjugation and de-conjugation, there is an extensive interplay between the modifications. First, SUMO can act as recognition signal for ubiquitin ligases, for example RNF4 and Mip1, which process SUMOylated substrates for ubiquitination and degradation [167, 168]. Consequently, inhibition of the proteasome results in the accumulation of ubiquitin as well as SUMO conjugates [169, 170]. Second, some SUMO targets are also ubiquitinated and vice versa, acting in concert or in opposition in determining substrate fate. For example, under hypoxic conditions, SUMOylation of HIF-1α is required for its ubiquitination by VHL [171]. In contrast, in case of Mdm2 and USP25 substrates, SUMOylation and ubiquitination at the same lysine residue have opposing effects on stability and activity, respectively [172, 173]. Finally, there is increasing evidence of mixed SUMO2/3 and ubiquitin chains with degradative and non-degradative function. On the one hand, heterologous SUMO-ubiquitin chains enhance ΙκΒα degradation to initiate NF-κB activation [174]; on the other hand, such hybrid chains promote DNA repair in case of BRCA1 and RAP80 [175].
SUMO and Ubiquitin Are Related After Ischemia
The role of SUMOylation in hypoxia and ischemia has been reviewed very recently and will not be discussed in detail here [176]. Rather, I will focus on the relationship of SUMO to ubiquitin after ischemia since this could give further clues on how ubiquitination could impact the ischemic outcome. SUMO conjugation predominantly consisting of SUMO2/3 chains is neuroprotective after global and focal transient ischemia [177,178,179,180,181,182], and there are several indications that post-stroke SUMO and ubiquitin might be connected (Fig. 5). First, ischemia-reperfusion injury increases SUMO conjugation with an identical time course as ubiquitination [100, 183]. SUMO and ubiquitin are not only present at the same time in the same brain areas after stroke. They physically interact. Proteomics revealed that SUMO pull-down precipitates ubiquitin after OGD in neuroblastoma cells [184]. Our lab confirmed these data in a mouse focal ischemia model where we co-precipitated SUMO with ubiquitin and vice versa [100]. This was surprising to us as contrary to SUMO, ubiquitin was mostly found to exit the nucleus after stroke. However, recent data indicate that at least a fraction of ubiquitin localizes to the nucleus after ischemia [183]. Finally, there is some evidence that ubiquitination is dependent on prior SUMO2/3 conjugation [184] and attaches partially to the same substrates after stroke [183]. At this time, it is definitely a stretch to extrapolate the protective findings of post-stroke SUMOylation to ubiquitin; however, it is tempting to speculate that both modifications might act in concert to protect neurons from ischemic damage.
Ubiquitin and SUMO converge after cerebral ischemia. Several scenarios of how ubiquitin and SUMO interact after ischemia are conceivable and backed up by existing experimental data. First, despite temporal similarities, substrate proteins could be ubiquitinated and SUMOylated independently of each other. The majority of SUMOylated proteins are modified with SUMO2/3, only a minority with SUMO1. In this scenario, the consequences of SUMO and ubiquitin conjugation may be different for individual substrates (pathway 1). Second, ubiquitin and SUMO could be attached to the same substrate, conjointly deciding the substrate’s fate. Thereby, SUMO and ubiquitin may be attached simultaneously (pathway 2) or sequentially (pathway 3). Finally, ubiquitin and SUMO2/3 may form mixed chains on certain targets, again affecting a substrates fate together (pathway 4)
Challenges and Future Directions
The ultimate research goal in the field is certainly to provide information on how the ubiquitin system after stroke can be targeted for therapeutic and drug development. To achieve this, two major questions need to be answered. The first is how is ubiquitination induced by ischemia. It was long assumed that elevated ubiquitination is purely a consequence of proteasomal inhibition; however, recent data on ubiquitin types and timelines of ubiquitination versus proteasome function after ischemia raise doubt about whether this is indeed the case [98, 100, 123]. The view that ubiquitin conjugates serve purely degradative functions is outdated and should be revisited in stroke research as well. The second question is whether post-ischemic ubiquitination is neuroprotective or detrimental. Data collected so far suggest that changed levels of both proteasomal function and/or ubiquitination may influence stroke outcome, thus attesting causality [94, 95, 146,147,148]. However, all models used outcome-modifying treatments; therefore, an altered ubiquitin-proteasome system may be secondary to alteration of the ischemic damage. Targeted modulation of ubiquitination and proteasomal activity during or after stroke must be carried out to conclusively address causality. Considering that ubiquitin targets many proteins that will be differently regulated, it would be naïve to think that looking at global changes in tissue or cell ubiquitination levels is a good indicator of whether the modification is beneficial or detrimental. Consequently, the path forward would be to identify and characterize ubiquitination substrates and substrate-specific enzymes that are specifically active after stroke. Potential therapeutic approaches could be designed to modulate any of the enzymatic activities central to the ubiquitination system. However, since ubiquitination is essential to cell homeostasis and survival, pharmacological intervention at a broader level would likely be harmful. It would be preferable to target enzymes that confer specificity to the system, like the E2, E3, or DUB enzymes, and with that limit the negative effects of global modulation of ubiquitination. Clinical results for cancer and neurodegenerative therapy support this approach, since the most successful ubiquitin pathway drugs have targeted E3 ligases and DUB enzymes (reviewed in [185]). Another appealing approach might be the targeting of individual substrates themselves, for example by inhibition or promotion of binding to ubiquitination enzymes, depending on outcome prediction.
Conclusions
Post-ischemic protein ubiquitination is rapidly and transiently increased in neurons at the periphery of the ischemic territory, a process dependent on reperfusion. Given the importance of ubiquitination for cell homeostasis and stress responses, the impact of this change for neuronal survival and functioning must be significant. There are precedents for therapies targeting the ubiquitination system in cancer and neurodegeneration, which suggests that such interventions may also be applicable to stroke. However, currently, we know little about how ubiquitination is induced after cerebral ischemia, what enzymes are involved and what is the impact of ubiquitination on neuronal fate and ultimately stroke outcome. A better understanding of the roles of ubiquitin in ischemic stroke is necessary before the exploration of modulating ubiquitination for therapeutic purposes can begin.
References
Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–40.
Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980;77(3):1365–8.
Hershko A, Eytan E, Ciechanover A, Haas AL. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982;257(23):13964–70.
Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4(3):192–201.
Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4(11):855–64.
Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3(4):255–61.
Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–5.
Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–86.
Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72.
Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci U S A. 2004;101(32):11707–12.
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34.
Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–86.
Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25(20):4877–87.
Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29.
Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24(5):701–11.
Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127(7):1401–13.
Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282(24):17375–86.
Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–6.
Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195–201.
Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270(29):17442–56.
Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–45.
Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37(Pt 5):937–53.
Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–73.
Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9(10):865–75.
Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279(8):7055–63.
Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324(4):637–47.
Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 a resolution. Nature. 1997;386(6624):463–71.
Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, et al. The structure of the mammalian 20S proteasome at 2.75 a resolution. Structure. 2002;10(5):609–18.
Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299(5605):408–11.
Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;3(151):ra88.
Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94(5):615–23.
Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118(1):99–110.
Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–5.
Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–7.
Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1(4):221–6.
Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem. 1992;267(31):22369–77.
Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21(13):3516–25.
Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, et al. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem. 1994;115(2):257–69.
Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432(3):585–94.
Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2(3):179–87.
Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, Gomez-Ramos P, Moran MA, Castano JG, et al. Neuronal induction of the immunoproteasome in Huntington's disease. J Neurosci. 2003;23(37):11653–61.
Orre M, Kamphuis W, Dooves S, Kooijman L, Chan ET, Kirk CJ, et al. Reactive glia show increased immunoproteasome activity in Alzheimer's disease. Brain. 2013;136(Pt 5):1415–31.
Puttaparthi K, Elliott JL. Non-neuronal induction of immunoproteasome subunits in an ALS model: possible mediation by cytokines. Exp Neurol. 2005;196(2):441–51.
Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140(4):567–78.
Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2009;284(39):26655–65.
Jarome TJ, Kwapis JL, Ruenzel WL, Helmstetter FJ. CaMKII, but not protein kinase a, regulates Rpt6 phosphorylation and proteasome activity during the formation of long-term memories. Front Behav Neurosci. 2013;7:115.
Lin JT, Chang WC, Chen HM, Lai HL, Chen CY, Tao MH, et al. Regulation of feedback between protein kinase a and the proteasome system worsens Huntington's disease. Mol Cell Biol. 2013;33(5):1073–84.
Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282(31):22460–71.
Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44(42):13893–901.
Liu K, Paterson AJ, Zhang F, McAndrew J, Fukuchi K, Wyss JM, et al. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J Neurochem. 2004;89(4):1044–55.
Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115(6):715–25.
Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci U S A. 1999;96(11):6223–8.
Wang D, Fang C, Zong NC, Liem DA, Cadeiras M, Scruggs SB, et al. Regulation of acetylation restores proteolytic function of diseased myocardium in mouse and human. Mol Cell Proteomics. 2013;12(12):3793–802.
Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33(10):1159–76.
Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38(5):733–45.
Bianchetta MJ, Lam TT, Jones SN, Morabito MA. Cyclin-dependent kinase 5 regulates PSD-95 ubiquitination in neurons. J Neurosci. 2011;31(33):12029–35.
Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–35.
Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010;30(49):16718–29.
Zemoura K, Trumpler C, Benke D. Lys-63-linked ubiquitination of gamma-aminobutyric acid (GABA), type B1, at multiple sites by the E3 ligase mind bomb-2 targets GABAB receptors to lysosomal degradation. J Biol Chem. 2016;291(41):21682–93.
Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–61.
Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–48.
Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51.
Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–9.
Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6(3):231–42.
Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74(6):1023–30.
Lopez-Salon M, Alonso M, Vianna MR, Viola H. Mello e Souza T, Izquierdo I et al. the ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14(11):1820–6.
Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40(3):595–607.
Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M, et al. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci. 2012;15(12):1655–66.
Lussier MP, Nasu-Nishimura Y, Roche KW. Activity-dependent ubiquitination of the AMPA receptor subunit GluA2. J Neurosci. 2011;31(8):3077–81.
Jurd R, Thornton C, Wang J, Luong K, Phamluong K, Kharazia V, et al. Mind bomb-2 is an E3 ligase that ubiquitinates the N-methyl-D-aspartate receptor NR2B subunit in a phosphorylation-dependent manner. J Biol Chem. 2008;283(1):301–10.
Saliba RS, Michels G, Jacob TC, Pangalos MN, Moss SJ. Activity-dependent ubiquitination of GABA(a) receptors regulates their accumulation at synaptic sites. J Neurosci. 2007;27(48):13341–51.
Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32(6):1013–26.
Watts RJ, Hoopfer ED, Luo L. Axon pruning during drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38(6):871–85.
Bond U, Schlesinger MJ. Ubiquitin is a heat shock protein in chicken embryo fibroblasts. Mol Cell Biol. 1985;5(5):949–56.
Fornace AJ Jr, Alamo I Jr, Hollander MC, Lamoreaux E. Ubiquitin mRNA is a major stress-induced transcript in mammalian cells. Nucleic Acids Res. 1989;17(3):1215–30.
Kaneko M, Iwase I, Yamasaki Y, Takai T, Wu Y, Kanemoto S, et al. Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci Rep. 2016;6:30955.
Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–5.
Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9(2):543–50.
Shang F, Gong X, Taylor A. Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently up-regulated. J Biol Chem. 1997;272(37):23086–93.
Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843(1):182–96.
Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–9.
Pickart CM. Ubiquitin and the stress response. In: Latchman DS, editor. Stress Proteins. Berlin, Heidelberg: Springer Berlin Heidelberg; 1999. p. 133–52.
Preston GM, Brodsky JL. The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem J. 2017;474(4):445–69.
Arnason T, Ellison MJ. Stress resistance in Saccharomyces Cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14(12):7876–83.
Silva GM, Finley D, Vogel C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol. 2015;22(2):116–23.
Hayashi T, Takada K, Matsuda M. Changes in ubiquitin and ubiquitin-protein conjugates in the CA1 neurons after transient sublethal ischemia. Mol Chem Neuropathol. 1991;15(1):75–82.
Hayashi T, Takada K, Matsuda M. Post-transient ischemia increase in ubiquitin conjugates in the early reperfusion. Neuroreport. 1992;3(6):519–20.
Hu BR, Janelidze S, Ginsberg MD, Busto R, Perez-Pinzon M, Sick TJ, et al. Protein aggregation after focal brain ischemia and reperfusion. J Cereb Blood Flow Metab. 2001;21(7):865–75.
Hochrainer K, Jackman K, Anrather J, Iadecola C. Reperfusion rather than ischemia drives the formation of ubiquitin aggregates after middle cerebral artery occlusion. Stroke. 2012;43(8):2229–35.
Gubellini P, Bisso GM, Ciofi-Luzzatto A, Fortuna S, Lorenzini P, Michalek H, et al. Ubiquitin-mediated stress response in a rat model of brain transient ischemia/hypoxia. Neurochem Res. 1997;22(1):93–100.
Vannucci SJ, Mummery R, Hawkes RB, Rider CC, Beesley PW. Hypoxia-ischemia induces a rapid elevation of ubiquitin conjugate levels and ubiquitin immunoreactivity in the immature rat brain. J Cereb Blood Flow Metab. 1998;18(4):376–85.
Hu BR, Martone ME, Jones YZ, Liu CL. Protein aggregation after transient cerebral ischemia. J Neurosci. 2000;20(9):3191–9.
Liu CL, Martone ME, Hu BR. Protein ubiquitination in postsynaptic densities after transient cerebral ischemia. J Cereb Blood Flow Metab. 2004;24(11):1219–25.
Liu JJ, Zhao H, Sung JH, Sun GH, Steinberg GK. Hypothermia blocks ischemic changes in ubiquitin distribution and levels following stroke. Neuroreport. 2006;17(16):1691–5.
Liu C, Chen S, Kamme F, Hu BR. Ischemic preconditioning prevents protein aggregation after transient cerebral ischemia. Neuroscience. 2005;134(1):69–80.
Morimoto T, Ide T, Ihara Y, Tamura A, Kirino T. Transient ischemia depletes free ubiquitin in the gerbil hippocampal CA1 neurons. Am J Pathol. 1996;148(1):249–57.
Hayashi T, Takada K, Matsuda M. Subcellular distribution of ubiquitin-protein conjugates in the hippocampus following transient ischemia. J Neurosci Res. 1992;31(3):561–4.
Iwabuchi M, Sheng H, Thompson JW, Wang L, Dubois LG, Gooden D, et al. Characterization of the ubiquitin-modified proteome regulated by transient forebrain ischemia. J Cereb Blood Flow Metab. 2014;34(3):425–32.
Kamikubo T, Hayashi T, Ohkawa K. Lack of effect of transient ischemia on ubiquitin conjugation. Neurochem Res. 1995;20(4):391–4.
Hochrainer K, Jackman K, Benakis C, Anrather J, Iadecola C. SUMO2/3 is associated with ubiquitinated protein aggregates in the mouse neocortex after middle cerebral artery occlusion. J Cereb Blood Flow Metab. 2015;35(1):1–5.
Bedford L, Layfield R, Mayer RJ, Peng J, Xu P. Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurones. Neurosci Lett. 2011;491(1):44–7.
Noga M, Hayashi T. Ubiquitin gene expression following transient forebrain ischemia. Brain Res Mol Brain Res. 1996;36(2):261–7.
Gautam V, Trinidad JC, Rimerman RA, Costa BM, Burlingame AL, Monaghan DT. Nedd4 is a specific E3 ubiquitin ligase for the NMDA receptor subunit GluN2D. Neuropharmacology. 2013;74:96–107.
Hung AY, Sung CC, Brito IL, Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 2010;5(3):e9842.
Kato A, Rouach N, Nicoll RA, Bredt DS. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci U S A. 2005;102(15):5600–5.
Cabral-Miranda F, Nicoloso-Simoes E, Adao-Novaes J, Chiodo V, Hauswirth WW, Linden R, et al. rAAV8-733-mediated gene transfer of CHIP/stub-1 prevents hippocampal neuronal death in experimental brain ischemia. Mol Ther. 2017;25(2):392–400.
Lackovic J, Howitt J, Callaway JK, Silke J, Bartlett P, Tan SS. Differential regulation of Nedd4 ubiquitin ligases and their adaptor protein Ndfip1 in a rat model of ischemic stroke. Exp Neurol. 2012;235(1):326–35.
Howitt J, Lackovic J, Low LH, Naguib A, Macintyre A, Goh CP, et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J Cell Biol. 2012;196(1):29–36.
Mengesdorf T, Jensen PH, Mies G, Aufenberg C, Paschen W. Down-regulation of parkin protein in transient focal cerebral ischemia: a link between stroke and degenerative disease? Proc Natl Acad Sci U S A. 2002;99(23):15042–7.
Tang J, Hu Z, Tan J, Yang S, Zeng L. Parkin protects against oxygen-glucose deprivation/reperfusion insult by promoting Drp1 degradation. Oxidative Med Cell Longev. 2016;2016:8474303.
Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40(6):1542–7.
Kumar R, Jangir DK, Verma G, Shekhar S, Hanpude P, Kumar S, et al. S-nitrosylation of UCHL1 induces its structural instability and promotes alpha-synuclein aggregation. Sci Rep. 2017;7:44558.
Zhang M, Cai F, Zhang S, Zhang S, Song W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer's progression in vivo. Sci Rep. 2014;4:7298.
Liu H, Li W, Ahmad M, Miller TM, Rose ME, Poloyac SM, et al. Modification of ubiquitin-C-terminal hydrolase-L1 by cyclopentenone prostaglandins exacerbates hypoxic injury. Neurobiol Dis. 2011;41(2):318–28.
Shen H, Sikorska M, Leblanc J, Walker PR, Liu QY. Oxidative stress regulated expression of ubiquitin carboxyl-terminal hydrolase-L1: role in cell survival. Apoptosis. 2006;11(6):1049–59.
Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111(2):209–18.
Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12(16):1945–58.
Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84.
Lee JH, Shin SK, Jiang Y, Choi WH, Hong C, Kim DE, et al. Facilitated tau degradation by USP14 aptamers via enhanced proteasome activity. Sci Rep. 2015;5:10757.
Min JW, Lu L, Freeling JL, Martin DS, Wang H. USP14 inhibitor attenuates cerebral ischemia/reperfusion-induced neuronal injury in mice. J Neurochem. 2017;140(5):826–33.
Asai A, Tanahashi N, Qiu JH, Saito N, Chi S, Kawahara N, et al. Selective proteasomal dysfunction in the hippocampal CA1 region after transient forebrain ischemia. J Cereb Blood Flow Metab. 2002;22(6):705–10.
Caldeira MV, Curcio M, Leal G, Salazar IL, Mele M, Santos AR, et al. Excitotoxic stimulation downregulates the ubiquitin-proteasome system through activation of NMDA receptors in cultured hippocampal neurons. Biochim Biophys Acta. 2013;1832(1):263–74.
Ge P, Luo Y, Liu CL, Hu B. Protein aggregation and proteasome dysfunction after brain ischemia. Stroke. 2007;38(12):3230–6.
Kamikubo T, Hayashi T. Changes in proteasome activity following transient ischemia. Neurochem Int. 1996;28(2):209–12.
Keller JN, Huang FF, Zhu H, Yu J, Ho YS, Kindy TS. Oxidative stress-associated impairment of proteasome activity during ischemia-reperfusion injury. J Cereb Blood Flow Metab. 2000;20(10):1467–73.
Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Modulation of p53 degradation via MDM2-mediated ubiquitylation and the ubiquitin-proteasome system during reperfusion after stroke: role of oxidative stress. J Cereb Blood Flow Metab. 2005;25(2):267–80.
Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci 2010;3:12.
Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–78.
Aiken CT, Kaake RM, Wang X, Huang L. Oxidative stress-mediated regulation of proteasome complexes. Mol Cell Proteomics. 2011;10(5):R110 006924.
Huang Q, Wang H, Perry SW, Figueiredo-Pereira ME. Negative regulation of 26S proteasome stability via calpain-mediated cleavage of Rpn10 subunit upon mitochondrial dysfunction in neurons. J Biol Chem. 2013;288(17):12161–74.
Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276(32):30057–63.
Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch Biochem Biophys. 2006;453(1):135–42.
Li Y, Luo Y, Luo T, Lu B, Wang C, Zhang Y, et al. Trehalose inhibits protein aggregation caused by transient ischemic insults through preservation of proteasome activity, not via induction of autophagy. Mol Neurobiol. 2016; https://doi.org/10.1007/s12035-016-0196-5.
Liu Y, Lu L, Hettinger CL, Dong G, Zhang D, Rezvani K, et al. Ubiquilin-1 protects cells from oxidative stress and ischemic stroke caused tissue injury in mice. J Neurosci. 2014;34(8):2813–21.
Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18(5):538–43.
Buchan AM, Li H, Blackburn B. Neuroprotection achieved with a novel proteasome inhibitor which blocks NF-kappaB activation. Neuroreport. 2000;11(2):427–30.
Doeppner TR, Kaltwasser B, Kuckelkorn U, Henkelein P, Bretschneider E, Kilic E, et al. Systemic proteasome inhibition induces sustained post-stroke neurological recovery and neuroprotection via mechanisms involving reversal of peripheral immunosuppression and preservation of blood-brain-barrier integrity. Mol Neurobiol. 2016;53(9):6332–41.
Doeppner TR, Mlynarczuk-Bialy I, Kuckelkorn U, Kaltwasser B, Herz J, Hasan MR, et al. The novel proteasome inhibitor BSc2118 protects against cerebral ischaemia through HIF1A accumulation and enhanced angioneurogenesis. Brain. 2012;135(Pt 11):3282–97.
Phillips JB, Williams AJ, Adams J, Elliott PJ, Tortella FC. Proteasome inhibitor PS519 reduces infarction and attenuates leukocyte infiltration in a rat model of focal cerebral ischemia. Stroke. 2000;31(7):1686–93.
Williams AJ, Hale SL, Moffett JR, Dave JR, Elliott PJ, Adams J, et al. Delayed treatment with MLN519 reduces infarction and associated neurologic deficit caused by focal ischemic brain injury in rats via antiinflammatory mechanisms involving nuclear factor-kappaB activation, gliosis, and leukocyte infiltration. J Cereb Blood Flow Metab. 2003;23(1):75–87.
Zhang L, Zhang ZG, Buller B, Jiang J, Jiang Y, Zhao D, et al. Combination treatment with VELCADE and low-dose tissue plasminogen activator provides potent neuroprotection in aged rats after embolic focal ischemia. Stroke. 2010;41(5):1001–7.
Qiu JH, Asai A, Chi S, Saito N, Hamada H, Kirino T. Proteasome inhibitors induce cytochrome c-caspase-3-like protease-mediated apoptosis in cultured cortical neurons. J Neurosci. 2000;20(1):259–65.
Meller R, Thompson SJ, Lusardi TA, Ordonez AN, Ashley MD, Jessick V, et al. Ubiquitin proteasome-mediated synaptic reorganization: a novel mechanism underlying rapid ischemic tolerance. J Neurosci. 2008;28(1):50–9.
Zhang X, Yan H, Yuan Y, Gao J, Shen Z, Cheng Y, et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9(9):1321–33.
Liu C, Gao Y, Barrett J, Hu B. Autophagy and protein aggregation after brain ischemia. J Neurochem. 2010;115(1):68–78.
Yamashita K, Eguchi Y, Kajiwara K, Ito H. Mild hypothermia ameliorates ubiquitin synthesis and prevents delayed neuronal death in the gerbil hippocampus. Stroke. 1991;22(12):1574–81.
Ide T, Takada K, Qiu JH, Saito N, Kawahara N, Asai A, et al. Ubiquitin stress response in postischemic hippocampal neurons under nontolerant and tolerant conditions. J Cereb Blood Flow Metab. 1999;19(7):750–6.
Liang J, Yao J, Wang G, Wang Y, Wang B, Ge P. Ischemic Postconditioning protects neuronal death caused by cerebral ischemia and reperfusion via attenuating protein aggregation. Int J Med Sci. 2012;9(10):923–32.
Ouyang YB, Xu L, Giffard RG. Geldanamycin treatment reduces delayed CA1 damage in mouse hippocampal organotypic cultures subjected to oxygen glucose deprivation. Neurosci Lett. 2005;380(3):229–33.
Hayashi T, Tanaka J, Kamikubo T, Takada K, Matsuda M. Increase in ubiquitin conjugates dependent on ischemic damage. Brain Res Mol Brain Res. 1993;620(1):171–3.
Velickovska V, van Breukelen F. Ubiquitylation of proteins in livers of hibernating golden-mantled ground squirrels, Spermophilus Lateralis. Cryobiology. 2007;55(3):230–5.
Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19(3):350–5.
Cubenas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell. 2013;24(1):1–12.
Ptak C, Wozniak RW. SUMO and nucleocytoplasmic transport. Adv Exp Med Biol. 2017;963:111–26.
Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276(38):35368–74.
Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–9.
Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinformatics. 2008;9:8.
Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 Pt 1):1457–70.
Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ET. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273(18):11349–53.
Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5(12):2298–310.
Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9(6):769–79.
Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275(9):6252–8.
Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2(72):ra24.
Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131(2):309–23.
Kang X, Li J, Zou Y, Yi J, Zhang H, Cao M, et al. PIASy stimulates HIF1alpha SUMOylation and negatively regulates HIF1alpha activity in response to hypoxia. Oncogene. 2010;29(41):5568–78.
Shao R, Zhang FP, Tian F, Anders Friberg P, Wang X, Sjoland H, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569(1–3):293–300.
Sobko A, Ma H, Firtel RA. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev Cell. 2002;2(6):745–56.
Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10(5):538–46.
Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics. 2008;7(11):2107–22.
Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282(47):34167–75.
Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131(3):584–95.
Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101(7):753–62.
Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30(5):610–9.
Aillet F, Lopitz-Otsoa F, Egana I, Hjerpe R, Fraser P, Hay RT, et al. Heterologous SUMO-2/3-ubiquitin chains optimize IkappaBalpha degradation and NF-kappaB activity. PLoS One. 2012;7(12):e51672.
Guzzo CM, Berndsen CE, Zhu J, Gupta V, Datta A, Greenberg RA, et al. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal. 2012;5(253):ra88.
Peters M, Wielsch B, Boltze J. The role of SUMOylation in cerebral hypoxia and ischemia. Neurochem Int. 2017;107:66–77.
Bernstock JD, Lee YJ, Peruzzotti-Jametti L, Southall N, Johnson KR, Maric D, et al. A novel quantitative high-throughput screen identifies drugs that both activate SUMO conjugation via the inhibition of microRNAs 182 and 183 and facilitate neuroprotection in a model of oxygen and glucose deprivation. J Cereb Blood Flow Metab. 2016;36(2):426–41.
Cimarosti H, Ashikaga E, Jaafari N, Dearden L, Rubin P, Wilkinson KA, et al. Enhanced SUMOylation and SENP-1 protein levels following oxygen and glucose deprivation in neurones. J Cereb Blood Flow Metab. 2012;32(1):17–22.
Datwyler AL, Lattig-Tunnemann G, Yang W, Paschen W, Lee SL, Dirnagl U, et al. SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab. 2011;31(11):2152–9.
Lee YJ, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS One. 2011;6(10):e25852.
Yang W, Sheng H, Warner DS, Paschen W. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab. 2008;28(5):892–6.
Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28(2):269–79.
Yang W, Sheng H, Thompson JW, Zhao S, Wang L, Miao P, et al. Small ubiquitin-like modifier 3-modified proteome regulated by brain ischemia in novel small ubiquitin-like modifier transgenic mice: putative protective proteins/pathways. Stroke. 2014;45(4):1115–22.
Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, et al. Analysis of oxygen/glucose-deprivation-induced changes in SUMO3 conjugation using SILAC-based quantitative proteomics. J Proteome Res. 2012;11(2):1108–17.
Huang X, Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26(4):484–98.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares that she has no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by the author.
Rights and permissions
About this article
Cite this article
Hochrainer, K. Protein Modifications with Ubiquitin as Response to Cerebral Ischemia-Reperfusion Injury. Transl. Stroke Res. 9, 157–173 (2018). https://doi.org/10.1007/s12975-017-0567-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-017-0567-x