Abstract
Weissella confusa has recently received attention for its probiotic potential. Some W. confusa and Weissella cibaria strains isolated from fermented foods show favorable probiotic effects. However, the probiotic properties of W. confusa isolated from giant panda remain unreported to date. Thus, this study isolated a W. confusa strain from giant panda feces and then investigated its characteristics and probiotic properties. A lactic acid bacteria strain was isolated from giant panda fecal samples. The isolated strain was screened by in vitro probiotic property tests, including in vitro antimicrobial test, antioxidant test, surface hydrophobicity, and stress resistance. On the basis of biochemical identification and 16S rDNA sequencing, the W. confusa strain was identified as BSP201703. This Weissella confusa strain can survive at pH 2 and 0.3% (w/v) concentration of bile salt environment and inhibit common intestinal pathogens. It also possesses an in vitro antioxidant capacity, a high auto-aggregation ability, and a high surface hydrophobicity. BSP201703 might serve as a probiotic to giant pandas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Giant panda (Ailuropoda melanoleuca) is a rare, vulnerable species that is extremely popular worldwide. As a herbivore species, giant panda retains a typical carnivorous digestive system that is easily afflicted by various gastrointestinal diseases [1]. Antibiotics are commonly used in animal disease treatment, but their negative effects have been gradually recognized. Therefore, probiotic therapy is an ideal alternative to antibiotic therapy for treating various diseases, including gastrointestinal diseases. Probiotics are live microorganisms that provide many health benefits, including pathogen growth inhibition, digestive enzyme secretion, intestinal barrier protection, and keep intestinal stability [2].
Weissella confusa was first identified by Collins in 1993 [3, 4], which belongs to Weissella spp. which was a kind of lactic acid bacteria (LAB). W. confusa was isolated from human clinical specimens, such as blood and feces sample of patients with bacteremia [5,6,7,8]. However, whether W. confusa plays a role in human infections remains unclear.
Weissella spp. can also be isolated from fermented food and animal intestinal flora, including in giant pandas. Given that Weissella cibaria is always present in the early stage of fermentation [9], Weissella spp. may serve as a starter culture. W. confusa and W. cibaria have recently attracted attention because of their probiotic properties. Lee et al. [10] showed that W. confusa isolated from human feces features probiotic properties. Hyeran et al. [11] found that W. confusa PL9001 can bind to gastric cells and prevent Heliobacter pylori adherence. A previously isolated W. cibaria strain also shows a probiotic potential in periodontal treatment by controlling periodontal pathogens and bacteriocin production [12].
These studies prove that W. confusa is a valuable strain to isolate, screen, and evaluate for its probiotics potential. In the present studies, we isolated a W. confusa strain from giant panda feces and evaluate the probiotic properties.
Materials and Methods
Samples
Fresh giant panda feces from different panda individuals were collected at Cheng Du Zoo, putted in ice pack, and taken back to the laboratory. The fecal sample was stored at − 80 °C.
Isolation of Bacteria
Composite fecal samples from different giant panda were homogenized with sterile normal saline in a constant temperature shaker at 37 °C for 30 min then diluted by ten-times step dilution. Diluted sample was spread on deMan-Rogosa-Sharpe (MRS) agar plates and incubated at 37 °C. After incubation, bacteria in colony were selected to purification. Preliminary identification of selected strain was measured through Gram stain and catalase activity.
Catalase activity of selected strain was detected by using hydrogen peroxide. Purified bacteria colony was moved to a clean glass slide, hydrogen peroxide was added on glass slide, and bubble formation was observed. Bacteria colony that generates bubbles was regarded as catalase positive.
Antimicrobial Assay
Antimicrobial ability of isolated strains was measured by the method described by reference [13,14,15,16,17] with some modifications. Six pathogen strains our laboratory preserved before were adopted in antimicrobial assay to evaluate the antimicrobial activity of isolated LAB strains: Escherichia coli ATCC25922, Salmonella SC06, Staphylococcus saureus BJ216, Enterotoxigenic Escherichia coli (ETEC) CVCC196, Shigella flexneri, and Escherichia fergusonii. Each pathogen was coating on the LB agar plate. After pathogens were coated, carefully put the oxford cup in surface of the plate, 100 μl of overnight bacteria culture was putted inside the oxford cup, the plate was incubated at 37 °C for 12 h, and the diameter of inhibition zones was measured.
Antioxidant Capacity
Sample Preparation
Selected bacteria strains were inoculated in MRS broth at 37 °C for 24 h, then centrifuged (5000×g, 10 min, 4 °C), discard the supernatant, resuspend the sediment with sterilized phosphate buffer solution (PBS, pH 7.4), the concentration was adjusted to 109 cfu/ml. Cell-free supernatant (CFS) was prepared by ultrasonication method (4 s, 5 min) at same concentration bacteria suspension. After ultrasonication, centrifuged at 8000×g for 10 min, collect the supernatant.
DPPH Scavenging Activity
DPPH (2, 2-diphenyl-1-picrylhydrazyl) radical scavenging activity of selected LAB and their cell-free supernatant (CFS) were examined by the spectrophotometric method described by reference [18,19,20] with some modification. LAB on concentration of 109 cfu/ml and same concentration CFS were reacted with 0.2 mM DPPH solution (dissolved by absolute ethyl alcohol, freshly prepared). The reaction was left in dark and at room temperature for 30 min. Absorbance was measured at 517 nm by the spectrophotometer. Distilled water was served as negative control. Each sample was performed in triplicate. DPPH scavenging percentage was computed according to the following formula:
Ferrous Ion (Fe2+) Chelation Ability
The ability to chelating ferrous ion of samples and CFS were determined by using method described by reference [21], 0.5 ml of suspension (or CFS) was putted in a test tube, mixed with 0.1 ml of FeSO4 (0.4% w/v), 1 ml of NaOH (0.2 M), 0.1 ml of ascorbate (0.1%, w/v), then mixture was incubated at 37 °C water bath for 20 min, in order to precipitate proteins, 0.2 ml of trichloroacetic acid (TCA, 10%, w/v) was added after incubation. Centrifuged at 3000×g for 10 min, then 2 ml O-phenanthroline (0.1%, w/v) was added. Reaction for 10 min, distilled water was served as negative control. Absorbance was measured at 510 nm. Each sample was performed in triplicate. Chelating percentage was computed according to the following formula:
Superoxide Anion (O2 2−) Scavenging Activity
The ability to scavenge superoxide anion was tested by the method described by Xing [22] with some modification. First, 0.1 ml of sample or CFS was mixed with 4.5 ml of Tris-HCl (pH 8.0), incubated at 25 °C water bath for 20 min, then 0.4 ml of pyrogallic acid (25 mM) was added, with 5-min reaction at 25 °C water bath, a few HCl (8 M) was added to terminate reaction, distilled water was served as negative control. Absorbance at 325 nm was measured. Each sample was performed in triplicate. O2−scavenging percentage was computed according to the following formula:
Lipid Preoxidation (ILAP) Assay
The lipid preoxidation ability of bacteria suspensions and CFS was examined by TBA (thiobarbituric acid) method with some modifications; 1 ml of linoleic acid and 0.2 ml of FeSO4, 0.2 ml of ascorbic acid (0.02%, w/v), 0.5 ml of sample were mixed and incubated at 37 °C for 12 h. Then, 2 ml of the reaction mixture was mixed with 0.2 ml trichloroacetic acid (TCA, 4%, w/v) and 0.2 ml of butylated hydroxytoluene(0.4%, w/v), 2 ml of TBA (0.8%, w/v), then the mixture was reacted at 100 °C water bath for 30 min. After cooling on ice, 2 ml of chloroform was added for extraction. The upper extract was obtained and absorbance was determined at 532 nm. Distilled water was served as negative control. Every sample was performed in triplicate. The inhibition rate was calculated by using the following formula:
Reducing Power Assay
The ability of selected LAB strains and CFS to reduce iron (III) was examined according to method described by Fakruddin [21], 0.5 ml of LAB/CFS was mixed with 0.5 ml of potassium ferricyanide (1%, w/v), 0.5 ml of PBS (0.2 M, pH 6.6). The mixture was incubated at 50 °C for 20 min. After incubation, 0.5 ml of 10% trichloroacetic acid was added and centrifuged at 3000×g for 5 min. Then, 1 ml of upper layer reaction mixture was mixed with 1 ml of ferric chloride and 1 ml of distilled water, the absorbance was measured at 700 nm, every sample was performed in triplicate. Sample that has higher absorbance possesses better reducing power.
Hydroxyl Radical (OH−) Scavenging Ability
One milliliter of LAB suspension was mixed with 1 ml O-phenanthroline (2.5 mM) and 1 ml of PBS (0.2 M, pH 7.4), with sufficient mixing, 1 ml of FeSO4 (2.5 mM) and 1 ml of hydrogen peroxide was added. After being incubated at 37 °C water bath for 1 h, distilled water was served as negative control. Every sample was performed in triplicate. The absorbance was measured at 536 nm. The scavenging percentage of hydroxyl radical was computed according to the following formula:
Auto-Aggregation
The auto-aggregation ability of selected bacteria was tested using the method described by Campana R [23] with some modifications. The bacteria were grown in MRS broth for 16–18 h and centrifuged at 6000×g for 10 min. Precipitate was washed twice by distilled PBS, then re-suspended in PBS, and adjust the absorbance of suspension to 0.5 at 600 nm. The suspension was incubated at 37 °C for 5 h. Each hour, 1 ml of the upper phase was removed to measure OD600. Cell auto-aggregation percentage was calculated by following formula.
Cell Surface Hydrophobicity
Cell surface hydrophobicity of selected bacteria was measured by method described by Yadav R [16], the bacteria culture was grown in MRS broth for 24 h, and centrifuged at 6000×g for 10 min, then pellets were washed twice in distilled PBS, pellets were re-suspended in distilled PBS, and concentration was adjusted to 109 cfu/ml, measure the absorbance of this suspension at 600 nm, 3 ml of the suspension was mixed with 1 ml hydrocarbon (xylene, n-hexadecane, and chloroform). Incubated at room temperature for 20 min, after separation of the aqueous phase and the organic phase, the aqueous phase was carefully removed and the absorbance was measured at 600 nm, percent hydrophobicity was calculated by the following formula:
16S rDNA Gene Sequencing
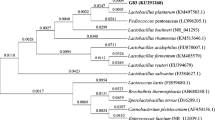
After probiotic property test, DNA of selected strain was extracted as template. The universal primer 27Fand 1492R [24] were used to amplify the 16S rDNA gene by PCR method, the PCR mixture (25 μl) contained 12.5 μl PCR Master Mix, 9.5 μl ddH2O, 1 μl forward primer, 1 μl reverse primer, and 1 μl DNA sample, performed under the standard PCR procedure. PCR amplicons were checked on 2% agarose gel electrophoresis with golden view; the PCR sample was sent to BGI to sequencing. According to 16S rRNA sequencing result, use the BLAST tool at NCBI (http: //www. ncbi. nih. gov) to compare the sequence similarities. Ten different strains from BLAST result were selected for phylogenetic analyses. Phylogenetic tree was established by MEGA5 software.
Biochemical Identification
Biochemical identification of selected strains was measured by biochemical tube following the Bergey’s Manual of Determinative Bacteriology. The ability to ferment different sugars of selected strain was tested; other examinations including gelatin liquefaction and sulfuretted hydrogen production were also determined.
Resistance to Bile Salt
Bile salt resistance of strain X3 and generally accepted probiotics LGG were tested by the method described by reference with some modifications [25, 26]. Active cultures of X3 and LGG were inoculated in MRS broth containing 0, 0.1, 0.2, and 0.3% (w/v) bile salt for 2 h, and after inoculation, 0.5 ml of samples was serially diluted in phosphate buffer saline, sample of different dilution gradient was plated on MRS agar plate and incubated at 37 °C for 12 h. Viable cells were enumerated. No bile salt group was served as control group. Survival rate in different bile salt concentration was calculated and compared the tolerance to bile salt of X3 and LGG.
Resistance to Low pH
Active cultures of strain X3 and LGG were centrifuged at 6000×g for 10 min and discard the supernatant; precipitate was washed twice in PBS (pH 7.4), then resuspended in MRS broth on different pH (pH 2, pH 3, pH 7) for 2 h, 0.5 ml of samples was serially diluted in phosphate buffer saline, sample of different dilution gradient was plated on MRS agar plate and incubated at 37 °C for 12 h. Viable cells were enumerated. The pH 7 group was served as a control. Survival rate in low pH was calculated and compared the tolerance to low pH of X3 and LGG.
Antibiotic Susceptibility
Antibiotic susceptibility of selected LAB strains was tested by agar diffusion methods [18]; the antibiotics used in this experiment were ampicillin, streptomycin, neomycin, and vancomycin. Four different concentrations (30 μg/ml, 300 μg/ml, 3 mg/ml, and 30 mg/ml) were tested. Selected strain was coated on MRS agar plate; plate was punching four-hole use puncher. Add four different concentrations of antibiotics on the holes; plate was incubated at 37 °C overnight. Record the sensitivity or resistance of selected strain. The area shows inhibition zone record as sensitive, and no inhibition zones record as resistant.
Results
Bacteria Isolation
A total 48 strains of LAB (Gram positive and catalase negative) were isolated from MRS agar plate. The strains were numbered X1 to X48. These 48 LAB strains were initially screened by antimicrobial activity test.
Antimicrobial Activity
The inhibition zones of all 48 strains are shown in Fig. 1. Except for X5, X12, X19, and X22, all the isolated LAB strains exerted different antagonic effects on pathogens. Strains that can inhibit all six pathogens were selected to further antioxidant assay.
Inhibition zone diameter of isolated 32 strains to 6 pathogens, values are mean ± SD of three different observations (n = 3). a Inhibition zone diameter of salmonella SC06. b Inhibition zone diameter of E. coli ATCC 25922. c Inhibition zone diameter of staphylococcus aureus BJ216. d Inhibition zone diameter of Enterotoxigenic Escherichia coli CVCC196. e Inhibition zone diameter of Shigella flexneri. f Inhibition zone diameter of Escherichia fergusonii
Antioxidant Activity
Probiotics that scavenge free radicals can alleviate oxidative stress of the human body. In this test, six indexes were chosen to evaluate the antioxidant property of the selected strains. After antimicrobial test, seven LAB strains (X1, X2, X3, X4, X27, X36, and X40) that can inhibit six pathogens were selected. The antioxidant capacities, including DPPH scavenging, Fe2+chelation, superoxide anion scavenging, lipid preoxidation, and reducing power and hydroxyl radical scavenging capacities of the bacterial suspension and CFS were measured (Tables 1 and 2). Suspension of live LGG showed the best DPPH scavenging rate (39.1%), followed by X1 (36.1%) and X3 (30.1%). The superoxide onion scavenging rate of strain X3 was the best of the test strain (17.1%). Meanwhile, live cells of strain X27 and X36 did not show O22− scavenging ability. The hydroxyl radical scavenging rate of X40 was the best (65.9%), followed by X3 (64%). Strain X3 showed best metal ion chelating rate (37.9%), followed by LGG (37.1%), while those of other strains ranged from 29.7 to 10%. The best lipid preoxidation inhibitory rate was exhibited by X3 (66.4%). Relative to Lactobacillus rhamnosus GG, X3 possesses a comprehensive antioxidant property; it can scavenge all types of free radicals. Moreover, CFS has a certain extent radical scavenge ability. The antioxidant ability of the whole cell culture was generally better than that of CFS.
Auto-Aggregation
The cell auto-aggregation abilities of the seven strains of LAB and LGG are shown in Table 3. The selected strains had different auto-aggregation rates after stewing. Strain LGG reached its maximum auto-aggregation rate (39%) after 5 h standing. The LAB strains isolated from giant panda feces also showed good auto-aggregation abilities after 5 h, with X2 having the highest rate at 28.1% compared with the 7.9 to 25.2% range of the other strains.
Cell-Surface Hydrophobicity
The surface hydrophobicity of the selected strain is shown in Table 4. In different hydrocarbons, the selected LAB strains showed different degrees of hydrophobicity. In chloroform, LGG showed the best hydrophobicity (44.7%), followed by X3 (39.1%). The other strains also demonstrated different degrees of hydrophobicity ranging from 2.9 to 34.8%. In xylene, X2 had the best hydrophobicity (31.0%), followed by X3 (25.4%). In cetane, the selected strains showed no significant hydrophobicity, with X3 having the highest rate (17.1%), followed by LGG (16.3%).
16S rDNA Sequencing
On the basis of the results of probiotic property tests, X3 was selected for 16S rDNA sequencing. The target band of the strain was checked by agarose gel electrophoresis (AGE). The PCR product was also sent for sequencing. Blast results showed that X3 is highly similar to W. confusa strain JCM1093 (99%). A phylogenetic tree is shown in Fig. 2.
Biochemical Identification
The biochemical identification result of strain X3 is shown in Table 5. In accordance with Bergey’s Manual of Determinative Bacteriology, strain X3 was consistent with all biochemical characteristics of W. confusa.
X3 was identified as W. confusa strain BSP201703 on the basis of the combined results of 16S rDNA sequencing and biochemical identification.
Resistance to Bile Salt
The survival rate of BSP201703 at different bile salt concentrations is shown in Table 6; on the basis of valuable count, BSP201703 has a survival rate of 3.3% in a 0.3% bile salt environment and thus can be generally accepted as a probiotic. Conversely, LGG could not survive at 0.3% bile salt concentration.
Resistant to Low pH
The result of survival rate at low pH is shown in Table 6. BSP201703 has survival rates of 67.78 and 0.32% at pH 3 and 2, respectively. Compared with LGG, BSP201703 has a much lower survival rate, but could survive at low pH. This result indicates that BSP201703 has a great chance of adhering to intestinal viscera than LGG because of its tolerance to acidic environment.
Antibiotic Susceptibility
The susceptibility of BSP201703 to four antibiotics is listed on Table 7. BSP201703 was sensitive to three commonly used antibiotics (ampicilin, streptomycin, and neomycin). Interestingly, BSP201703 was resistant to low concentrations of vancomycin (30 and 300 μg/ml) but sensitive to high concentrations of this antibiotic (3 and 30 mg/ml).
Discussion
As a vulnerable species, the giant panda (Ailuropoda melanoleuca) is considered a “national treasure” in China. Only about 375 pandas were in captivity in 2013, and 2549 pandas were in the wild in 2015 across the country [27]. Various diseases especially intestinal disease threaten the health of these captive pandas. Antibiotic therapy is the most commonly used in animal disease treatment, but this strategy is limited by its side effects, which include antibiotic resistance and antibiotic resistance gene transfer. Moreover, long-term use of antibiotics could lead to intestinal micro-flora imbalance, which could increase the sensitivity of the intestines to enteric pathogens.
Therefore, probiotics have received attention as a potential replacement to antibiotics. Probiotics applied on animal disease models show therapeutic effect while avoiding all the side effects of antibiotics abuse. Lactobacillus plantarum BSGP201683 isolated from giant panda feces shows protective effect such as attenuate inflammation and improve gut microflora on mice challenged with enterotoxigenic Escherichia coli (ETEC) [1]. Lactobacillus johnsonii BS15 can promote growth performance, lower fat deposition, and prevent subclinical necrotic enteritis in broiler chickens by improving lipid metabolism and gut microflora [28, 29]. Therefore, probiotic therapy is a better choice than antibiotic therapy for disease treatment and health improvement of the giant panda. Probiotics applied on giant pandas have broad prospects, and probiotics isolated from giant panda itself would be better.
Weissella spp. are the dominant microorganisms founded in the food and intestinal walls of human and animals; Weissella spp. play an important role in fermentation and served as a starter culture [9, 30]. Recently, the probiotic potential for Weissella spp. has become research hotspots. Kang Wook Lee et al. have isolated W. confusa from human fecal samples and evaluated its probiotic properties. Similar to probiotics, W. confusa can inhibit pathogen growth and can tolerate high bile salt concentration and low pH. The survival rate in 0.3% concentration bile salt of W. confusa strains isolated from human ranges from 2.2 to 128.8%, and its acid tolerance ranges from 2.4 to 20.2% [10]. Lim et al. [31] have studied W. cibaria WIKIM28 applied in a mouse atopic dermatitis model and found that this strain can ameliorate AD-like symptoms by suppressing allergic Th2 responses. Monique Suela Silva et al. [12] have evaluated the probiotic properties of W. cibaria isolated from the Mexican beverage tejuino and found that the strain does not show a high adhesion capacity to HT-29 cells, but can produce short-chain fatty acids and exert an antagonistic activity toward pathogens that contribute to eubiosis. Amandine et al. [32] reported that lactic fermentation of juice by different lactic acid bacteria, W. cibaria 64 used in this research can moderate acidification ability and enhance antioxidant activity. Moreover, W. confusa strains can produce non-digestible oligosaccharides, mainly β-dextran, which was applied as prebiotics. Ilkka et al. [33] have cloned and characterized a dextransucrase gene of W. confusa VTTE90932 and explored its application in high fiber-baking. Some W. cibaria and W. confusa strains can produce the bacteriocin called weissellin. The bacteriocin produced by W. confusa A3 shows broad spectrum of inhibitory effect against Bacillus cereus, E. coli, Pseudomonas aeruginosa, Micrococcus luteus, Lactococcus lactis, and Enterococcus faecium [34].
In the present study, we isolated a W. confusa strain and measured its probiotic properties by performing antimicrobial test, in vitro antioxidant capacity test, cell surface hydrophobicity test, auto-aggregation test, and GI environment resistance test. The antibiotic susceptibility of the strain was also measured.
Colonization in the intestinal tract is the most important step for probiotics to develop beneficial effects. Probiotics should overcome the acidic gastric conditions and bile salt conditions in the gastrointestinal tract. The small intestine and the colon contain relatively high concentrations of bile salt which is toxic to living cells [35]. In the present study, X3 showed a survival rate of 67.78% at pH 3.0 after 2 h of exposure. Aside from low pH resistance, X3 could also survive at 0.3% bile salt concentration. Cell surface hydrophobicity and auto-aggregation are important selection criteria for probiotic screening. Multiple reports have demonstrated that probiotics with a high surface hydrophobicity and co-aggregation rate could have a strong adherence to the gastrointestinal tract [36,37,38,39], which was essential for probiotics to colonize and play a beneficial role. In the present study, these two criteria of selected LAB strains were measured. X3 demonstrated strong hydrophobicity in three different organic solvents and high auto-aggregation rate. Therefore, strain X3 shows potential to be a probiotic.
Probiotics keep intestinal health through inhibiting intestinal pathogen growth. In the present study, we have tested the antagonistic effect on common intestinal pathogens. The 48 selected LAB strains inhibited intestinal pathogens including E. coli and ETEC. X3 exerted inhibitory effects on all test pathogens, suggesting its potential to improve intestinal health. Antioxidant property allows probiotics to relieve body oxidative stress through various mechanisms, including secretion of antioxidant enzymes and mediation of antioxidant signaling pathways. Previous studies [40, 41] reported that potential probiotics usually possess desirable antioxidant properties in vivo or in vitro. In the present study, the in vitro antioxidant properties of the selected LAB strains and their cell-free suspension were measured. Results showed that the antioxidant property of full bacteria cell was better than that of CFS. X3 demonstrated the best in vitro antioxidant property among all selected strains. Therefore, strain X3 could be considered as a probiotic.
Antibiotic susceptibility is another concern for bacteria to be probiotics; bacteria that contain antibiotic resistance genes may induce horizon gene transfer. In the present study, we tested the antibiotic susceptibility of strain X3 to four kinds of antibiotics. Result showed that X3 was sensitive to commonly used antibiotics. Therefore, strain X3 could be considered safe.
In conclusion, strain X3, which was identified as BSP201703, was isolated from giant panda feces and found to have favorable probiotic properties. However, further research is needed to explore additional functional aspects of this strain.
References
Liu Q, Ni X, Wang Q, Peng Z, Niu L, Wang H, Zhou Y, Sun H, Pan K, Jing B, Zeng D (2017) Lactobacillus plantarum BSGP201683 Isolated from Giant panda feces attenuated inflammation and improved gut microflora in mice challenged with Enterotoxigenic Escherichia coli. Front Microbiol 8:1885. https://doi.org/10.3389/fmicb.2017.01885
Ibrahem MD (2015) Evolution of probiotics in aquatic world: potential effects, the current status in Egypt and recent prospectives. J Adv Res 6(6):765–791. https://doi.org/10.1016/j.jare.2013.12.004
Collins MD, Samelis J, Metaxopoulos J, Wallbanks S (1993) Taxonomic studies on some Leuconostoc-like organisms from fermrnted sausages:description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J Appl Bacteriol 75:595–603. https://doi.org/10.1111/j.1365-2672.1993.tb01600.x
Abriouel H, Lerma LL, Casado Munoz Mdel C, Montoro BP, Kabisch J, Pichner R, Cho GS, Neve H, Fusco V, Franz CM, Galvez A, Benomar N (2015) The controversial nature of the Weissella genus: technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front Microbiol 6:1197. https://doi.org/10.3389/fmicb.2015.01197
Kumar A, Augustine D, Sudhindran S, Kurian AM, Dinesh KR, Karim S, Philip R (2011) Weissella confusa: a rare cause of vancomycin-resistant gram-positive bacteraemia. J Med Microbiol 60(Pt 10):1539–1541. https://doi.org/10.1099/jmm.0.027169-0
Flaherty JD, Levett PN, Dewhirst FE, Troe TE, Warren JR, Johnson S (2003) Fatal case of endocarditis due to Weissella confusa. J Clin Microbiol 41(5):2237–2239. https://doi.org/10.1128/jcm.41.5.2237-2239.2003
Kamboj K, Vasquez A, Balada-Llasat JM (2015) Identification and significance of Weissella species infections. Front Microbiol 6:1204. https://doi.org/10.3389/fmicb.2015.01204
Ana I, Vela CP (2003) Weissella confusa infection in primate (Cercopithecus mona). Emerq Infect Dis 9(10):1307–1309
Kang BK, Cho MS, Park DS (2016) Red pepper powder is a crucial factor that influences the ontogeny of Weissella cibaria during kimchi fermentation. Sci Rep 6:28232. https://doi.org/10.1038/srep28232
Lee KW, Park JY, Jeong HR, Heo HJ, Han NS, Kim JH (2012) Probiotic properties of Weissella strains isolated from human faeces. Anaerobe 18(1):96–102. https://doi.org/10.1016/j.anaerobe.2011.12.015
Nam H, Ha M, Bae O, Lee Y (2002) Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl Environ Microb 68(9):4642–4645. https://doi.org/10.1128/aem.68.9.4642-4645.2002
Silva MS, Ramos CL, González-Avila M, Gschaedler A, Arrizon J, Schwan RF, Dias DR (2017) Probiotic properties of Weissella cibaria and Leuconostoc citreum isolated from tejuino – a typical Mexican beverage. LWT Food Sci Technol 86:227–232. https://doi.org/10.1016/j.lwt.2017.08.009
Abdel-Daim A, Hassouna N, Hafez M, Ashor MS, Aboulwafa MM (2013) Antagonistic activity of Lactobacillus isolates against Salmonella typhi in vitro. Biomed Res Int 2013:680605–680612. https://doi.org/10.1155/2013/680605
Adeniyi BA, Adetoye A, Ayeni FA (2015) Antibacterial activities of lactic acid bacteria isolated from cow faeces against potential enteric pathogens. Afr Health Sci 15(3):888–895. https://doi.org/10.4314/ahs.v15i3.24
Ilavenil S, Park HS, Vijayakumar M, Arasu MV, Kim DH, Ravikumar S, Choi KC (2015) Probiotic potential of Lactobacillus strains with antifungal activity isolated from animal manure. Sci World J 2015:802570–802510. https://doi.org/10.1155/2015/802570
Yadav R, Puniya AK, Shukla P (2016) Probiotic properties of Lactobacillus plantarum RYPR1 from an indigenous fermented beverage Raabadi. Front Microbiol 7:1683. https://doi.org/10.3389/fmicb.2016.01683
Owusu-Kwarteng J, Tano-Debrah K, Akabanda F, Jespersen L (2015) Technological properties and probiotic potential of Lactobacillus fermentum strains isolated from west African fermented millet dough. BMC Microbiol 15:261. https://doi.org/10.1186/s12866-015-0602-6
Belaqziz M, Tan SP, El-Abbassi A, Kiai H, Hafidi A, O'Donovan O, McLoughlin P (2017) Assessment of the antioxidant and antibacterial activities of different olive processing wastewaters. PLoS One 12(9):e0182622. https://doi.org/10.1371/journal.pone.0182622
Jemil N, Ben Ayed H, Manresa A, Nasri M, Hmidet N (2017) Antioxidant properties, antimicrobial and anti-adhesive activities of DCS1 lipopeptides from Bacillus methylotrophicus DCS1. BMC Microbiol 17(1):144. https://doi.org/10.1186/s12866-017-1050-2
Kumar A, Kumar D (2016) Development of antioxidant rich fruit supplemented probiotic yogurts using free and microencapsulated Lactobacillus rhamnosus culture. J Food Sci Technol 53(1):667–675. https://doi.org/10.1007/s13197-015-1997-7
Fakruddin M, Hossain MN, Ahmed MM (2017) Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complem Altern M 17(1):64. https://doi.org/10.1186/s12906-017-1591-9
Xing J, Wang G, Zhang Q, Liu X, Gu Z, Zhang H, Chen YQ, Chen W (2015) Determining antioxidant activities of lactobacilli cell-free supernatants by cellular antioxidant assay: a comparison with traditional methods. PLoS One 10(3):e0119058. https://doi.org/10.1371/journal.pone.0119058
Campana R, van Hemert S, Baffone W (2017) Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog 9:12. https://doi.org/10.1186/s13099-017-0162-4
Lee A, Cheng KC, Liu JR (2017) Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PLoS One 12(8):e0182220. https://doi.org/10.1371/journal.pone.0182220
Martin R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, Sokol H, Bermudez-Humaran LG, Thomas M, Langella P (2017) Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front Microbiol 8:1226. https://doi.org/10.3389/fmicb.2017.01226
Adnan M, Patel M, Hadi S (2017) Functional and health promoting inherent attributes of Enterococcus hirae F2 as a novel probiotic isolated from the digestive tract of the freshwater fish Catla catla. Peer J 5:e3085. https://doi.org/10.7717/peerj.3085
Zhou S, Qu Y, Huang J, Huang Y, Li D, Zhang H (2017) A summary of researches on the wild Giant panda population dynamics (in Chinese). J Sichuan For Sci Technol 38(2):19–33. (周世强,屈元元,黄金燕,黄强,李德生 and 张和民 (2017) 野生大熊猫种群动态的研究综述. 四川林业科技 38(2):19-33). https://doi.org/10.16779/j.cnki.1003-5508.2017.02.004
Qing X, Zeng D, Wang H, Ni X, Liu L, Lai J, Khalique A, Pan K, Jing B (2017) Preventing subclinical necrotic enteritis through Lactobacillus johnsonii BS15 by ameliorating lipid metabolism and intestinal microflora in broiler chickens. AMB Express 7(1):139. https://doi.org/10.1186/s13568-017-0439-5
Wang H, Ni X, Qing X, Zeng D, Luo M, Liu L, Li G, Pan K, Jing B (2017) Live probiotic Lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front Microbiol 8:1073. https://doi.org/10.3389/fmicb.2017.01073
Kang BK, Cho MS, Ahn T-Y, Lee ES, Park DS (2015) The influence of red pepper powder on the density of Weissella koreensis during kimchi fermentation. Sci Rep 5(1):15445. https://doi.org/10.1038/srep15445
Lim SK, Kwon MS, Lee J, Oh YJ, Jang JY, Lee JH, Park HW, Nam YD, Seo MJ, Roh SW, Choi HJ (2017) Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci Rep 7:40040. https://doi.org/10.1038/srep40040
Fessard A, Kapoor A, Patche J, Assemat S, Hoarau M, Bourdon E, Bahorun T, Remize F (2017) Lactic fermentation as an efficient tool to enhance the antioxidant activity of tropical fruit juices and teas. Microorganisms 5(2):E23. https://doi.org/10.3390/microorganisms5020023
Yan Y-B, Kajala I, Shi Q, Nyyssölä A, Maina NH, Hou Y, Katina K, Tenkanen M, Juvonen R (2015) Cloning and characterization of a Weissella confusa Dextransucrase and its application in high fibre baking. PLoS One 10(1):e0116418. https://doi.org/10.1371/journal.pone.0116418
Goh HF, Philip K (2015) Purification and characterization of Bacteriocin produced by Weissella confusa A3 of dairy origin. PLoS One 10(10):e0140434. https://doi.org/10.1371/journal.pone.0140434
Tokatli M, Gulgor G, Bagder Elmaci S, Arslankoz Isleyen N, Ozcelik F (2015) In Vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. Biomed Res Int 2015:315819–315818. https://doi.org/10.1155/2015/315819
Shin J-H, Lee J-S, Seo J-G (2015) Assessment of cell adhesion, cell surface hydrophobicity, autoaggregation, and lipopolysaccharide-binding properties of live and heat-killed Lactobacillus acidophilus CBT LA1. Korean J Microbiol 51(3):241–248. https://doi.org/10.7845/kjm.2015.5029
Grigoryan S, Bazukyan I, Trchounian A (2017) Aggregation and adhesion activity of lactobacilli isolated from fermented products in vitro and in vivo: a potential probiotic strain. Probiotics Antimicrob Proteins. https://doi.org/10.1007/s12602-017-9283-9
Gupta A, Tiwari SK (2014) Probiotic potential of Lactobacillus plantarum LD1 isolated from batter of Dosa, a south Indian fermented food. Probiotics Antimicrob Proteins 6(2):73–81. https://doi.org/10.1007/s12602-014-9158-2
Son SH, Jeon HL, Yang SJ, Lee NK, Paik HD (2017) In vitro characterization of Lactobacillus brevis KU15006, an isolate from kimchi, reveals anti-adhesion activity against foodborne pathogens and antidiabetic properties. Microb Pathog 112:135–141. https://doi.org/10.1016/j.micpath.2017.09.053
Tan LT, Chan KG, Khan TM, Bukhari SI, Saokaew S, Duangjai A, Pusparajah P, Lee LH, Goh BH (2017) Streptomyces sp. MUM212 as a source of antioxidants with radical scavenging and metal chelating properties. Frontiers in. Pharmacology 8:276. https://doi.org/10.3389/fphar.2017.00276
Kadaikunnan S, Rejiniemon T, Khaled JM, Alharbi NS, Mothana R (2015) In-vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann Clin Microbiol Antimicrob 14:9. https://doi.org/10.1186/s12941-015-0069-1
Acknowledgements
The present study was supported by National Natural Science Foundation of China (No. 31672318) and the Funded Project of Chengdu giant panda Breeding Research Foundation (No. CPF2014-15 and CPF2015-16).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Lvchen Xiong, Xueqin Ni, and Lili Niu are the joint first authors.
Rights and permissions
About this article
Cite this article
Xiong, L., Ni, X., Niu, L. et al. Isolation and Preliminary Screening of a Weissella confusa Strain from Giant Panda (Ailuropoda melanoleuca). Probiotics & Antimicro. Prot. 11, 535–544 (2019). https://doi.org/10.1007/s12602-018-9402-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-018-9402-2