Abstract
Cowpea is one of the most important indigenous food and forage legumes in Africa. It serves as a primary source of protein for poor farmers in drought-prone areas of Ethiopia. The crop is used as a source of food, and insurance crop during the dry season. Cowpea is adaptable to a wide range of climatic conditions. Despite this, the yield of the crop is generally low due to lack of stable and drought-tolerant varieties. In this study, 25 cowpea genotypes were evaluated in five environments using a lattice design during the 2017 and 2018 main cropping seasons. The objectives of this study were to estimate the magnitude of genotype by environment interaction (GEI) and grain yield stability of selected drought-tolerant cowpea genotypes across different environments. The additive main effect and multiplicative interaction (AMMI) model indicated the contribution of environment, genotype and GEI as 63.98, 2.66% and 16.30% of the total variation for grain yield, respectively. The IPCA1, IPCA2 and IPCA3 were all significant and explained 45.47%, 28.05% and 16.59% of the GEI variation, respectively. The results from AMMI, cultivar superior measure, genotype plus genotype-by-environment biplot yield stability index, and AMMI stability value analyses identified NLLP-CPC-07-145-21, NLLP-CPC-103-B and NLLP_CPC-07-54 as stable and high yielding genotypes across environments. Thus, these genotypes should be recommended for release for production in drought-prone areas. NLLP-CPC-07-143, Kanketi and CP-EXTERETIS were the least stable. The AMMI1 biplot showed that Jinka was a high potential and favorable environment while Babile was an unfavorable environment for cowpea production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea [Vigna unguiculata (L.) Walp.] is one of the most important food and forage legumes grown in the semi-arid tropics and some temperate regions of the world (Timko and Singh 2008). Cowpea, at all stages of growth, serves as food source (Ahenkora et al. 1998; Singh et al. 2003). Poor people in developing countries of the tropics derive their protein, animal feed and cash income from the production of the crop (Diouf 2011). The young leaves, green pods and green grains are used as vegetables, and the dry grains are used in various preparations for both human food and livestock feed (Filho et al. 2017; Owade et al. 2020). The cowpea grain is highly nutritious and contains about 22.8–28.9% protein with an average of 25.6% (Weng et al. 2019). The portion of the cowpea crop above ground (except for pods) serves as a useful source of nutrient-rich fodder for livestock in many areas of the world (Singh et al. 2003; Weng et al. 2019). Cowpea can withstand harsh growing conditions, particularly temperature and moisture stress in comparison to other crops (Agbicodo et al. 2009; Goufo et al. 2017; Fatokun et al. 2012).
Environment, growing season, and rainfall distribution and intensity may have positive or negative impacts on cowpea genotypes (Marina et al. 2017). Plant breeders evaluate genotypes in multi-environments, representing favorable and unfavorable growing conditions, to estimate and understand the complexity of the genotype across environments (Mohammadi and Amri 2008).
Cowpea is greatly influenced by seasonal environmental fluctuations and shows large genotype by environment interaction (GEI), which is a major challenge in obtaining a full understanding of genetic control of varieties when compared across a series of environments (Kuruma et al. 2019; Olajide and Ilori 2017; Gerrano et al. 2020; Simion 2018; Simion et al. 2018). Studying and understanding of GEI is important to plant breeding programs for improving yield and yield components (Yan and Tinker 2006) and is also used for identifying the basic causes of differences between genotypes for yield stability (Yan and Tinker 2006). GEI exists when the ranks of genotypes show an obvious shift from one environment to another (Leflon et al. 2008). Measuring GEI is important to determine the optimum breeding strategy for releasing genotypes with adequate adaptation to target environments (Das et al. 2019; Yan 2016).
The methods commonly used for quantifying GEI and stability include; principal component analysis (PCA) (Zobel et al. 1988), additive main effect and multiplicative interaction (AMMI) (Zobel et al. 1988), genotype plus genotype-by-environment (GGE) biplot analysis (Yan and Tinker 2006), cultivar superiority measure (Pi) (Lin and Binns 1988), AMMI stability value (ASV) (Purchase et al. 2000) and yield stability index (YSI) (Tumuhimbise et al. 2014). Understanding the magnitude of effects of the environment, genotype and their interaction on yield and stability performance of cowpea genotypes across environments are important because it reduces the efficiency of the genetic gain through the development of high yielding genotypes with desirable agronomic traits (Hall et al. 2003; Simion et al. 2018). Ethiopia is a victim of repeated droughts that cause partial or total crop failure, and subsequently, famine in the country. Cowpea can be used to reduce the consequence of drought.
Knowledge on the effect of genotype, environment, and their interactions on drought tolerant cowpea grain yield is limited in Ethiopia. The objectives of the present study, therefore, were to estimate the magnitude of GEI and grain yield stability of selected drought tolerant cowpea genotypes across drought prone environments.
Materials and methods
Study area
The field experiments were conducted during the 2017 and 2018 main cropping season at five environments namely Babile, Melkassa, Miesso, Jinka and Sirnka (Kobo) (Table 1).
Plant materials
A total of 25 genotypes were used for this study (Table 2). Of these, two released varieties were used as standard checks and 23 genotypes were selected as drought tolerant from a drought stress experiment, which included 324 genotypes (data not shown).
Experimental Design and Procedures
The experiments were laid out using a 5 × 5 lattice design with three replications. The plot size was 4 m long, 0.75 m between rows and 0.2 m between plants. Plots consisted of four rows with 20 plants per row. The distance between plots, intra blocks and replications was 1 m, 1.5 m and 2 m, respectively. The data were collected from the middle two rows.
Data Collection and Analyses
According to the descriptors of cowpea (IBPGR 1983), yield on a plot basis was collected and converted to grain yield per hectare (kg ha−1). SAS (SAS 2013) software was used for combined analysis of variance (ANOVA) over environments and seasons. A mixed linear model was used for ANOVA (Gomez & Gomez 1984). Environment and season were considered as random and genotypes as fixed effects (Hartley 1950). F-Max ratio was used to test the homogeneity of error variances before analyzing the combined data. GEA-R was used for AMMI, GGE biplot, and Pi analysis (Pacheco et al. 2016).
The AMMI model analysis proposed by Zobel et al., (1988) was used for analyzing GEI. AMMI partitions the sum of squares into interaction principal component (IPC) axes.
GGE biplots were constructed from the data (Yan et al. 2000, 2007; Yan and Rajcan 2002; Yan 2011). The GGE biplot has many visual interpretations, which the AMMI does not have. It also allows visualization of crossover GEI (Yan et al. 2007). Moreover, the GGE biplot is more logical for biological objectives in terms of explaining the first PC score, which represents genotypic level rather than additive level (Yan et al. 2000). The GGE biplot is based on the first two major components of a PCA using the Site Regression (SREG) model. When the first component is highly correlated with the genotype main effect, the proportion of the yield is considered to be due only to the characteristics of the genotype. The second component represents the variation in the yield due to the GEI (Yan 2011). The GGE biplots were generated using a singular value decomposition model of the first two principal components (Yan 2002; Yan and Rajcan 2002).
GEA-R was used for AMMI, GGE biplot analysis and Pi (Pacheco et al. 2016). According to Purchase et al. (2000) and Farshadfar et al. (2011) the ASV would be essential to quantify and rank genotypes according to their yield stability (Farshadfar et al. 2011).
where ASV = AMMI’s stability value, IPCA1 = interaction of first principal component, IPCA2 = interaction of second principal component.
YSI incorporates both mean yield and stability in a single criterion. Low values of both parameters show desirable genotypes with high mean yield and stability (Tumuhimbise et al. 2014). The YSI was calculated using the following formula:
where: RASV is the ranking of the AMMI stability value and R the ranking of genotypes in all environments.
Pi measures the deviation from the yield of a given genotype in relation to the maximum in each environment. The significant difference of Pi was compared by computing a cutoff point for each value. Even though distributional properties of the cultivar superiority measure is not exactly known, the cut-off point was calculated by multiplying the 5% or 1% significant F-values for Pi at environment (E) and genotype (E-2) degrees of freedom by the deviation from regression mean squares, where G and E denote the number of genotypes and environments respectively (Lin and Binns 1988). The Pi measures were calculated using the following formula:

where: \({\dot{\text{X}}\text{i}}\) = is the mean of genotype i in the environments, \(\mathop M\limits^{\prime }\) = is the genotype with maximum response among all genotypes in the jth environment, Xij = is the response of the ith genotype in the jth environment, Mj = is the genotype with maximum response among all genotypes in the jth environment.
Results
Analysis of variance for grain yield across environments
The results of the combined ANOVA across the tested genotypes showed that environment (E) and season (Y) main effects, G × E, G × Y, E × Y, G × E × Y were all highly significant (p < 0.0001) for grain yield, and the genotype main effect (G) was significant (p < 0.05) for grain yield (Table 3).*P ≤ 0.05, ** P ≤ 0.01, DF = degree of freedom.
Grain yield performance across the environments
G2, G8, G10, G3 and G 18 had high yield performance at specific environments Sirnka, Jinka, Melkassa, Miesso, and Babile, respectively. However, G20, G9, G12, G15, G11, and G9 were the poorest performers at Sirnka, Jinka, Melkassa, Miesso, and Babile, respectively (Table 4). The highest grain yielding was recorded at Jinka (2445.7 kg ha−1), while the poorest grain yield was recorded at Babile (555.81 kg ha−1). The highest yielding genotype across the environments and seasons was G8 with a yield of 1641.25 kg ha −1 while the poorest yielding genotype was G9 with a yield of 1267.62 kg ha −1.
Additive main effect and multiplicative interaction analysis (AMMI)
AMMI analysis (Table 5) showed that environment and GEI effects were highly significant (p < 0.0001), and genotype effect was significant (p < 0.05) for grain yield. The test environments contributed 63.98% of the total variation in yield. Genotype and GEI accounted for 2.66% and 16.30% of the total variation for grain yield, respectively. The ratio of genotype effect to genotype + genotype × environment (G + G × E) was 0.14. The magnitude of the GEI sum of squares was 6.12 times that of the genotype sum of squares for grain yield. The AMMI model extracted three highly significant (p < 0.0001) IPCA’s from the interaction PC axes (Table 5). Those three IPCA’s accounted for a total of 90.11% of the observed variation due to GEI. IPCA1, IPCA2, and IPCA3 captured 45.47%, 28.05%, and 16.59% of the sum of squares, respectively.
Lin and Binns Cultivar superiority measure (Pi)
According to this stability model, the three most stable genotypes with the lowest Pi values were G8, G17 and G13, which ranked 1st, 2nd and 3rd for grain yield (Table 6). The most unstable genotypes were G9, G2, and G25 which ranked 25th, 20th and 24th for grain yield.
GGE biplots
Which-won-where and what
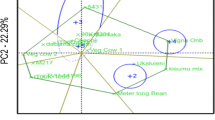
The polygon view of the genotypes in the GGE biplot for 25 genotypes is presented in Fig. 1. The cumulative variation contributed by PC1 (AXIS 1) and PC2 (AXIS2) was 69.16%, both of which were highly significant. The biplot showed that two environments (Babile and Sirnka) grouped in the same mega-environments. The other three environments (Jinka, Melkassa, and Miesso) each fell in a different mega-environment. The plot showed that G8, G13, G3, G16 and G4 recorded the highest grain yield in Jinka, Melkassa, Miesso, Sirnka, and Babile. On the other hand, G9, G25, and G23 did not fall in a specific environment and were poor yielders.
Ideal genotypes (ranking genotypes)
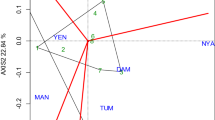
The first two principal components (PC1 and PC2) were highly significant (p < 0.0001) and explained 42.31% and 26.85% of the yield variation among the genotypes, respectively (Fig. 2). The GGE biplot, which identifies ideal genotypes that are high yielding and stable across the test environments, identified G8, G6 and G1, which fell close to the center of the concentric circle as ideal genotypes. Based on the average environment coordination (AEC) method, G9, G25 and G23 were the most unstable and low yielding genotypes across the environments (Fig. 2).
Ideal environment (ranking environment)
Jinka was close to the concentric circle and provided the most ideal environment and the most powerful to discriminate performance of the tested genotypes (Fig. 3). In contrast, Babile was located far from the center of the concentric circle, indicating poor discriminating power.
Mean vs stability
The GGE biplot visualizes performance and effectively identifies the best performing genotypes across environments with the help of the AEC. The mean of PC1 and PC2 scores of the tested environments is represented by the arrowhead, and the AEC ordinate is the line that passes through the biplot origin and it is perpendicular to the AEC abscissa (Fig. 4).
The length of the abscissa discriminates the grain yield of genotypes that are above and below average yield if right and left of the biplot origin, respectively (Fig. 4). The length of the ordinate approximates the GEI associated with the genotype stability, and a longer ordinate corresponds to higher variability and lower stability and vice-versa. NLLPP-CPC-07-10 (G1), NLLP-CPC-07-57 (G20), ACC-216-747 (G6), NLLP-CPC-07-145-21 (G8), NLLP-CPC-07-28 (G14), NLLP-CPC-07-36 (G15), and NLLP-CPC-103-B (G11) were above average yielders with higher stability, however, NLLP-CPC-07-169 (G3), ACC-215-821 (G5), NLLP-CPC-07-156 (G12), NLLP-CPC-07-54 (G13), and NLLP-CPC-07-03-B (G17) had above average yield but with lower stability. Furthermore, CP-EXTERETIS, Dass 001 (G4), NLLP-CPC-07-143 (G9), NLLP-CPC-07-166 (G19), NLLP-CPC-07-167 (G18), NLLP-CPC-07-157 (G7), and NLLP-CPC-07-140 () were stable but their yield below average, and NLLP-CPC-07-139 (), ACC-215-762 (G10), NLLP-CPC-07-19 (G16), ACC-211-490 (G23), Bole (G24) and Kanketi (G25) were not stable and their yield also below the average.
AMMI stability value (ASV) and yield stability index (YSI)
Utilizing YSI, the combination of AMMI stability values and average grain yield was estimated to quantify and classify the genotype (Table 7). According to the YSI model, G14, G17, and G21 were the most stable genotypes across environments and high grain yielders. On the other hand, genotypes G10, G2, and one check (Bole) were unstable as indicated by high YSI values of 40, 41, and 45 respectively, and those genotypes had poor productivity and lower stability.
Discussion
The majority of cowpea growing environments are extremely vulnerable to moisture stress, and the farmers use this crop as an insurance crop as they experience prolonged drought in this area. The higher yield variation contributed by the environment over genotype and GEI, indicates that the test environments were highly variable and had a great impact on genotypes. The significant GEI necessitates the need to identify adaptable genotypes with consistent high grain yield (Yan and Tinker 2006).
In addition, the response of genotypes varied considerably for grain yield due to the genetic makeup of the materials and the interaction between genetic constitution and environmental influences. This is in agreement with Gerrano et al. (2020) who reported that genotype and environment directly affect the yield potential of cowpea. From this study, the effect of environment, season, and environment × season was high as it was responsible for 33.98%, 16.23%, and 29.27% of the total variation for grain yield, respectively. Thus, the environment and the season effects were very high, contributing to diverse cowpea grain yield. Cowpea grain yield will therefore also be largely affected by climate change. A previous study also reported variation in responses of genotype across environments in different seasons (Kuruma et al. 2019).
Generally, the existing variation due to environment, season, genotype performance and GEI in relation to genotype effect suggested to there was the possibility of mega-environment effects for different genotypes. Therefore, based on the variable response of genotypes, it would help to map the mega-environments suitable for the improvement of grain yield to combat the rapid climate change.
Grain yield increment is the goal of cowpea for any stressed environment because yield is governed by multi traits with different levels of expression for various environments and their interaction. In the present study, 52% of the tested genotypes recorded a higher yield than the standard checks (Bole and Kanketi). Genotype NLLP-CPC-07–145-21 (G8) had a yield advantage of 15.26% and 22.25% compared to the worst genotype NLLP-CPC-07–143 (G9), and the standard checks, respectively. Therefore identification of the highest yielding and adaptable genotypes for the specific range of environments is important for the selection and evaluation of superior genotypes in multi-environment studies and are the main targets of cowpea breeding programs. Kuruma et al. (2019) and Muranaka et al. (2016) reported that high grain yield variation could be due to greater differences between the genotypes.
AMMI analysis showed that the environmental effects accounted for the most (63.98%) of the total variation compared to the other components, implying that differential cowpea yield performance was typically caused by environmental changes. This is in agreement with the findings of Gerrano et al. (2020) and Simion (2018) that indicated the environment made the largest contribution to grain yield variation in cowpea. The magnitude of the GEI sum of squares was 6.12 times that of the genotype sum of squares for grain yield of cowpea, indicating that there were considerable differences in genotypic responses across environments.
In the present investigation, the three IPCA's accounted for 90.11% of the interaction sum of squares. Zobel et al. (1988) stated that AMMI with the first two multiplicative terms was the best predictive model. In this study, the high (45.47%) and significant contribution of IPCA1 to the total variation across the tested environments implies that IPCA1 could identify stable and unstable genotypes based on the value scores or nearest or furthest to zero, which is in line with the findings of previous investigations (Muranaka et al. 2016; Gerrano et al. 2020; Simion 2018; Yaw et al. 2020). The positive and negative IPCA scores of genotypes in AMMI analysis are the best indicators of stability or adaptation over environments. High positive interaction of the genotypes like NLLP-CPC-07–169 (G3) in IPCA1 in an environment can exploit the agro-ecological conditions of the specific environment (Sirnka). Therefore, it would be possible to identify adaptable and suitable genotype/s for the specific environment. Kandus et al. (2010) and Yan et al. (2007) reported that the different high yielding genotypes fall in a specific environment, and it shows crossover GEI, suggesting that the test environment could be classified into mega-environments.
Using Pythagorean Theorem the distance from the origin (0:0) in a two-dimensional scattergram indicates the most stable genotypes (Purchase et al. 2000). In the ASV method, a genotype with the lowest ASV score is the most stable; accordingly, G14, G17, and G21 were stable and these genotypes were the highest yielding among the tested genotypes, indicating that the yield performance and stability showed a similar trend. Oliveira et al. (2014) noted that the dynamics of stable genotype and yield response are always parallel to the mean response of the tested environments. Unstable genotypes like Bole, CP-EXTERETIS (G2), and ACC-215–762 (G10) had high ASV values, and they were adapted to a specific favorable environment. Likewise, Oladosu et al. (2017) reported that the higher the IPCA score, and ASV the more specifically adapted a genotype is to a certain environment. In this study, crossover stability and yield did not have the same trend for all genotypes across season and environment. Jadhav et al. (2019) and Mohammadi and Amri (2008) suggested that principle stability per se should, however, not be the only selection parameter because the most stable genotypes would not necessarily give the best yield performance. Therefore, there is a need for approaches that incorporate both mean yield and stability in a single index.
The lowest YSI value is considered as the most stable, with high grain yield (Bose et al. 2014). NLLP-CPC-07–28 (G14), NLLP-CPC-07–145-21 (G8) and NLLP-CPC-07–54 (G13) were the most stable genotypes with good yield performance. Thus, according to the YSI method, the most desirable genotypes can be considered as widely adapted and with grain yield above the grand mean among 25 genotypes. Similarly Zali et al. (2012) also indicated that both yield and stability of performance should be considered simultaneously to exploit the useful effect of GEI and to select genotypes for diverse environments.
The polygon view of the “which -won-where and what” GGE-biplot (Fig. 1) showed that genotypes NLLP-CPC-07–143 (G9), ACC-215–762 (G10), NLLP-CPC-07–156 (G12), NLLP-CPC-07–54 (G13), NLLP-CPC-07–145-21 (G8) and ACC-211–490 (G23) were genotype markers located farthest from the biplot origin in various directions and it shows that the genotypes were well adapted to specific environments. If the environment markers fall in different sectors it shows that different cultivars won in different environments (Oladosu et al. 2017; Gerrano et al. 2020). In addition, because of long environment vectors, the rest of the genotypes scattered around the biplot origin. This clearly explained that the environment effect was higher than the genotype effect. This implies that the genotype had less response for GEI because of high environment exertion to the GEI. The GGE-biplot showed that the tested environments occurred in different sectors, indicating that the particular environment had different high yielding genotypes for those sectors, this indicating the existence of crossover GEI. It also indicated the possibility to classify the environments into mega-environments for cowpea production. Yan and Rajcan (2002) stated that the presence or absence of crossover GEI indicates the existence of different mega-environments. Generally, the GGE biplot effectively identified the best performing genotypes across environments and best genotypes for specific environments, whereby specific genotypes can be recommended for specific environments and can be used to evaluate the yield and stability of genotypes, which is not possible with AMMI analysis (Kaya et al. 2006; Yan and Tinker 2006).
GGE biplots help visualize and compare the distance between each genotype and the ideal genotype located at the center of the concentric circle (Yan and Rajcan 2002). The ideal genotypes, based on proximity to the center of the concentric circle of the GGE biplot were ACC-216-747 (G6) and NLLP-CPC-07-145-21 (G8), with high yield and stability (Fig. 2). In addition, NLLP-CPC-07-10 (G1) was located on the next homocentric circle and might be considered as a desirable genotype. In principle, the ideal genotype should have the longest vector, highest mean performance and with zero GEI, and/or it should perform consistently in all environments. Because of the genetic background or nature of the traits (yield) and level of expression, the ideal genotype does not always exist in reality. Therefore, such like genotypes the breeders can be used as a reference for genotype for further study. Genotypes which were high yielding but were not stable across environments could be recommended for a particular environment.
Yan and Rajcan (2002) and Yan et al. (2007) specified that the environments with long vectors (PC1 scores) and relatively small angles or absolute with the AEC abscissa are valuable for greater discriminatory capacity (in terms of the genotype main effect) and is representative of the other environments. Therefore, Jinka was in the epicenter of the concentric circle, and it was identified as a highly discriminating environment for these genotypes, thus this environment is considered as an ideal environment for developing high yielding genotypes, or for identify ideal genotypes. Hence, Jinka allowed the genotype to express genetic potential, minimizing population development expenses by discriminating the worst genotypes at an early stage.
Conclusions
The present study showed that genotype, environment, and genotype × environment, genotype × season and environment × season interaction effects were significant for grain yield. The performance of tested genotypes was largely affected by the environment. Jinka was the most ideal environment and it had discriminatory power for genotype performance in all seasons. The AMMI showed that the environmental effects accounted for 63.98% of the total yield variation. The GGE biplot showed that the tested environments occurred in different sectors which could be used for classifying the environments into mega-environments for cowpea production. In terms of ideal genotypes (having a long vector), good yielding capacity and stability, NLLP-CPC-07-145-21 (G8), NLLP-CPC-103-B (G11) and NLLP_CPC-07-54 (G13) were identified as ideal genotypes for drought-prone environments for the country and could be proposed for release for production. The results of this study confirmed that the ideal genotype should have a long vector, high yield performance, but such genotypes do not always exist in reality.
References
Agbicodo EM, Fatokun CA, Muranaka S et al (2009) Breeding drought tolerant cowpea: constraints, accomplishments, and future prospects. Euphytica 167:353–370. https://doi.org/10.1007/s10681-009-9893-8
Ahenkora K, Adu Dapaah HK, Agyemang A (1998) Selected nutritional components and sensory attributes of cowpea (Vigna unguiculata [L.] Walp) leaves. Plant Foods Hum Nutr 52:221–229. https://doi.org/10.1023/A:1008019113245
Bose LK, Jambhulkar NN, Pande K, Singh ON (2014) Use of AMMI and other stability statistics in the simultaneous selection of rice genotypes for yield and stability under direct-seeded conditions. Chil J Agric Res 74:1–9. https://doi.org/10.4067/S0718-58392014000100001
Das A, Parihar AK, Saxena D, Singh D, Singha KD, Kushwaha KPS, Chand R, Bal RS, Chandra S, Gupta S (2019) Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L). Front Plant Sci 10:825. https://doi.org/10.3389/fpls.2019.00825
De OEJ, Paulo J, Freitas XD, Jesus ON (2014) AMMI analysis of the adaptability and yield stability of yellow passion fruit varieties. Sci Agricola 71:139–145
Diouf D (2011) Recent advances in cowpea [Vigna unguiculata (L.) Walp.] “omics” research for genetic improvement. Afr J Plant Biotech 10:2803–2810. https://doi.org/10.5897/AJBx10.015
Farshadfar E, Mahmodi N, Yaghotipoor A (2011) AMMI stability value and simultaneous estimation of yield and yield stability in bread wheat (Triticum aestivum L.). Aust J Crop Sci 5:1837–1844
Fatokun CA, Boukar O, Muranaka S (2012) Evaluation of cowpea (Vigna unguiculata ( L.) Walp) germplasm lines for tolerance to drought. Plant Genet Resour Charact Util 10:171–176. https://doi.org/10.1017/S1479262112000214
Filho JT, Dos Santos Oliveira CNG, da Silveira LM, de Sousa Nunes GH, da Silva AJR, da Silva MFN (2017) Genotype by environment interaction in green cowpea analyzed via mixed models. Rev Caatinga, Mossoro 30:687–697
Gerrano A, Jansen van Rensburg WS, Mathew I, Shayanowako AIT, Bairu MW, Venter SL, Swart W, Mofokeng A, Mellem J, Labuschagne M (2020) Genotype and genotype × environment interaction effects on the grain yield performance of cowpea genotypes in dryland farming system in South Africa. Euphytica 216:1–11. https://doi.org/10.1007/s10681-020-02611-z
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research, 2nd edn. John Wiley and Sons Inc., New York, USA
Goufo P, Moutinho-pereira JM, Jorge TF, Correia CM (2017) Cowpea (Vigna unguiculata L. Walp.) metabolomics : osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front Plant Sci 8:1–22. https://doi.org/10.3389/fpls.2017.00586
Hall AE, Cisse N, Thiaw S et al (2003) Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crop Res 82:103–134. https://doi.org/10.1016/S0378-4290(03)00033-9
Hartley O (1950) The maximum F-ratio as a short-cut test for heterogeneity of variance. Biometrika 37:308–312
IBPGR (1983) Cowpea descriptors, 1st edition. international board for plant genetic resources, Rome, Italy
SAS II (2013) Statistical Analysis System, Version 9.4 SAS institute Inc. 476
Jadhav S, Balakrishnan D, Shankar VG, Beerelli K, Chandu G, Neelamraju S (2019) Genotype by environment interaction study on yield traits in different maturity groups of rice. J Crop Sci Biotech 22:425–449. https://doi.org/10.1007/s12892-018-0082-0
Kandus M, Almora D, Ronceros RB, Salerno JCS (2010) Statistical models for evaluating the genotype-environment interaction in Statistical models for evaluating the genotype-environment interaction in maize (Zea mays L.) Modelos estadísticos para evaluar la interacción genotipo-ambiente en maíz (Zea mays). Phyton (B Aires) 79:39–46
Kaya Y, Akçura M, Taner S (2006) GGE-Biplot analysis of multi-environment yield trials in bread wheat. Turkish J Agric for 30:325–337
Kuruma RW, Sheunda P, Muriuki C (2019) Yield stability and farmer preference of cowpea (Vigna unguiculata) lines in semi-arid eastern Kenya. Afrika Focus 32:65–82
Leflon M, Lecomte C, Barbottin A, Jeuffroy MH, Robert N, Brancourt-Hulmel M (2008) Characterization of environments and genotypes for analyzing genotype × environment interaction. J Crop Improv 14:249–298. https://doi.org/10.1300/J411v14n01
Lin CS, Binns MR (1988) A superiority measure of cultivar performance for cultivar x location data. Can J Plant Sci 68:193–198
Marina M, Fernández J, Ochoa J et al (2017) Genotype by environment interactions in cowpea (Vigna unguiculata L. Walp) grown in the Iberian Peninsula. Crop Pasture Sci 68:924–931
Mohammadi R, Amri ÆA (2008) Comparison of parametric and non-parametric methods for selecting stable and adapted durum wheat genotypes in variable environments. Euphytica 159:419–432. https://doi.org/10.1007/s10681-007-9600-6
Muranaka S, Shono M, Myoda T, Takeuchi J (2016) Genetic diversity of physical, nutritional and functional properties of cowpea grain and relationships among the traits. Plant Genet Resour Charact Util 14:67–76. https://doi.org/10.1017/S147926211500009X
Oladosu Y, Rafii MY, Abdullah N, Magaji U, Miah G, Hussin G, Ramli A (2017) Genotype × Environment interaction and stability analyses of yield and yield components of established and mutant rice genotypes tested in multiple locations in Malaysia. Acta Agric Scan 67:590–606. https://doi.org/10.1080/09064710.2017.1321138
Olajide AA, Ilori CO (2017) Effects of drought on morphological traits in some cowpea genotypes by evaluating their combining abilities. Adv Agric 2017:1–10
Owade JO, Abong G, Okoth M, Mwang AW (2020) A review of the contribution of cowpea leaves to food and nutrition security in East Africa. Food Sci Nutr 8:36–47. https://doi.org/10.1002/fsn3.1337
Pacheco A, Vargas M, Alvarado G, Rodriguez F, Crossa J, Burgueno J (2016) User's Manual GEA-R (Genotype by Environment Analysis with R ). GEA-R 1–42
Purchase JL, Hatting H, Van DCS (2000) Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa : I. AMMI analysis of yield performance. South Afr J Plant Soil 17:95–100. https://doi.org/10.1080/02571862.2000.10634877
Simion T (2018) Adaptability performances of cowpea [Vigna unguiculata ( L.) Walp ] genotypes in Ethiopia. Food Sci Qual Manag 72:43–47
Simion T, Mohammed W, Fenta BA (2018) Genotype by environment interaction and stability analysis of cowpea [Vigna unguiculata (L.) Walp] genotypes for yield in Ethiopia. J Plant Breed Crop Sci 10:249–257. https://doi.org/10.5897/JPBCS2018.0753
Singh BB, Ajeigbe HA, Tarawali SA et al (2003) Improving the production and utilization of cowpea as food and fodder. Field Crop Res 84:169–177. https://doi.org/10.1016/S0378-4290(03)00148-5
Timko, M.P. and Singh BB (2008) Cowpea, a multifunctional legume. In: Paul, H. Moore and Ray M (ed) genomics of tropical crop plants, 1st edn. Springer Science+Business Media, LLC, pp 227–258
Tumuhimbise R, Melis R, Shanahan P, Kawuki R (2014) Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava. Crop J 2:329–337. https://doi.org/10.1016/j.cj.2014.04.008
Weng Y, Jun Q, Eaton S, Yang Y, Ravelombola WS, Shi A (2019) Evaluation of seed protein content in USDA cowpea germplasm. HortScience 54:814–817
Yan W (2002) Singular-value partitioning in biplot analysis of multi environment trial data. Agron J 94:990–996. https://doi.org/10.2134/agronj2002.0990
Yan W (2011) GGE Biplot vs. AMMI Graphs for genotype-by-environment data analysis. Journal Indian Soc Agric Stat 65:183–193
Yan W (2016) Analysis and handling of G × E in a practical breeding program. Crop Sci 56:2106–2118. https://doi.org/10.2135/cropsci2015.06.0336
Yan W, Rajcan I (2002) Biplot Analysis of test sites and trait relations of soybean in Ontario. Crop Sci 42:11–20. https://doi.org/10.2135/cropsci2002.0011
Yan W, Tinker NA (2006) Biplot analysis of multi-environment trial data: principles and applications. Can J Plant Sci 86:623–645. https://doi.org/10.4141/P05-169
Yan W, Hunt LA, Sheng Q, Szlavnics Z (2000) Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci 40:597–605. https://doi.org/10.2135/cropsci2000.403597x
Yan W, Kang MS, Ma B, Woods S, Cornelius PL (2007) GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci 47:643–655. https://doi.org/10.2135/cropsci2006.06.0374
Yaw E, Isaac O, Amegbor K, Mohammed H, Kusi F, Atpkle I, Sie EK, Ishahku M, Zakaria M, Iddrisu S, Kendey HA, Boukar O, Fatokun C, Nutsugah SK (2020) Genotype × environment interactions of yield of cowpea (Vigna unguiculata ( L.) Walp ) inbred lines in the Guinea and Sudan Savanna ecologies of Ghana. J Crop Sci Biotechnol 23:453–460. https://doi.org/10.1007/s12892-020-00054-5
Zali H, Farshadfar E, Sabaghpour SH (2012) Evaluation of genotype × environment interaction in chickpea using measures of stability from AMMI model. Ann Biol Res 3:3126–3136
Zobel RW, Wright MJ, Gauch HG (1988) Statistical analysis of a yield trial. Agron J 80:388–393. https://doi.org/10.2134/agronj1988.00021962008000030002x
Acknowledgements
The authors sincerely acknowledge the contribution of Wolkite University, Melkassa Agricultural Research Center (EIAR), and The McKnight Foundation USA, for providing the research funds to complete this project. The author acknowledges the staff of national lowland pulse improvement program of Melkassa Agricultural Research Center (MARC) is highly appreciated for their comprehensive support.
Funding
This study was supported by The McKnight Foundation, USA through Collaborative Crop Research Program (CCRP) for Ethiopian Agricultural research Institute (EIAR).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by TWM, FM and BA. The first draft of the manuscript was written by TWM and FM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mekonnen, T.W., Mekbib, F., Amsalu, B. et al. Genotype by environment interaction and grain yield stability of drought tolerant cowpea landraces in Ethiopia. Euphytica 218, 57 (2022). https://doi.org/10.1007/s10681-022-03011-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03011-1