Abstract
Four lactic acid bacteria, Leuconostoc mesenteroides subsp. mesenteroides, were isolated from aguamiel the sap obtained from Agave salmiana from México and identified by 16S rRNA gene sequence analysis. The probiotic potential of these strains was evaluated and compared with a commercial probiotic (Lactobacillus plantarum 299v) from human origin. All the strains survived the in vitro gastrointestinal simulation conditions: the stomach simulation (3 h, pH 2, 37 °C) and the intestinal simulation (4 h, bile salts 0.5 %, 37 °C). All the strains showed a strong hydrophilic character with n-hexadecane and chloroform assays, and all the strains showed a mucin adhesion rate similar to that of L. plantarum 299v. The strains of L. mesenteroides subsp. mesenteroides exhibited similar antimicrobial activity against some pathogens in comparison with L. plantarum 299v. Some antibiotics inhibited the growth of the strains. L. mesenteroides subsp. mesenteroides exhibited in vitro probiotic potential, and it could be better characterized through future in vivo tests.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A high percent of the population has gastrointestinal disorders as a consequence of a poor diet, unhealthy lifestyle, and stress [1]. Probiotics are conceived as a strategy to restore composition and function of gut microbiota, which may contribute to decrease gastrointestinal disorders [2, 3]. The most important genera of probiotics commercialized to date are Lactobacillus and Bifidobacterium [4]. Probiotics may provide a relief from lactose intolerance and prevent episodes of diarrheas of different etiologies [5]. Most commercialized probiotic bacteria have been isolated from dairy fermented products and human feces [6, 7]. However, evidence for efficacy of existing probiotics in humans is less strong than expected [8], which has encouraged the selection of strains with improved functions from unconventional sources [9]. In 2001 and 2002, a group of experts working under the umbrella of FAO (Food and Agriculture Organization of the United Nations) and the WHO (World Health Organization) defined probiotics as those microorganisms that administered in adequate amounts confer a health benefit to the host. This group of experts also established the criteria for evaluation of probiotics used in foods and food supplements [7, 10–12]. According to which, probiotics should be safe and, therefore, not be pathogenic or toxigenic or harbor antibiotic resistance genes that can be transferred. Additionally, it is also desirable that probiotic strains are resistant to gastrointestinal conditions (e.g., stomach acid pH, bile acids, etc.); adhere to intestinal epithelial cells [13, 14], produce antimicrobial compounds and exclude pathogens by positively enhancing immune functions [15–21]. Furthermore, effectiveness of probiotic strains should be confirmed in human studies. Considering the importance of probiotics in health, there is great interest in studies on fermented food products as a source of new probiotic isolates [22, 23]. Moreover, the strains isolated from dairy products are candidates for inclusion in foods as probiotics because they are adapted to the conditions [24]. In this study, the aim was to evaluate the probiotic potential of four isolates of Leuconostoc mesenteroides subsp. mesenteroides isolated from the aguamiel of Agave salmiana, in order to select good candidates for further studies. Aguamiel is the sap obtained from A. salmiana, A. mapisaga, A. atrovirens, A. americana, and A. ferox which is fermented to produce pulque, a traditional alcoholic beverage from Mexico [25].
Materials and Methods
Four isolates of L. mesenteroides from the aguamiel of A. salmiana were employed in the present research: SD1, SD23, SF2, and SF3. In addition, one commercial strain of probiotic from human origin was used as positive probiotic control: Lactobacillus plantarum 299v was isolated from Protransitus® (Laboratorios Salvat, Barcelona, Spain). All strains were cultured at 30 °C for 24 h in lactobacilli MRS broth (Dibico, Mexico City, México). Pathogenic strains of Salmonella enterica subsp. enterica Serovar. Thyphimurium ATCC 14028, Listeria monocytogenes ATCC 19115, Escherichia coli ATCC 43895, Staphylococcus aureus ATCC 25923, and S. aureus FRI 184 were used in the antimicrobial activity assay. All pathogenic strains were grown in Müeller-Hinton broth (Difco, Detroit, USA) at 37 °C under aerobic conditions. All the strains were preserved frozen at −20 °C with glycerol as a cryoprotectant until used.
Bacterial Strains and Growth Conditions
The aguamiel of A. salmiana from Nopaltepec, México was used for the isolation of L. mesenteroides. For isolation of Leuconostoc, 10 mL of aguamiel was homogenized in 90 mL of sterile water; aliquots of serially diluted suspensions were pour-plated De Man-Rogosa-Sharpe (MRS, Difco Laboratories, MI, USA) isolated agar. Plates were incubated at 30 °C for 48 h. After incubation, lenticular colonies were picked up, and the isolates were purified three times with repeated pour-plating with MRS agar. Purity was checked by plating on corresponding agar media (MRS) and microscopic examination. Cultural characteristics of colonies were observed directly on the plates. For morphological characteristics, Gram staining was carried out by using a crystal violet/iodine solution (Hycel, México) following the procedure as described by Murray et al. [26].
Isolates Identification
The four isolates were cultivated in lactobacilli MRS broth. The total genomic DNA for each isolated strain was extracted according to the Kit Promega Cat#A1125 wizard Genomic DNA purification. The identities of the strains were determined with the use of 16S rRNA PCR amplification [27]. Each PCR was performed with the following components: 5 μL Buffer 10×; 4 μL MgSO4; 4 μL of each dNTP; 0.5 μL of each primer: LeuR (TTTGTCTCCGAAGAGAACA) and LeuF (CGAAAGGTGCTTGCACCTTTCAAG) [28] 2.5 μL of genomic DNA; 0.5 μL of KOD (DNA polymerase from Thermococcus kodakarensis) [29] and adjusted to 50 μL with distilled water. The PCR were carried out in a DNAThermal Cycler (Labnet model: TC020-24) using the following amplification conditions: 30 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 20 s at 70 °C; the cycles were preceded by denaturation at 94 °C for 5 min and were followed by extension at 70 °C for 5 min. The reaction products were analyzed by electrophoresis through 1 % (w/v) agarose in 0.5× TAE (Tris-Acetate-EDTA buffer). PCR products were purified using to the Kit WizardSV cell and PCR clean-up system. After these PCR products were sequenced in Macrogen Inc. (Seoul, Korea), using the same primers. Sequences from isolated bacteria were compared with BLASTsearch v. 2.2.3 [30] (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Survival under Conditions Simulating the Human Gastrointestinal (GI) Tract
The strains were grown in MRS broth for 24 h, subsequently 2 × 109 CFU/mL of each strain were added to a flask containing 50 mL of MRS broth adjusted to pH 2 with 6 N HCl and incubated at 37 °C for 3 h (stomach conditions). The strains were also inoculated in MRS broth supplemented with 0.5 % Oxgall (w/v) and incubated at 37 °C for 4 h (intestinal conditions). Both experiments were performed in triplicate. The viability was calculated as percent viability = [Log (CFUfinal/mL)/Log (CFUinitial/mL)] × 100. The initial value corresponds to plate counts of inoculated bacteria in fresh medium initially, and the final value corresponds to the bacterial counts obtained after incubation in simulated GI conditions [9].
Survival after the successive passages through gastric juice and intestinal juice was determined according to Vizoso et al, [31] with some modifications. A bacterial suspension (10 %) was added to an artificial gastric fluid, consisting of 3 mg/mL pepsin (Sigma, St. Louis, MO, USA) in sterile electrolyte solution at pH 2. After 1 h of incubation at 37 °C, 1 mL aliquot was removed, serially diluted in PBS (phosphate-buffered saline), and spread-plated onto MRS agar. A 10 % of the bacterial suspension of the gastric fluid was added to an artificial intestinal juice to pH 6.5, 0.5 % bile salts (Oxgall), and 1.9 mg/mL pancreatin (Sigma, St. Louis, MO, USA). One milliliter aliquots were again removed after 30 min, 90 min, and 3 h, serially diluted in PBS and spread-plated onto MRS agar to determine the CFU/ml and the total survival could be calculated.
Adherence to the Intestinal Mucosa
The adhesion of the strains to intestinal cells was evaluated using the intestine of Wistar male rats of 3 months of age as model, according to Brink et al. [32] with additional slight modifications. The intestine was washed three times in PBS buffer solution at 10 °C and then inoculated with previously activated strains (1.5 × 109 CFU/mL) in MRS broth. After incubation at 37 °C for 3 h, the tissue was scraped to count the viable bacteria in MRS agar. The adhesion was expressed as percent of bacteria adhered to the tissue respect to total bacteria initially present in the adhesion assay: % ad = [Log (CFUadhered/mL)/Log (CFUinitial/mL)] × 100.
Scanning Electron Microscopy (SEM)
Preparations of the samples for SEM analysis were performed according to the method of Panyarachum et al. [33]: The strains were fixed in 2.5 % glutaraldehyde for at least 2 h. They were washed three times with 0.1 M sodium cacodylate buffer, pH 7.2, at 4 °C, and post-fixed in 1 % osmium tetroxide in 0.1 M sodium cacodylate buffer, pH 7.2, at 4 °C for 1 h. After washing in three changes of distilled water, they were dehydrated through increasing concentrations of ethanol, and dried in a Hitachi HCP-2 critical point drying machine using liquid carbon dioxide as a transitional medium. Thereafter, the samples were mounted on aluminum stubs and coated with gold in anion-sputtering apparatus, SPI-Model sputter coater for 15 min, and they were examined in a JEOL JSM-5800LV.
Hydrophobicity
Microbial adhesion to organic solvents was determined by the MATH method (microbiological adhesion to hydrocarbons) developed by Rosenberg et al. [34] and Doyle and Rosenberg [35]. The strains were cultured at 30 °C for 24 h in lactobacilli MRS broth, harvested by centrifugation (13,000 rpm/5 min, 20 °C), and washed twice with PBS buffer pH 7.4. Three ml of bacterial suspension (OD 560 nm = 0.23 ± 0.07) was put in contact with 0.75 mL of n-hexadecane (a nonpolar organic solvent) or chloroform (an acidic polar organic solvent). The mixture was gently mixed in a vortex for 2 min. Tubes were allowed to stand for 1 h in a water bath at 37 °C, and the absorbance at 560 nm of the aqueous phase was measured. The hydrophobicity to each solvent was calculated as H % = [(A i − A f)/A i] × 100, where A i is the initial optical density (without the solvent) and A f is the optical density of the aqueous phase at the end of the experiment. Hydrophobicity was measured at least in triplicate during three different days.
Adhesion to Mucin Assay
Adhesion of the strains to mucin was determined according to Melgar-Lalanne et al. [22] with additional slight modifications. Crude porcine gastric type II mucin (Sigma-Aldrich) was dissolved in PBS at pH 7.2 to a final concentration of 10 mg/mL, and 300 μL were placed in 96-well polystyrene plates and immobilized by overnight incubation at 4 °C. The wells were washed twice with 150 μL of PBS and saturated with 2.0 % (w/v) bovine serum albumin (BSA) solution for 4 h at 4 °C. Finally, the wells were washed twice with 150 μL of PBS. The strains were grown for 16 h at 30 °C in MRS broth. Bacterial suspensions were centrifuged (6000 rpm, 15 min, 20 °C), washed twice with PBS, and the concentration of each strain was approximately 2 × 108 CFU/mL. 150 μL of the bacterial suspension was added to each well and incubated for 1 h at 37 °C. The wells were washed with 150 μL of PBS and taken under agitation (50 rpm, 5 min). This procedure was repeated three times to remove all the unbound bacteria. The wells were then treated with 100 μL of a 0.05 % (v/v) solution of Triton X-100 solution to release the bound bacteria. Plates were then incubated 40 min at room temperature at 100 rpm. An aliquot of 100 μL of the content of each well was removed, diluted in PBS, and plated on MRS agar (after and before the adhesion to mucin). Previous studies had shown that 0.05 % Triton X-100 does not affect cell viability [36].
Antimicrobial Activity
To evaluate the inhibitory activity of all strains, these were incubated in MRS broth at 30 °C for 24 h. After incubation, supernatants were prepared by centrifugation (6000 rpm, 20 min, 20 °C) and sterilized by syringe filtering (0.42 µm). To neutralize the inhibitory effect of lactic acid, the supernatants were adjusted to pH 6.5 with NaOH 1 N and catalase (1 mg/mL) was added to exclude the inhibition due to hydrogen peroxide production. The compounds were tested using the diffusion agar assay. Pathogenic bacteria were grown in Müeller-Hinton broth overnight at 37 °C (1.2 × 108 CFU/mL), and they were extended in Müeller-Hinton, and 0.85 cm diameter wells were punched into the surface of each plate. Subsequently, 25 μL of each supernatant was added to each well and incubated at 37 °C for 24 h. Antimicrobial activity was evaluated by measuring the formation of inhibition zones around the wells. The experiments were performed in triplicate [37].
The proteinaceous nature of the antimicrobial agents produced by the strains tested in this research was shown by treating separately their supernatants (pH 6.5) with three proteolytic enzymes [22]: protease type VIII from Bacillus licheniformis (Sigma, Novozyme Corp, Denmark) (1 µL/mL), trypsin type III from bovine pancreas (Sigma, St. Louis, MO, USA)(1 mg/mL), and pronase E from Streptomyces griseous (Sigma, St Louis, MO, USA) (1 mg/mL). The enzymes were dissolved in 0.1 M potassium phosphate buffer (pH 8.0) before being added to the supernatants. Each mixture of the supernatants plus the proteolytic enzyme was incubated for 2 h at 37 °C for the hydrolysis to take place. The enzymatic reaction was stopped by heating at 90 °C for 10 min in a water bath. After that, the residual antimicrobial activity of the supernatants with the hydrolyzed proteinaceous compounds was tested using the well-diffusion agar assay as described above.
Antibiotic Resistance
To evaluate the resistance to antibiotics, each bacteria was incubated in MRS broth at 30 °C for 24 h, and 100 µL of the microbial suspensions of each bacteria (109 CFU/mL) was inoculated into MRS agar at 30 °C and Antibiotic discs (multiplate Bio-RAD, Gram positive) were placed on the surface of the agar [38]. Diameters of the inhibition zones were measured. Both experiments were performed in triplicate, and the results were expressed as sensitive (S) or resistant (R) depending on the presence or absence of inhibition halos, respectively.
Hemolytic Activity
Fresh bacterial cultures were streaked in triplicate on Columbia agar plates, containing 5 % (w/v) human blood, and incubated for 48 h at 30 °C. Blood agar plates were examined for signs of β-hemolysis (clear zones around colonies), α-hemolysis (green-hued zones around colonies) or γ-hemolysis (no zones around colonies) [9].
Statistical Analysis
The survival rate in stomach and intestine was calculated as the logarithmic difference between final and initial colony counts as follows: percent viability = [Log (CFUfinal/mL)/Log (CFUinitial/mL)] × 100. Arithmetic mean and standard derivation were calculated both in adhesion (%) and hydrophobicity (%) tests. All statistical analyses were performed with the MS Excel software.
Results
PCR Amplification of 16S rRNA Sequence
The bands of the PCR amplification can be observed in Fig. 1, the length of PCR products was approximately of 1000 bp (Fig. 1). The strain of highest similarity was L. mesenteroides subsp. mesenteroides (KJ026680.1 in GenBank). Based on 16S rRNA sequences analysis, the isolates were identified as strains of the lactic acid bacteria, L. mesenteroides subsp. mesenteroides.
Survival under Conditions Simulating the Human GI Tract
Table 1 shows the viability expressed as the percent viability = [Log (CFUfinal/mL)/Log (CFUinitial/mL)] × 100, indicating that under conditions of pH 2, the four strains of L. mesenteroides suffered a decrease in their percent viability. L. plantarum had slight changes in their numbers. The strain SD1 was the most sensitive, but all strains could survive the acidic conditions after an incubation period of 3 h. Significant differences in survival between L. plantarum 299v and the strains of L. mesenteroides were observed under acidic conditions. Under in vitro intestinal simulation conditions (0.5 % w/v of bile salts), all the strains tested could survive after the physiological time (4 h). L. plantarum 299v kept its number almost constant, while the most sensitive strain was SD1, which presented the lowest percent of viability. All the strains of L. mesenteroides could survive to the conditions of the GI tract compared with the control strain from human origin. Under the effect of the successive passages through gastric and intestinal juices, all the strains tested could survive after the physiological time of 90 min, but they could not survive after 3 h. SD23 had the highest survival with 27.43 % after 30 min and 16.75 % after 90 min. The strains SD1, SF3, and SF2 showed percentages of survival ranking between 23.35 and 25.87 % after 30 min and ranking between 11.68 and 13.49 % after 90 min.
Adherence to the Intestinal Mucosa
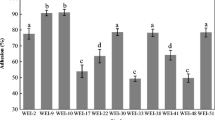
The percent of adherence to the intestinal mucosa values of all strains is shown in Table 2. The strains of L. mesenteroides presented higher adhesion values than the control strain L. plantarum 299v, after 3 h at 37 °C. SEM analyses revealed that the adherence to the intestinal mucosa was presented in all the strains (see Fig. 2).
Hydrophobicity
All strains of L. mesenteroides presented higher hydrophobicity values than the control strain (see Table 3), measured using the interaction with chloroform and n-hexadecane to simulate the ability to adhere to the intestinal epithelium [35]. Microbial adhesion to the nonpolar solvent (n-hexadecane) ranking between 13.06 and 85.19 % indicating a hydrophilic surface. The percentage of hydrophobicity to chloroform was between 25.39 and 84.14 %.
Adhesion to Mucin Assay
Adhesion to mucin of all strains of L. mesenteroides was compared to that of L. plantarum 299v (see Table 4). The strain L. mesenteroides SD23 presented the highest adhesion value (95.78 %) of all the strains, and the L. mesenteroides SF3 strain from aguamiel of A. salmiana had an adhesion value similar to that of L. plantarum 299v.
Antimicrobial Activity
The supernatant of the isolated showed inhibition to three enteropathogenic strains: E. coli ATCC43895 (EC), S. enterica ATCC 14028 (ST), and L. monocytogenes ATCC 19115 (LM) as well as two enterotoxigenic strains: S. aureus [ATCC 25923 (SA) and FRI 184 (SAI)]. These inhibition zones were similar to values previously reported [39], high inhibition zones between 11 and 13 mm were only found with the supernatant from L. mesenteroides SD23, SD1, and SF2 that inhibited L. monocytogenes. Low inhibition zones (7 mm) were found with the supernatant from L. plantarum 299v that inhibited S. enterica and E. coli, the supernatant from L. mesenteroides SD23 that inhibited S. enterica and S. aureus FRI 184, the supernatant from L. mesenteroides SD1 that inhibited S. aureus FRI 184, and the supernatant from L. mesenteroides SF2 that inhibited S .aureus FRI 184. The rest of the supernatants showed medium levels of inhibition against the five pathogens tested (see Table 5). A complete inactivation or at least a significant reduction in the antimicrobial activity of the agents produced by the supernatants of the strains tested against the pathogenic strains was observed after treatment of the supernatants with protease type VIII, trypsin, and pronase E indicating the possible production of bacteriocins by the Leuconostoc strains.
Antibiotic Resistance
The isolates were sensitive to common antibiotic used in clinical applications, however, presented resistance against dicloxacillin, pefloxacin, trimethoprim, and ceftazidime (see Table 6). The observations indicated that the isolates showed low probability of antibiotic resistance. Moreover, the strain L. plantarum 299v was resistant to nine out of twelve antibiotics.
Hemolytic Activity
None of the examined strains exhibited β-Hemolytic activity when grown in Columbia human blood agar. All the strains were γ-Hemolytic (no hemolysis). Similar observations were made for strains isolated from dairy products which showed γ-hemolysis except of few that showed α-hemolysis [40].
Discussion
In Vitro Gastrointestinal Simulation
This study showed that all strains of L. mesenteroides subsp. mesenteroides SD1, SD23, SF2 and SF3 can survive against the stress conditions assayed in this study. A potential probiotic should have some properties, including the ability to tolerate and survive the acidic environment of the stomach and the bile salts in the small intestine. Previous studies reported that L. mesenteroides subsp. mesenteroides isolated from different sources survived better under neutral conditions present in the small intestine [23, 41]. Other studies also indicated that LAB were able to grow up and survive at low pH levels [42, 43], but it is important to point out that the in vitro trials involving pH, and bile salts cannot predict all the patterns of behavior in the human body, due to the existence of other factors that affect the survival of microorganisms in the upper gastrointestinal tract such as the presence of different enzymes and the peristaltic movements [11, 36, 44]. Bile salts play an important role in the defense mechanisms of the gut; its inhibitory effects depend on the concentrations of this [45], the physiological concentrations of human bile range from 0.3 to 0.5 % [13, 46]. In this study, all strains of L. mesenteroides subsp. mesenteroides and L. plantarum 299v survived in the presence of 0.5 % bile salts. According to Argyri et al. [9], Vizoso et al. [31], LAB isolated from fermented olives and traditional African fermented milk products could survive the stress conditions found in the gastrointestinal tract. The values reported by Argyri et al. [9] and Vizoso et al. [31] were similar to the values obtained in this study. This positive behavior can increase the possibility of these microorganisms to colonize and grow in gut condition.
In Vitro Adhesion Abilities
The ability to adhere to the intestinal mucosa is a criterion important for probiotic strains. The colonization of the intestine by probiotic strains may increase the beneficial biological responses to the host [47, 48]. The process of mucosal cell adhesion is complex and may involve Van der Waals and electrostatic forces between the bacterial cell and the mucosal surface, explained by the DLVO theory (Derjaguin, Landau, Verwey, and Overbeek) [49]. Investigation of the specific bacterial molecules involved in adhesion comparing the proteome of high and low adherent strains of L. plantarum also led to the identification of possible proteins involved [50]. We found that the four strains of L. mesenteroides satisfied the most important criteria for the selection of probiotics, the ability to adhere to the intestinal mucosa (Tables 2, 3, 4). Different methods have been used to evaluate probiotics adhesion as adhesion to intestinal mucus, and the adhesion ability to human colon carcinoma cells (Caco-2) [9]. However, the intestinal mucus tissue models allow for the testing of host-specific factors such as health status or age in adhesion studies, providing further information and is probably the most realistic option to test the adhesion because the normal microbiota present in the intestine mucosa is taken into account in the assay [51–53]. The hydrophobicity might be the first contact between the microorganisms and the host cells [23, 54]. The results obtained in the hydrophobicity to the nonpolar solvent (n-hexadecane) for the strains of L. mesenteroides subsp. mesenteroides SD1, and SF3 were higher than some other strains reported in the literature [55, 56]: LAB isolated from fermented vegetables, sourdough, milk products, and sheep and human excreta (23.0–73.0 %), Pediococcus pentosaceus CFRR38 and CFRR35, and Lactobacillus rhamnosus GG ATCC 53510 (44.8–59.0 %), and Leuconostoc paramesenteroides isolated from cheddar cheese (46.11 %) [54]. Hydrophobicity may depend on different compounds commonly used (n-hexadecane, xylene, chloroform and toluene), and on the strains evaluated that can lead to different results. It is known that the ability to adhere to mucus is a requirement of a probiotic microorganism. The mucus layer is a viscous material that coats the intestinal tract and consists mainly of glycosylated proteins (mucins) and glycolipids as well as antibodies, ions, dietary products, and water [57], recently some researches indicate that a mucin adhesion method could be used to simulate adhesion [58]. In our study, the strains of L. mesenteroides SD1, SD23, and SF2 presented the highest adhesion values to mucin (see Table 4) and the strain L. mesenteroides SF3 had an adhesion value similar to that of the probiotic strain from human origin L. plantarum 299v.
Antimicrobial Activity
The most important property by which probiotic bacteria exert their protective and beneficial physiological effects is through antagonistic activity against pathogenic bacteria [59]. This antimicrobial effect can be mediated by the ability of the strains to lower the pH of the medium by fermentation and generation of organic acids [60] (especially lactic and acetic acids), which have bactericidal or bacteriostatic effects. The ability to produce various antimicrobial compounds can be one of the main characteristics for effective competitive exclusion of pathogen survival in the gut and the development of a probiotic effect [61]. The acidic conditions in the stomach can even improve the antimicrobial activity of these compounds [62]. Furthermore, these probiotic characteristics may be due in part to the production of relevant concentrations of lactic acid that in combination with a detergent such as bile salts inhibits the growth of gram-negative pathogens [63]. In this study, neutralized supernatant from all strains exhibited strong inhibitory activity; this activity may be due to the production of antimicrobial substances such as bacteriocins or bacterion-like substances. The proteinaceous nature of the antimicrobial agents produced by the strains tested in this research was verified by treatment with three proteolytic enzymes: protease type VIII, trypsin, and pronase E that are able to break peptide bonds at different amino acid residue positions [64], this caused the loss of activity in the supernatants peptides of the strains tested.
Antibiotic Resistance
In this study, different groups of antibiotics were used: cell wall inhibitors (Penicillin, Dicloxacillin and Ampicillin); inhibitors of protein synthesis (tetracycline, gentamicin, erythromycin, Cephalothin, Cefotaxime, Ceftazidime, Cefuroxime) and inhibitors of DNA and RNA synthesis (Pefloxacin and Trimethoprim) [39]. The frequent consumption of these types of antibiotics may cause imbalance in the intestinal sensitive microbiota. The antibiotic resistance in probiotics usually does not constitute a safety issue when mutations or intrinsic resistance mechanisms are accountable for the resistance phenotype and so the microbial balance can be preserved [65]. A single bacterial strain may possess several types of resistance mechanisms: intrinsic or innate, acquired and mutational [66]. Generally, the bacteria are capable of developing transmissible mechanisms of antimicrobial resistance, including the production of bacterial enzymes that inactive the antibiotics [67, 68]. In the case of gram-positive bacteria, they may produce β-lactamase [69]. The strains isolated of A. salmiana were resistant to dicloxacillin, pefloxacin, trimethoprim, and ceftazidime and could be useful for restoring gut microbiota in patients who are under these antibiotics treatment.
Hemolytic Activity
Absence of hemolytic activity is considered as a safety prerequisite for the selection of a probiotic strain [10]. None of the examined strains exhibited β-hemolytic activity when grown in Columbia human blood agar. Similar observations were made for all the strains of L. paracasei subsp. paracasei, Lactobacillus spp. and L. casei isolated from dairy products which showed γ-hemolysis except of few that showed α-hemolysis [40].
Conclusion
The four strains isolated from aguamiel of A. salmiana, identified as L. mesenteroides subsp. mesenteroides presented interesting probiotic characteristics, especially greater pH and bile tolerance, in vitro adhesion to intestinal mucus, and suppressed pathogen growth under in vitro conditions, which were found in vitro to possess desirable probiotic properties similar or superior to the reference probiotic strain L. plantarum 299v. These strains are good candidates for further investigation with in vivo studies to elucidate their potential health benefits.
References
Douglas LC, Sanders ME (2008) Probiotics and prebiotics in dietetics practice. J Am Diet Assoc 108(3):510–521. doi:10.1016/j.jada.2007.12.009
Correia MITD, Liboredo JC, Consoli MLD (2012) The role of probiotics in gastrointestinal surgery. Nutrition 28(3):230–234. doi:10.1016/j.nut.2011.10.013
Guarner F, Khan AG, Garisch J, Eliakim R, Gangl A, Thomson A, Krabshuis J, Le Mair T (2009) World Gastroenterology Organisation practice guideline: probiotics and prebiotics. Arab J Gastroenterol 10(1):33–42. doi:10.1016/j.ajg.2009.03.001
Geier MS, Butler RN, Howarth GS (2007) Inflammatory bowel disease: current insights into pathogenesis and new therapeutic options; probiotics, prebiotics and synbiotics. Int J Food Microbiol 115(1):1–11. doi:10.1016/j.ijfoodmicro.2006.10.006
Kaur IP, Chopra K, Saini A (2002) Probiotics: potential pharmaceutical applications. Eur J Pharm Sci 15(1):1–9. doi:10.1016/S0928-0987(01)00209-3
Conway PL, Gorbach SL, Goldin BR (1987) Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J Dairy Sci 70(1):1–12. doi:10.3168/jds.S0022-0302(87)79974-3
Fooks LJ, Fuller R, Gibson GR (1999) Prebiotics, probiotics and human gut microbiology. Int Dairy J 9(1):53–61. doi:10.1016/S0958-6946(99)00044-8
Neef A, Sanz Y (2013) Future for probiotic science in functional food and dietary supplement development. Curr Opin Clin Nutr Metab Care 16(6):679–687. doi:10.1097/MCO.0b013e328365c258
Argyri AA, Zoumpopoulou G, Karatzas K-AG, Tsakalidou E, Nychas G-JE, Panagou EZ, Tassou CC (2013) Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol 33(2):282–291. doi:10.1016/j.fm.2012.10.005
FAO/WHO (2002) Guidelines for the evaluation of probiotics in food. Report of a Joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in foods. FAO/WHO, London
Lilly DM, Stillwell RH (1965) Probiotics: growth-promoting factors produced by microorganisms. Science 147(3659):747–748. doi:10.1126/science.147.3659.747
Stanton C, Gardiner G, Meehan H, Collins K, Fitzgerald G, Lynch PB, Ross RP (2001) Market potential for probiotics. Am J Clin Nutr 73(2):476s–483s
Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 73(2):386s–392s
Kailasapathy K, Chin J (2000) Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol 78(1):80–88. doi:10.1046/j.1440-1711.2000.00886.x
Gomes AMP, Malcata FX (1999) Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci Technol 10(4–5):139–157. doi:10.1016/S0924-2244(99)00033-3
Mathara JM, Schillinger U, Guigas C, Franz C, Kutima PM, Mbugua SK, Shin HK, Holzapfel WH (2008) Functional characteristics of Lactobacillus spp. from traditional Maasai fermented milk products in Kenya. Int J Food Microbiol 126(1–2):57–64. doi:10.1016/j.ijfoodmicro.2008.04.027
Moraes PM, Perin LM, Todorov SD, Silva A, Franco BDGM, Nero LA (2012) Bacteriocinogenic and virulence potential of Enterococcus isolates obtained from raw milk and cheese. J Appl Microbiol 113(2):318–328. doi:10.1111/j.1365-2672.2012.05341.x
Ranadheera RDCS, Baines SK, Adams MC (2010) Importance of food in probiotic efficacy. Food Res Int 43(1):1–7. doi:10.1016/j.foodres.2009.09.009
Sanders ME, Klaenhammer TR (2001) Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci 84(2):319–331. doi:10.3168/jds.S0022-0302(01)74481-5
Schillinger U, Guigas C, Heinrich Holzapfel W (2005) In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int Dairy J 15(12):1289–1297. doi:10.1016/j.idairyj.2004.12.008
Todorov S, LeBlanc J, Franco BGM (2012) Evaluation of the probiotic potential and effect of encapsulation on survival for Lactobacillus plantarum ST16 Pa isolated from papaya. World J Microbiol Biotechnol 28(3):973–984. doi:10.1007/s11274-011-0895-z
Melgar-Lalanne G, Rivera-Espinoza Y, Reyes Méndez A, Hernández-Sánchez H (2013) In vitro evaluation of the probiotic potential of halotolerant lactobacilli isolated from a ripened tropical Mexican cheese. Probiotics Antimicro Prot 5(4):239–251. doi:10.1007/s12602-013-9144-0
de Paula A, Jeronymo-Ceneviva A, Silva L, Todorov S, Franco BM, Penna A (2014) Leuconostoc mesenteroides SJRP55: a potential probiotic strain isolated from Brazilian water buffalo mozzarella cheese. Ann Microbiol. doi:10.1007/s13213-014-0933-9
Turchi B, Mancini S, Fratini F, Pedonese F, Nuvoloni R, Bertelloni F, Ebani V, Cerri D (2013) Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J Microbiol Biotechnol 29(10):1913–1922. doi:10.1007/s11274-013-1356-7
Ortiz-Basurto RI, Pourcelly G, Doco T, Williams P, Dornier M, Belleville M-P (2008) Analysis of the main components of the aguamiel produced by the maguey-pulquero (Agave mapisaga) throughout the harvest period. J Agric Food Chem 56(10):3682–3687. doi:10.1021/jf072767h
Murray RGE, Doetsch DR, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 21–41
Persing DH (1993) In vitro nucleic acid amplification techniques. In: Persing DH, Smith TF, Tenover FC (eds) Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC, pp 51–87
Jang J, Kim B, Lee J, Han H (2003) A rapid method for identification of typical Leuconostoc species by 16S rDNA PCR-RFLP analysis. J Microbiol Methods 55(1):295–302. doi:10.1016/S0167-7012(03)00162-3
Nishioka M, Mizuguchi H, Fujiwara S, Komatsubara S, Kitabayashi M, Uemura H, Takagi M, Imanaka T (2001) Long and accurate PCR with a mixture of KOD DNA polymerase and its exonuclease deficient mutant enzyme. J Biotechnol 88(2):141–149. doi:10.1016/S0168-1656(01)00275-9
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. doi:10.1093/nar/25.17.3389
Vizoso Pinto MG, Franz CMAP, Schillinger U, Holzapfel WH (2006) Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int J Food Microbiol 109(3):205–214. doi:10.1016/j.ijfoodmicro.2006.01.029
Brink M, Todorov SD, Martin JH, Senekal M, Dicks LMT (2006) The effect of prebiotics on production of antimicrobial compounds, resistance to growth at low pH and in the presence of bile, and adhesion of probiotic cells to intestinal mucus. J Appl Microbiol 100(4):813–820. doi:10.1111/j.1365-2672.2006.02859.x
Panyarachun B, Sobhon P, Tinikul Y, Chotwiwatthanakun C, Anupunpisit V, Anuracpreeda P (2010) Paramphistomum cervi: surface topography of the tegument of adult fluke. Exp Parasitol 125(2):95–99. doi:10.1016/j.exppara.2009.12.020
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9(1):29–33. doi:10.1111/j.1574-6968.1980.tb05599.x
Doyle RJ, Rosenberg M (1995) Measurement of microbial adhesion to hydrophobic substrata. In: Doyle RJ, Ofek I (eds) Methods in enzymology, vol 253. Academic Press, Waltham, pp 542–550. doi:10.1016/S0076-6879(95)53046-0
Vinderola CG, Reinheimer JA (2003) Lactic acid starter and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res Int 36(9–10):895–904. doi:10.1016/S0963-9969(03)00098-X
Reller LB, Weinstein M, Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755. doi:10.1086/647952
Jeronymo-Ceneviva A, de Paula A, Silva L, Todorov S, Franco BM, Penna A (2014) Probiotic properties of lactic acid bacteria isolated from water-buffalo mozzarella cheese. Probiotics Antimicro Prot 6(3–4):141–156. doi:10.1007/s12602-014-9166-2
Zhang W, Liu M, Dai X (2013) Biological characteristics and probiotic effect of Leuconostoc lactis strain isolated from the intestine of black porgy fish. Braz J Microbiol 44(3):685–691. doi:10.1590/s1517-83822013005000053
Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, Tsakalidou E (2006) Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J 16(3):189–199. doi:10.1016/j.idairyj.2005.02.009
Allameh SKDH, Yusoff FM, Saad CR, Ideris A (2012) Isolation, identification and characterization of Leuconostoc mesenteroides as a new probiotic from intestine of snakehead fish (Channa striatus). Afr J Biotechnol 11(16):3810–3816. doi:10.5897/AJB11.1871
Divya J, Varsha K, Nampoothiri K (2012) Newly isolated lactic acid bacteria with probiotic features for potential application in food industry. Appl Biochem Biotechnol 167(5):1314–1324. doi:10.1007/s12010-012-9561-7
Mishra V, Prasad DN (2005) Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics. Int J Food Microbiol 103(1):109–115. doi:10.1016/j.ijfoodmicro.2004.10.047
Koseki S, Mizuno Y, Sotome I (2011) Modeling of pathogen survival during simulated gastric digestion. Appl Environ Microbiol 77(3):1021–1032. doi:10.1128/aem.02139-10
Charteris Kelly, Morelli Collins (1998) Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol 84(5):759–768. doi:10.1046/j.1365-2672.1998.00407.x
Zavaglia AG, Kociubinski G, Pérez P, De Antoni G (1998) Isolation and characterization of Bifidobacterium strains for probiotic formulation. J Food Prot 61(7):865–873
Collado MC, Meriluoto J, Salminen S (2007) Development of new probiotics by strain combinations: is it possible to improve the adhesion to intestinal mucus? J Dairy Sci 90(6):2710–2716. doi:10.3168/jds.2006-456
Gueimonde M, Salminen S (2006) New methods for selecting and evaluating probiotics. Dig Liver Dis 38:S242–S247. doi:10.1016/s1590-8658(07)60003-6
Collado MC, Gueimonde M, Salminen S (2010) Chapter 23—probiotics in adhesion of pathogens: mechanisms of action. In: Preedy RRWR (ed) Bioactive Foods in Promoting Health. Academic Press, Boston, pp 353–370. doi:10.1016/B978-0-12-374938-3.00023-2
Izquierdo E, Horvatovich P, Marchioni E, Aoude-Werner D, Sanz Y, Ennahar S (2009) 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis 30(6):949–956. doi:10.1002/elps.200800399
Haller D, Colbus H, Gänzle MG, Scherenbacher P, Bode C, Hammes WP (2001) Metabolic and functional properties of lactic acid bacteria in the gastro-intestinal ecosystem: a comparative in vitro studybetween bacteria of intestinal and fermented food origin. Syst Appl Microbiol 24(2):218–226. doi:10.1078/0723-2020-00023
Ouwehand AC, Parhiala R, Salminen S, Rantala A, Huhtinen H, Sarparanta H, Salminen E (2004) Influence of the endogenous mucosal microbiota on the adhesion of probiotic bacteria in vitro. Microb Ecol Health Dis 16(4):202–204. doi:10.1080/08910600410021774
Ouwehand AC, Tuomola EM, Tölkkö S, Salminen S (2001) Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int J Food Microbiol 64(1–2):119–126. doi:10.1016/S0168-1605(00)00440-2
Shobharani P, Agrawal R (2011) A potent probiotic strain from cheddar cheese. Indian J Microbiol 51(3):251–258. doi:10.1007/s12088-011-0072-y
Aswathy R, Ismail B, John R, Nampoothiri K (2008) Evaluation of the probiotic characteristics of newly isolated lactic acid bacteria. Appl Biochem Biotechnol 151(2–3):244–255. doi:10.1007/s12010-008-8183-6
Raghavendra P, Halami PM (2009) Screening, selection and characterization of phytic acid degrading lactic acid bacteria from chicken intestine. Int J Food Microbiol 133(1–2):129–134. doi:10.1016/j.ijfoodmicro.2009.05.006
Ofek I, Hasty DL, Sharon N (2003) Anti-adhesion therapy of bacterial diseases: prospects and problems. FEMS Immunol Med Microbiol 38(3):181–191. doi:10.1016/s0928-8244(03)00228-1
Sánchez B, Bressollier P, Urdaci MC (2008) Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol Med Microbiol 54(1):1–17. doi:10.1111/j.1574-695X.2008.00454.x
Ripamonti B, Agazzi A, Bersani C, De Dea P, Pecorini C, Pirani S, Rebucci R, Savoini G, Stella S, Stenico A, Tirloni E, Domeneghini C (2011) Screening of species-specific lactic acid bacteria for veal calves multi-strain probiotic adjuncts. Anaerobe 17(3):97–105. doi:10.1016/j.anaerobe.2011.05.001
Tejero-Sariñena S, Barlow J, Costabile A, Gibson GR, Rowland I (2012) In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18(5):530–538. doi:10.1016/j.anaerobe.2012.08.004
Ouwehand AC, Salminen SJ (1998) The health effects of cultured milk products with viable and non-viable bacteria. Int Dairy J 8(9):749–758. doi:10.1016/S0958-6946(98)00114-9
Gänzle MG, Weber S, Hammes WP (1999) Effect of ecological factors on the inhibitory spectrum and activity of bacteriocins. Int J Food Microbiol 46(3):207–217. doi:10.1016/S0168-1605(98)00205-0
Begley M, Gahan CGM, Hill C (2005) The interaction between bacteria and bile. FEMS Microbiol Rev 29(4):625–651. doi:10.1016/j.femsre.2004.09.003
Senbagam D, Gurusamy R, Senthilkumar B (2013) Physical chemical and biological characterization of a new bacteriocin produced by Bacillus cereus NS02. Asian Pac J Trop Med 6(12):934–941. doi:10.1016/S1995-7645(13)60167-4
Gueimonde M, Sánchez B, de los Reyes-Gavilán CG, Margolles A (2013) Antibiotic resistance in probiotic bacteria. Front Microbiol. doi:10.3389/fmicb.2013.00202
Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R (2014) Antibiotic resistance among commercially available probiotics. Food Res Int 57:176–195. doi:10.1016/j.foodres.2014.01.025
Courvalin P (2006) Antibiotic resistance: the pros and cons of probiotics. Dig Liver Dis 38:S261–S265. doi:10.1016/s1590-8658(07)60006-1
Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. Am J Med 119(6, Supplement 1):S3–S10. doi:10.1016/j.amjmed.2006.03.011
Hummel AS, Hertel C, Holzapfel WH, Franz CMAP (2007) Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl Environ Microbiol 73(3):730–739. doi:10.1128/aem.02105-06
Acknowledgments
The research leading to these results has received funding from CONACyT (435894/267594), the Instituto Politecnico Nacional (IPN-UPIBI) of Mexico, and the Central de Microscopía of ENCB, Mexico.
Conflict of interest
Castro-Rodríguez Diana, Hernández-Sánchez Humberto and Yáñez Fernández Jorge declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diana, CR., Humberto, HS. & Jorge, Y.F. Probiotic Properties of Leuconostoc mesenteroides Isolated from Aguamiel of Agave salmiana . Probiotics & Antimicro. Prot. 7, 107–117 (2015). https://doi.org/10.1007/s12602-015-9187-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-015-9187-5