Abstract
The variable cowpea productivity across different environments demands evaluating the performance of genotypes in a breeding program prior to their release. The aim of this study was to assess yield stability of eight cowpea advanced breeding lines selected from participatory varietal selection in multilocational trials, and to identify mega-environments for cowpea production in Ghana. The genotypes were evaluated across five environments in 2016 and 2017 in randomized complete block design with three replications. The GEA-R version 4.0 software was used for genotype main effect plus genotype by environment interaction (GGE) biplot analyses. Analysis of variance (PROC GLM of SAS using a RANDOM statement with the TEST option) detected significant variations for location, year, genotype, environment, and their interactions. The results showed that the yield performances of the cowpea genotypes were highly influenced by genotype × environment interaction effects. The principal component 1 (PC1) and PC2 were significant components which accounted for 46.75% and 22.84% of GGE sum of squares, respectively. We showed for the first time, two mega-environments for cowpea production and testing in the major cowpea production agro-ecologies in Ghana. The genotypes SARI-6-2-6 and IT07K-303-1 were adapted to Damongo, Nyankpala, and Tumu, whereas SARI-2-50-80 was adapted to Yendi and Manga. The best ranking location was Damongo followed by Tumu, and Nyankpala. The high-yielding genotypes, IT86D-610, IT10K-837-1, IT07K-303-1, and SARI-2-50-80 had significant higher grain yields than the check (Bawutawuta) and were recommended for release as cultivars (or as breeding lines) to boost cowpea production in Ghana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea (Vigna unguiculata (L.) Walp) is one of the most important grain legumes and a valuable component of the traditional cropping systems in the sub-Saharan Africa (SSA) (Singh et al. 2002). In Ghana, it is grown in diverse agro-ecological environments. However, the Guinea and Sudan savanna ecologies produce over 90% of total annual output for the nation, due to favorable environmental conditions. Cowpea plays a vital role in the livelihoods of the small-holder farmers through its contributions to their food and nutritional security, income generation, soil fertility enhancement and provision of biomass for crop–livestock integration (Boukar et al. 2016).
Despite the numerous importance of cowpea, its yield ranges below 0.6 t/ha on farmer fields in the savanna ecologies of West Africa, compared to its potential yield of over 2.0 t/ha (Boukar et al. 2018; Singh 2020). Owusu et al. (2018) attributed the abysmal performance of cowpea on farmer fields to inadequate improved varieties; they indicated that several farmers still cultivate landraces in Ghana. According to Padi (2004), yields of cowpea genotypes show specific responses to environmental conditions which increase local adaptation but limit their usefulness in other environments. Environmental differences affect crop growth, development and yield due to significant genotype × environment interactions (GE); hence, varieties developed for one particular environment may not perform well in other environments (Luo et al. 2013). The variable cowpea productivities across different environments demand assessing the performance of genotypes in a crop improvement program prior to their release.
Genotype (G) × environment (E) interaction is defined as a change in relative performance of a genotype from one environment to another. It has been used to identify responses of genotypes to different environments. The study of G × E interactions will guide breeders to develop strategies for testing and selecting genotypes most adapted to the target environments (Kimbeng et al. 2009). Genotypes which have stable mean yields across the testing environments are said to be adaptable. On the other hand, those with high-yielding genetic potential only in desirable environmental conditions but low-yielding potential in an undesirable one are genotypes with finite adaptability (Lin and Bins 1991). Genotype × environment interactions (GEI) are the differential responses of genotypes across different environments.
Genotype × environment interaction assessment is carried out using several methods including additive main effect and multiplicative interaction (AMMI), the genotype main effect and genotype–environment interaction (GGE biplot), Finlay–Wilkinson model, Eberhart and Russell model, and so on (Yan and Tinker 2006). AMMI analysis is commonly used to determine GEI for field trials primarily for yield. However, AMMI biplot’s application is limited. Yan et al. (2007) noted that the GGE biplot is more efficient than the AMMI graph in mega-environment analysis and genotype evaluation since it provides more information on G + GE and has the inner product property of the biplot. Their study also revealed that the discriminating power vs. representativeness view of the GGE biplot is effective in evaluating test environments, which cannot be achieved via AMMI analysis. The GGE biplot model utilizes multi-region data for environmental evaluation and provides better graphical illustration (Yan and Holland 2010). It provides a better understanding of complex G × E interactions in multi-environment trials of genotypes and agronomic experiments. GGE biplot has been used to identify the performance of crop cultivars under multiple stress environments, ideal cultivars, mega-environments, and core testing sites. It has also been successfully used in experiments for many crops such as peanut (Chen et al. 2009), soybean (Zhou et al. 2011), and sugarcane (Luo et al. 2015; Sousa et al. 2018).
Even though international and national cowpea improvement programs have developed and released some improved cowpea varieties, there is still the need to develop more varieties which are resilient to current climatic challenges and more adaptable to the various agro-ecologies with suitable consumer’s preferences for maximum returns.
In this study, eight advanced cowpea breeding lines were evaluated for their adaptability and yield stabilities in five major cowpea producing locations in the Guinea and Sudan savanna ecologies of Ghana in 2016 and 2017.
Materials and methods
Site description
The experiments were conducted during 2016 and 2017 cropping seasons at five locations in the Guinea and Sudan Savanna ecologies of Ghana under farmer field conditions. The locations of trials included Nyankpala (9.254° N, 0.584° W; 560 m.a.s.l), Yendi (9.535° N, 0.0091° W; 681 m.a.s.l), Damongo (9.014°N, 01.049° W; 189.1 m.a.s.l), Manga (10.273° N, 0.422° W; 712 m.a.s.l), and Tumu (10.879° N, 1.983° W; 1033 m.a.s.l).
Plant materials
Eight early-to-medium maturing cowpea advanced breeding lines, IT10K-837-1, IT86D-610, IT07K-303-1 (developed by the International Institute of Tropical Agriculture, (IITA), SARI-3-2-50-80, SARI-5-5-5, SARI-6-2-6, SARI-6-2-9 (developed by CSIR-Savanna Agricultural Research Institute (CSIR-SARI) and a check, Bawutawuta (a cultivar released by CSIR-SARI) were used in the present study. These genotypes were selected from a multi-location participatory variety selection trials conducted by the cowpea improvement program of CSIR-SARI.
Experimental layout
Each trial was established using a randomized complete block design with three replicates. The experimental plots were made up of four rows that were 5 m long and spaced 0.6 m between rows and 0.2 m within row. Field pests were controlled using the insecticide, K-Optimal (Cyhalothrin 15 g/l + Acetamiprid 20; EC) at the rate of 500 ml per ha at the vegetative, flowering, and podding stages of the crops. Manual weed control was carried out as and when necessary. No fertilizer was applied. At harvest, pods on the two inner rows were hand-picked, dried and threshed.
Data collected
Data were collected on maturity traits, namely number of days to 50% flowering (DFF), from day of planting to the day 50% of the plants on each plot flowered and the number of days to 90% pod maturity (DM) was determined from the day of planting to 90% of the pods on each plot change color. On the other hand, data on the following yield components were collected from five randomly tagged plants: number of pods per plant (Pods_PLT) was counted from the five plants; number of seeds per pod (Seed_pod), five pods were randomly selected from each of the five selected plants and the number of seeds was counted; pod length (Pod_L cm) of five randomly selected pods from each of the five plants was measured in cm using a tape measure; hundred-seed weight (HSW) was determined in grams from the weight of 100 randomly selected dried seeds; pod yield (Pod wt t/ha) and grain yield (GY t/ha) were determined as average weight of pods and seeds harvested in net plot, respectively.
Statistical analysis
Combined analysis of variance was performed for each location across years and consequently, combined analysis of variance for the data across environments was performed on plot means for traits measured with PROC GLM of SAS using a RANDOM statement with the TEST option (SAS Institute 2011).
The GEA-R (Genotype × Environment Analyses with R for Windows) version 4.0 (Pacheco et al. 2016) was used for stability analyses for grain yield. The model for genotype by trait (GT) biplot used is presented as
where Yij is the genetic value of the combination between inbred i and trait j; μ is the mean of all combinations involving trait j; βj is the main effect of trait j; λ1 and λ2 are the singular values for PC1 and PC2; gi1 and gi2 are the PC1 and PC2 eigenvectors, respectively, for inbred I; e1j and e2j are the PC1 and PC2 eigenvectors, respectively, for trait j: dj is the phenotypic standard deviation (with mean of zero and standard deviation of 1); and εij is the residual of the model associated with the combination of inbred i and trait j. The data were not transformed (‘Transform = 0’), but were standard deviation-standardized (‘Scale = 1’), and trait-centered (‘centering = 2’). Therefore, the outputs are appropriate for visualizing the relationships among genotypes and traits.
Results and discussion
Analysis of variance
The analysis of variance (ANOVA) for the cowpea genotypes studied varied for grain yield and most of the measured traits (Table 1). Similar to the results under the various locations, the ANOVA of the eight cowpea genotypes (G) for traits measured across five multi-environment tests (METs) revealed a significant mean square for location, year, genotype and environment for most of the traits examined (Table 2). Also, the interactions between genotypes and environments (G × E) were significant for most of the traits measured except for the number of pods per plant, seed per pod, pod length, biomass and hundred seed weight. The significant G × E observed for grain yield justified the use of stability analysis to determine genotypes with consistence performance of high yield (Yan and Tinker 2006).
The sum of squares for G × E interaction was less than that of genotype and environment (Table 2). This shows that genotypes and environments are both vital in governing the expression of grain yield (Gedif et al. 2014). Contrary to this study, other researches established that GEI effects were higher than those of genotype and the environment (Bhartiya et al. 2017) while Cravero et al. (2010) and Suwarto (2010) reported that environmental effect was three times larger than the genotype and genotype × environment effects.
Performance of genotypes across individual locations
The mean values of the genotypes varied significantly for all the variables measured. The genotypes SARI-5-5-5 and SARI-6-2-9 flowered earlier than the rest while the entry IT07K-303-1 flowered later. Days to maturity had a similar trend (Table 3). The days to flowering and days to maturity had an impact on the yields of cowpea genotypes evaluated. Even though the differences between the early and the late maturing genotypes were 5 days, they had implications on yields. The higher grain yields observed for the late flowering and days to maturity genotypes indicate that the late maturing genotypes used the extra days to accumulate more photosynthate which was partitioned into grain yield. This corroborates the findings of Kamai et al. (2014). The results further suggested that even though the early maturing genotypes provide food for “hunger period” and also mitigate the effect of terminal drought, this comes with significant yield penalties. Therefore, marker-assisted backcrossing is recommended for the development of early maturing varieties of cowpea to recover the genetic background of the high-yielding cultivars and also to reduce linkage drag that might be associated with earliness.
For grain yield, SARI-5-5-5 (1.30 t/ha) had the lowest yield while the genotype IT86D-610 (2.08 t/ha) had the highest yield followed by IT10K-873-1 (2.03 t/ha) and SARI-2-50-80 (1.90 t/ha). Genotypes IT86D-610, SARI-2-50-80, IT07K-303-1 and IT10K-837-1 were significantly higher than the check (Bawutawuta; 1.60 t/ha), implying some gain in grain yield has been achieved.
Genotype × environment interaction analysis using GGE biplot analysis
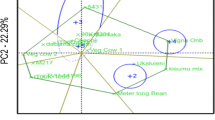
The significant genotype (G) × environment (E) mean squares for grain yield across the test locations (Fig. 1) implied that GGE biplot could be used to assess G × GE interaction effects as the genotypes performed differently across the study locations. The principal component axis 1 (PC1) accounted for 46.75% of total variation while the principal component axis 2 (PC2) accounted for 22.84%. The two principal components explained 69.59% of the total variations for grain yields (Fig. 1) which was relatively higher than the results obtained by Sousa et al. (2018). In their study, the first and second principal components (PC1 and PC2) accounted for 66% of the variation caused.
GGE biplot is an essential tool for addressing the mega-environment issues to show which genotype won in which environments. It is an effective visual tool in mega-environment identification (Yan et al. 2000). The term mega-environment analysis indicates the partition of a crop-growing region into different target agro-ecological zones.
The GGE biplot showing the mega-environments and their respective highest performing genotypes, and also displaying the “which-won-where pattern” as a concise summary of the GEI (Fig. 1). GGE biplot is an important tool used for addressing the mega-environment issues, by showing which genotype won in which environments, and mega-environment identification. The mega-environment differentiates and specifies adaptation of a genotype (Rakshit et al. 2012). The GGE biplot is made up of an irregular polygon and perpendicular lines drawn from the biplot origin (Gauch and Zobel 1996). These perpendicular lines divide the biplot into several sectors. In the present study, four lines in Fig. 1 divided the biplot into four distinct sectors, and the environments fall into only two of them. The vertex genotypes in this study were genotypes, SARI-5-5-5, SARI-6-2-9, IT10K-837-1, and IT07K-303-1. Yan and Tinker (2006) stated that the vertex genotypes were the most responsive genotypes because they are far away from the origin. Whereas Sbongeleni et al. (2019) indicated that varieties located at the vertex of the sector are considered the best-performing varieties in the mega-environments. Three environments (Damongo, Tumu and Nyankpala) fell into the first mega-environment. The vertex genotypes for this mega-environment were SARI-6-2-6 and IT07K-303-1 suggesting that these are the most responsive genotypes for these three environments (mega-environment). Two environments (Yendi and Manga) fell into the second mega-environment and the vertex genotype for this mega-environment was genotype SARI-5-5-5, while SARI-2-50-80 found in the same environments also performed well. On the other hand, genotypes (Bawutawuta, IT86D-610, IT10K-837-1, and SARI-6-2-9) were not adapted to any environment suggesting that those genotypes were poorly adapted in this study.
The ideal environment for cultivating any crop should have at least two factors of which one is to be highly discriminating of the cultivar while the other should be representative of the target location (Zhang et al. 2016). Discrimination is the situation whereby the locations used in the study can exploit the variance among candidate cultivars (Blanche and Myers 2006). On the other hand, representativeness displays the location which represents conditions of the other locations (Zhang et al. 2016). With efficient use of GGE biplot tool, genotype(s) that is (are) high yielding and stable can be identified from field trial experiments as was employed in the current study.
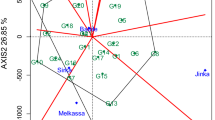
Discrimination and representativeness
The smaller circle represents the ideal environment which depends on the mean coordinates of all testing locations (Fig. 2). There was a positive correlation between the location vector length and the location discrimination ability, and negative correlation between the angle existing in location vector with ideal location and the location’s representativeness of the target environment, corroborating the study of Yan (2010). The study showed Damongo was the best-ranked location followed by Tumu and then Nyankpala. Though Nyankpala was identified as the most discriminating environment with the longest vector, Damongo presented the overall best location for cultivating cowpea in the Guinea and Sudan savanna ecologies.
Ranking of genotypes
The genotypes IT07K-303-1 and SARI-6-2-6 were the best ranking genotypes (Fig. 3). An ideal genotype is the one controlled by only one factor. The distance from ideal genotypes decreases either with mean yield or stability or both (Kumar et al. 2018). The distance is measured as an indicator of ranking genotypes under field evaluations.
Means vs. stability
Yield performance and stability of the eight tested cowpea genotypes were graphically presented through GGE biplot (Fig. 4). This could be evaluated by the average environmental coordinate (AEC) method (Yan 2002). The straight line passing through AEC with the biplot origin is as AEC abscissa and the straight line through the origin and perpendicular biplot is as AEC ordinate. Directions to the AEC ordinate that move away from the origin biplot showed increased stability. AEC ordinate divided the genotypes under and above the general yield average.
The two high-yielding cowpea genotypes, SARI-6-2-6 and IT07K-303-1, and performed above the general average yield were adapted to the same environment (Fig. 4). Even though genotypes IT86D-610 and IT10K-837-1 relatively performed above the average yield, they were not adapted to any specific environment.
These results showed that two high-yielding genotypes (SARI-6-2-6, and IT07K-303-1) out of eight cowpea genotypes evaluated were stable in their performances across the five environments. Previous studies conducted to investigate cowpea yield stability by Gurmu et al. (2009) and Sousa et al. (2018) found two ideal cowpea genotypes which exhibited both high mean yield and high-stability performances across the test environments. de Oliveira et al. (2017) also identified three cowpea genotypes, MNC02-675F-4-9, MNC02-675F-4-10, and MNC02-675F-9-2 with stable performance at the test locations. The genotypes SARI-6-2-9 and the check (Bawutawuta) were very stable, however; their yield performances did not exceed the average yield.
These findings will be of great interest not only to the cowpea breeders, but also to the seed companies, local and international NGOs and other partners who are into cowpea production and/or support cowpea farmers. Breeding lines, SARI-2-50-80, and IT07K-303-1 demonstrated stable performance with significantly high grain yield (at least 18.75%) than the check (Bawutawuta) and are recommended for release to farmers in Ghana, and other savanna agro-ecologies in West Africa in general. Even though, IT86D-610 and IT10K-837-1 were not stable across all the environments, they were among those with the highest yields. These two lines could, therefore, be used as breeding lines to improve well-adapted low-yielding cowpea cultivars.
References
Bhartiya A, Aditya JP, Singh K, Purwar JP, Agarwal A (2017) AMMI and GGE biplot analysis of multi environment yield trial of soybean in North Western Himalayan state Uttarakh and of India. Legume Res Int J 40(OF):306–312. https://doi.org/10.18805/lr.v0iOF.3548
Blanche SB, Myers GO (2006) Identifying discriminating locations for cultivar in Louisiana. Crop Sci 46:946–949
Boukar O, Fatokun CA, Huynh BL, Roberts PA, Close TJ (2016) Genomic tools in cowpea breeding programs: status and perspectives. Front Plant Sci 2016(7):1–13. https://doi.org/10.3389/fpls.2016.00757
Boukar O, Belko N, Chamarthi S, Togola A, Batieno J, Owusu E, Haruna M, Diallo S, Umar ML, Olufajo O, Fatokun C (2018) Cowpea (Vigna unguiculata): genetics, genomics and breeding. Plant Breed. https://doi.org/10.1111/pbr.12589
Chen SL, Li YR, Cheng ZS, Liu JS (2009) GGE-biplot analysis of effects of planting density growth and yield components of high oil peanut. Acta Agron Sin 35:1328–1335
Cravero V, Esposito MA, Anido FL, Garcia SM, Cointry E (2010) Identificación of an ideal test environment for asparagus evaluation by GGE-biplot analysis. Aust J Crop Sci 4(4):273–277
de Oliveira DSV, Franco LJD, de Menezes-Júnior JA, Damasceno-Silva KJ, de Moura RM, das Neves AC, de Sousa FS (2017) Adaptability and stability of the zinc density in cowpea genotypes through GGE-biplot method. Rev Cienc Agron 48:783–791. https://doi.org/10.5935/1806-6690.20170091
Gauch HG, Zobel RW (1996) AMMI analysis of yield trials. In: Kang MS, Gauch HG (eds) Genotype by environment interaction. CRC Press, Boca Raton, Florida, pp 85–122
Gedif M, Yigzaw D, Tsige G (2014) Genotype–environment interaction and correlation of some stability parameters of total starch yield in potato in Amhara region. Ethiopia J Plant Breed Crop Sci 6(3):31–40. https://doi.org/10.5897/JPBCS2013.0426
Gurmu F, Mohammed H, Alemaw G (2009) Genotype × environment interactions and stability of soybean for grain yield and nutrition quality. Afr Crop Sci J 17(2):87–99
Kamai N, Gworgwor NA, Wabekwa JW (2014) Varietal trials and physiological components determining yield differences among cowpea varieties in semiarid zone of Nigeria. ISRN Agron 2014:1–7. https://doi.org/10.1155/2014/925450
Kimbeng CA, Zhou MM, Da Silva JA (2009) Genotype x enviroment interactions and resources allocation in Suggarcane yield trials in the Rio Grande valley region of Texas. J Am Soc Suggar Cane Technol 29:11–24
Kumar B, Hooda E, Hooda BK (2018) GGE biplot analysis of multi-environment yield trials for wheat in Northern India. Adv Res 16:1–9. https://doi.org/10.9734/air/2018/43488
Luo J, Zhang H, Deng Z, Xu L, Yuan Z, Que Y (2013) Analysis of yield and quality traits in suggarcane varieties (lines) with GGE-biplot. Acta Agonomic Sin 39(1):142–152
Luo JYB, Pan YH, Que MP, Zhang L, Grisham (2015) Biplot evaluation of test environments and identification of mega environment for sugarcane cultivars in China. Sci Rep 5:15505
Lin C, Bins MR (1991) Genetics properties of four types of stability parameters. Theory Appl Genet 82:505–509
Owusu EY, Akromah R, Denwar NN, Adjebeng-Danquah J, Kusi F, Haruna M (2018) Inheritance of early maturity in Some Cowpea (Vigna unguiculata (L.) Walp.) genotypes under rain fed conditions in Northern Ghana. Adv Agric. https://doi.org/10.1155/2018/8930259(article ID 8930259)
Pacheco A, Rodriguea F, Alvarado M, Lopez M, Crossa J, Burgueno J (2016) GEA-R (genotype × environment analyses with R for Windows) version 4.0. https://doi.org/10.3389/fpls.2014.00217
Padi FK (2004) Relationship between stress tolerance and grain yield stability in cowpea. J Agric Sci Cambr 142:143–153
Rakshit S, Ganapathy KN, Gomashe SS, Rathore A, Gorade RB et al (2012) GGE biplot analysis to evaluate genotype, environment and their interactions in sorghum multi-location data. Euphytica 185(3):465–479. https://doi.org/10.1007/s10681-012-0648-6
SAS Institute Inc (2011) Base SAS 93 procedures guide. SAS Institute Inc, Cary
Sbongeleni WD, Hussein SSR, Admire ITS (2019) Genotype-by-region interactions of released sugarcane varieties for cane yield in the South African sugar industry. J Crop Improv. https://doi.org/10.1080/15427528.2019.1621974
Singh BB, Mohan DR, Dashiell KE, Jackai LEN (2002) Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Centre for Agricultural Sciences (JIRCAS0). IITA, Ibadan
Singh BB (ed) (2020) Cowpea: the food legume of the 21st century, vol 164. Wiley, New York
Sousa MBE, Damasceno-Silva KJ, Rocha MDM (2018) Genotype by environment interaction in cowpea lines using GGE biplot method. Rev Caatinga 31:64–71. https://doi.org/10.1590/1983-21252018v31n108rc
Suwarto D (2010) Genotype × environment interaction on rice Fe content. Gadjah Mada University, Yogyakarta
Yan W, Hunt LA, Sheng QL, Szlavnics Z (2000) Cultivar evaluation and mega-environment investigation based on GGE biplot. Crop Sci 40:596–605
Yan WK (2002) Singular-value partitioning in biplot analysis of multi-environment trial data. Agron J 94:990–996
Yan WK (2010) Optimal use of biplots in analysis of multi-location variety test data. Acta Agronomy Sinica 36:1805–1819
Yan W, Tinker NA (2006) Biplot analysis of multi-environment trial data: Principles and applications. Can J Plant Sci 86(3):623–645. https://doi.org/10.4141/P05-169
Yan W, Kang MS, Ma BL, Woods S, Cornelius PL (2007) GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci 47:643–655
Yan W, Holland JB (2010) A heritability-adjusted GGE biplot for test environment evaluation. Euphytica 171:355–369
Zhang PP, Song H, Ke XW (2016) GGE biplot analysis of yield stability and test location representativeness in proso millet (Panicum miliaceum L.) genotypes. J Integr Agric 15:1218–1227. https://doi.org/10.1016/S2095-3119(15)61157-1
Zhou CJ, Tian ZY, Li JY (2011) GGE-biplot analysis on yield stability and testing-site representativeness of soybean lines in multi-environment trials. Soybean Sci 30:318–322
Acknowledgements
Authors are very grateful to the Tropical Legumes III project of Bill and Melinda Gates Foundation for the financial support. We also express our heartfelt gratitude to Prof. Rajeev Kumar Varshney, Dr. Emmanuel Monyo and Dr. Chris Ojiewo for their contributions to this study. Finally, we thank the management of International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India, and International Institute of Tropical Agriculture (IITA), Nigeria for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Rights and permissions
About this article
Cite this article
Owusu, E.Y., Amegbor, I.K., Mohammed, H. et al. Genotype × environment interactions of yield of cowpea (Vigna unguiculata (L.) Walp) inbred lines in the Guinea and Sudan Savanna ecologies of Ghana. J. Crop Sci. Biotechnol. 23, 453–460 (2020). https://doi.org/10.1007/s12892-020-00054-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-020-00054-5